Abstract
The eutT gene of Salmonella enterica was cloned and overexpressed, and the function of its product was established in vivo and in vitro. The EutT protein has an oxygen-labile, metal-containing ATP:co(I)rrinoid adenosyltransferase activity associated with it. Functional redundancy between EutT and the housekeeping ATP:co(I)rrinoid adenosyltransferase CobA enzyme was demonstrated through phenotypic analyses of mutant strains. Lack of CobA and EutT blocked ethanolamine utilization. EutT was necessary and sufficient for growth of an S. enterica cobA eutT strain on ethanolamine as a carbon and energy or nitrogen source. A eutT+ gene provided in trans corrected the adenosylcobalamin-dependent transcription of a eut-lacZ operon fusion in a cobA strain. Cell extracts enriched for EutT protein contained strong, readily detectable ATP:co(I)rrinoid adenosyltransferase activity. The activity was only detected in extracts maintained under anoxic conditions, with complete loss of activity upon exposure to air or treatment with the Fe2+ ion chelator bathophenanthroline. While the involvement of another metal ion cannot be ruled out, the observed sensitivity to air and bathophenanthroline suggests involvement of Fe2+. We propose that the EutT protein is a unique metal-containing ATP:co(I)rrinoid adenosyltransferase. It is unclear whether the metal ion plays a structural or catalytic role.
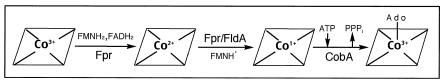
The biosynthesis of AdoCbl (coenzyme B12) is unique to some prokaryotes. Multiple functions (encoded by >25 genes) are required for the assembly of this complex molecule (36). The chief structural feature of AdoCbl is the presence of an adenosyl moiety liganded to the Co ion of cobalamin via a covalent Co-C bond. In the enterobacterium Salmonella enterica, corrinoid adenosylation is required for de novo synthesis and for the assimilation of incomplete precursors such as cobinamide (12). Insights into the corrinoid adenosylation pathway in S. enterica (Fig. 1) were recently reported, including the three-dimensional crystal structure of the ATP:co(I)rrinoid adenosyltransferase (CobA) enzyme responsible for the formation of the Co-C bond (2, 13-15, 35). The cobA gene in S. enterica is not part of the 17-gene cob operon (27, 33) and appears to be constitutively expressed (34). In this bacterium, CobA is the housekeeping adenosyltransferase needed for de novo biosynthesis and for the assimilation of exogenous corrinoids. However, CobA is not the only corrinoid adenosyltransferase present in S. enterica. Other large operons like the 1,2-propanediol utilization (pdu) and ethanolamine utilization (eut) operons appear to encode their own corrinoid adenosyltransferases. Johnson et al. recently reported evidence that the pduO gene of S. enterica encodes an ATP:cobalamin adenosyltransferase (19). The eutT gene of the eut operon was suggested to encode the corrinoid adenosyltransferase enzyme for this pathway, but support for this assignment was inconclusive (20). Interestingly, CobA, PduO, and EutT do not have an ancestor in common, suggesting that they evolved in response to a specific need (18).
FIG. 1.
The corrinoid adenosylation pathway in S. enterica. Fpr, NADP+:ferredoxin (flavodoxin) reductase; FAD/FADH2, flavin adenine dinucleotide/reduced flavin adenine dinucleotide; FMNH2, reduced flavin mononucleotide; FMNH · , flavin mononucleotide semiquinone; PPPi, inorganic triphosphate; Ado, 5-deoxyadenosine; FldA, flavodoxin A. The corrin ring is schematized as shown with the Co ion in the appropriate oxidation state.
In this paper we report biochemical and genetic evidence that the eutT gene of S. enterica encodes an ATP:cob(I)alamin adenosyltransferase. The data show that EutT enzyme activity requires the involvement of a metal ion(s) for function and is O2 sensitive.
MATERIALS AND METHODS
Bacteria, culture media, and growth conditions.
The genotypes of the strains used in these studies are listed in Table 1. The ability of strains to use ethanolamine as a nitrogen source was assessed on no-carbon, no-nitrogen medium (24) supplemented with glucose (11 mM), MgSO4 (1 mM), methionine (0.5 mM), cyanocobalamin (CNCbl; 15 nM), and ethanolamine (20 mM). The ability of strains to use ethanolamine as a carbon and energy source was assessed on no-carbon E (NCE) medium (4) supplemented with NH4Cl (50 mM), l-methionine (0.5 mM), MgCl2 (1 mM), ethanolamine (50 mM), adenosylcobalamin (AdoCbl; 0.2 μM), and hydroxycobalamin (HOCbl; 0.2 μM). When added to the medium, l-(+)-arabinose was at 0.2 mM. Trace mineral elixir was prepared as previously described (1) and used at 5 ml/liter of medium. Nutrient broth (NB; Difco) was used as rich medium. All chemicals were of high purity and were used without further purification. HOCbl, AdoCbl, and dicyanocobinamide [(CN)2Cbi] were purchased from Sigma. Ethanolamine hydrochloride (99.9% purity) was purchased from Aldrich.
TABLE 1.
Strains and plasmids used in this studya
| Strain | Genotype | Source or reference |
|---|---|---|
| TR6583 | metE205 ara-9 | K. Sanderson via J. Roth |
| Derivatives of TR6583 | ||
| JE1293 | cobA366::Tn10d(cat+) | |
| JE2236 | eutE18::MudA | |
| JE2257 | Δ299(hisG-cobT) Δ902(cobA-trp) | |
| JE3193 | Δ299(hisG-cobT) Δ902(cobA-trp) eut*-1149 | This study |
| JE3604 | Δ299(hisG-cob) Δ902(cobA-trp) | This study |
| JE6692 | STM2478-6377::Tn10d(tet+) eut*-1149/pKD46 bla+ | This study |
| JE7091 | eutT1144::cat+ | This study |
| JE7092 | ΔeutT1145 | This study |
| JE7145 | eutE18::MudI1734b | |
| JE7179 | eutE18::MudI1734 cobA366::Tn10d(cat+) | |
| JE7180 | cobA366::Tn10d(cat+) ΔeutT1145 | This study |
| JE7202 | eutE18::MudI1734 cobA366::Tn10d(cat+)/pBAD24 bla+ | This study |
| JE7203 | eutE18::MudI1734 cobA366::Tn10d(cat+)/pEUT7 ParaBAD-eutT+bla+ | This study |
| JE7204 | cobA366::Tn10d(cat+) ΔeutT1145/pBAD24 bla+ | This study |
| JE7205 | cobA366::Tn10d(cat+) ΔeutT1145/pEUT7 ParaBAD-eutT+bla+ | This study |
All strains are derivatives of S. enterica serovar Typhimurium LT2. Strains were from the laboratory collection unless otherwise stated.
Referred to in the text as MudJ (7).
Genetic techniques. (i) Transductions.
All of the crosses used bacteriophage P22 double mutant HT105/1 int-201 (28, 29). Transductions were performed as previously described (8, 10).
(ii) Isolation of a Tn10d(tet+) insertion element near the eut*-1149 gain-of-function mutation.
A pool of ∼100,000 S. enterica strains carrying Tn10d(tet+) elements inserted in their genome was obtained as previously described (11). Phage P22 grown on this pool of strains was used as the donor to transduce strain JE3193 [metE205 ara-9 Δ299(hisG-cobT) Δ902(cobA-trp) eut*-1149] to tetracycline resistance, screening for loss of the ability of the strain to grow on ethanolamine as an N source in minimal medium supplemented with CNCbl.
(iii) Mapping of the eut*-1149 mutation.
The general location of the eut*-1149 mutation on the chromosome was obtained by two different means. First, genetic crosses with the Mud-P22 mapping kit of Benson and Goldman (3) were performed. This method allows the location of mutations to regions of the chromosome approximately 40 kb in size. For this purpose, transducing lysates prepared on the set of lysogens in the mapping kit were used as donors in crosses that selected for loss of the Tn10d(tet+) element near the eut*-1149 mutation on Bochner plates (5, 21). Crude preparations of the phage P22 tailspike protein were obtained as previously described (30, 39). Approximately 35 μg of protein of a crude extract containing P22 tailspike protein was routinely spread onto the plate prior to dispensing Mud-P22 samples by means of a multiprong device. The physical location of the Tn10d(tet+) insertion element linked to the eut*-1149 mutation was determined by sequencing the DNA flanking the insertion element by protocols described elsewhere (6, 23).
(iv) Chemical mutagenesis.
The method described by Miller to mutagenize cells with N-methyl-N′-nitro-N-nitrosoguanidine (NG) (22) was used to isolate derivatives of cobA strain JE2257 that used ethanolamine as an N source. Briefly, strain JE2257 was grown in NB overnight at 37°C with shaking. A fresh culture of JE2257 was started by inoculating 0.1 ml of the overnight culture into 5 ml of fresh NB; the culture was allowed to reach mid-log phase (∼70 Klett units), and cells were washed twice with citrate buffer, pH 5.5, and resuspended in 5 ml of citrate buffer prior to the addition of NG to a final concentration of 25 μg/ml. Cells and mutagen were incubated at 37°C for 20 min; mutagenized cells were washed with phosphate buffer (pH 7.0), resuspended in 5 ml of fresh NB, and incubated at 37°C with shaking until the culture reached an A650 of 1.5. The culture was diluted, plated for single colonies on NB agar, and replica printed onto no-carbon, no-nitrogen minimal medium supplemented with ethanolamine as an N source. Clones able to grow on ethanolamine as an N source were restreaked on selection plates.
(v) Recombinant DNA techniques.
For construction of an in-frame, nonpolar deletion of eutT, the eutT gene was deleted by a previously described method (9). Briefly, primers EutTKOP1fwd (5′-TCATACGCTCAGCGAAGGATCGGAGATCCATCAGCCCGCTGACGCACGACTGACGTGTAGGCTGGAGCTGCTTC-3′) and EutTP2KOrev (5′-GGCGCGTACCGCCAGTTCGCGGGCGCGTTCAATGATCATGGCTTCTCTCCCAACCATATGAATATCCTCCTTAG-3′) were used to amplify the chloramphenicol cassette from plasmid pKD3 such that the 5′ and 3′ tails were homologous to the 5′ and 3′ regions of the S. enterica eutT gene. The linear PCR product was electroporated into strain JE6692 (metE205 ara-9/pKD46 bla+) selecting for chloramphenicol resistance and counterselecting against pKD46 by incubation at 37°C. The resulting strain was JE7091 (metE205 ara-9 eutT1144::cat+). Plasmid pCP20 was electroporated into strain JE7091 selecting for ampicillin resistance at 30°C. The resulting strain was incubated at 42°C to select against the plasmid. Clones sensitive to ampicillin and chloramphenicol were analyzed further. PCR amplification of the eutT gene was performed with primers Seeutup 500fwd (5′-GCGGCTCTCAGTGAACAGGA-3′) and Seeutdown 500rev (5′-CGCTGCAATCGGCGAACC-3′). The DNA sequence of the amplified product was determined by the nonradioactive ABI PRISM BigDye cycle sequencing method (PE Life Sciences) in accordance with the manufacturer's instructions with primers Seeutup 300seqfwd (5′-GTCATTGACGGCAGCAGCG-3′) and Seeutdown 300seqrev (5′-CATCAGCGGATCGCTAAGC-3′). The DNA sequence was determined at the Biotechnology Center of the University of Wisconsin—Madison. DNA sequencing confirmed the deletion of bases 91 to 786 in the eutT gene in strain JE7092 (metE205 ara-9 ΔeutT1145). No polar effects were observed under the growth conditions tested.
(vi) Plasmid constructions.
Plasmid pEUT7 was constructed by amplifying the eutT gene from S. enterica genomic DNA with primers EutTEcoRIf (5′-GTACGTCGCCTGGAATTCAAACTGGC-3′) and EutTXbaIr2 (5′-GCGCATCTAGAGAAAGACGACTCTGGC-3′). The 800-bp PCR product was cloned into the EcoRI and XbaI sites of plasmid pBAD24 (17), resulting in plasmid pEUT7. The presence of the eutT+ allele in plasmid pEUT7 was confirmed by DNA sequencing with primers pBADfwd (5′-CGCAACTCTCTACTGTTTCT-3′) and pBADrev (5′-GGCTGAAAATCTTCTCTCAT-3′).
Corrinoid adenosylation assays. (i) Preparation of cell extracts.
Strains JE7204 [metE205 ara-9 cobA366::Tn10d(cat+) ΔeutT1145/pBAD24] and JE7205 [metE205 ara-9 cobA366::Tn10d(cat+) ΔeutT1145/pEUT7] were grown in NCE minimal medium supplemented with ethanolamine as a carbon and energy source, AdoCbl (1 μM), and l-(+)-arabinose (0.2 mM) to activate transcription driven by the ParaBAD promoter in the plasmids. Cultures of the above strains were grown aerobically at 37°C for 22 h, cells we harvested by centrifugation at 4°C (12,096 × g for 10 min), cell paste was placed in a serum vial, the headspace was flushed with O2-free N2 gas for 15 min, and the vial was stored at −80°C until used. Cells were resuspended inside the anaerobic chamber in anoxic 0.2 M Tris Cl buffer (pH 8, 25°C) containing phenylmethylsulfonyl fluoride (1 μM) and 1× BugBuster reagent (Novagen) and stirred for 30 min at room temperature. Lysates were transferred into stainless steel tubes fitted with caps with an expanding O ring to maintain anoxic conditions; tubes were centrifuged at 43,667 × g and 4°C for 30 min in a Beckman Avanti J25I centrifuge equipped with a JA 25.50 rotor. Clarified extracts were placed into Pierce SnakeSkin 3.5-kDa MWCO bags and dialyzed for 1 h against 750 ml of anoxic Tris-Cl buffer (pH 8, 25°C); six changes of dialysis buffer were performed. Dialyzed, clarified cell extracts were transferred into serum vials, pressurized to 102 kPa with O2-free N2, and used within 24 h. Adenosylation assay conditions were as previously described (14, 35), except that 0.5 mM ATP was used.
(ii) Preparation of cob(II)alamin.
Cob(II)alamin was generated with the Fpr [ferredoxin (flavodoxin) NADP+ oxidoreductase] FldA system. HOCbl (25 μmol) was placed into a serum vial to which 30 ml of anoxic 0.2 M Tris Cl (pH 8, 37°C) was added under a stream of O2-free N2 gas; NADPH (50 μmol), Fpr (75 nmol), and FldA (25 nmol) were added anoxically. The reaction mixture was incubated for 1.5 h at 37°C in a water bath outside the anaerobic chamber. Cob(II)alamin was purified inside the chamber by binding to a deoxygenated C18 SepPak cartridge (Millipore), washing it with water, and eluting it with previously degassed 100% methanol. Samples were dried outside the chamber under a stream of O2-free nitrogen gas. Dried cob(II)alamin was placed in a serum vial pressurized at 100 kPa and maintained inside the anaerobic chamber until redissolved in the above-mentioned reaction buffer containing MnCl2 (0.8 mM).
Spectrofluorimetric β-galactosidase activity assays.
Whole-cell β-galactosidase assays were performed in 96-well microtiter dishes with the fluorogenic substrate 3-carboxyumbelliferyl-β-d-galactopyranoside (Molecular Probes) in accordance with the manufacturer's instructions. Two milliliters of NCE medium supplemented with glycerol, MgCl2, l-methionine, NH4Cl, and trace minerals was inoculated with 100 μl of an overnight NB culture of the appropriate strain. Cultures were grown at 37°C until the A650 was between 0.2 and 0.3 (∼2 h). At this time, 1 ml of the culture was centrifuged (18,000 × g, for 2 min) and cells were resuspended in 300 μl of sterile saline. A 10-μl sample was removed, diluted 1:100 in sterile saline, and used for viable counts. The remaining 290 μl was treated with chloroform (30 μl) to permeabilize the cell membrane. A 50-μl sample of cell suspension was used per well of a 96-well microtiter dish; measurement of β-galactosidase activity in each culture was performed in triplicate.
A 100-μl sample of the fluorogenic substrate 3-carboxy-umbelliferyl-β-d-galactopyranoside working solution (includes phosphate buffer, pH 7.3) was added, and the reaction mixture was incubated at room temperature for 30 min. The reaction was stopped by addition of stop buffer (50 μl), and the fluorescence was read at 460 nm (excitation at 390 nm). Fluorescence values in each well were normalized to a reference standard and compared to a standard curve to determine the number of picograms of β-galactosidase per well. The number of molecules of β-galactosidase (46.5 kDa) per cell was determined by taking into consideration that active β-galactosidase is a tetramer. Viable counts were used to calculate the number of active β-galactosidase tetramers per cell.
RESULTS AND DISCUSSION
Expression of the eut operon requires AdoCbl.
The EutR protein activates transcription of the eut operon in response to ethanolamine and cobalamin in the environment (25, 26). To determine the form of cobalamin sensed by EutR, we blocked the conversion of cobalamin to AdoCbl by the housekeeping CobA adenosyltransferase. A null allele of the cobA gene was introduced into strain JE7145 (metE205 ara-9 eutE18::MudJ) by phage P22-mediated transduction, resulting in strain JE7179 [metE205 ara-9 eutE18::MudJ cobA366::Tn10d(cat+)]. We measured expression of the eut-lacZ reporter in strains JE7145 and JE7179 as a function of the corrinoids used to supplement the medium. When nonadenosylated cobinamide or cobalamin was added to the culture medium, expression of the eut-lacZ reporter was five to eight times higher in strain JE7145 (cobA+) than in strain JE7179 (cobA) (Table 2). These results indicated that AdoCbl eut operon expression required AdoCbl as a coinducer. The need for CobA activity during ethanolamine utilization was confirmed by growth studies. A culture of a cobA strain displayed a long lag (>60 h) before the onset of logarithmic growth, and the final density of the culture was low when CNCbl was in the medium. Addition of AdoCbl in lieu of CNCbl abolished the lag, and the culture reached a cell density similar to that of a cobA+ strain (data not shown).
TABLE 2.
In vivo transcription evidence that EutT synthesizes AdoCbl
| Strain | Genotypec | β-Galactosidase (active tetramers/CFU) activitya in cells grown in minimal mediumb with the indicated supplement(s)
|
|||
|---|---|---|---|---|---|
| None | EA | EA, (CN)2Cbi | EA, HOCbl | ||
| JE7145 | cobA+ | 1,720 ± 100 | 1,960 ± 60 | 4,960 ± 400 | 6,330 ± 640 |
| JE7179 | cobA | 400 ± 30 | 440 ± 30 | 980 ± 60 | 810 ± 70 |
| JE7202 | cobA/pVOCd | NDe | ND | 770 ± 30 | 2,120 ± 80 |
| JE7203 | cobA/peutT+f | ND | ND | 4,750 ± 20 | 12,210 ± 450 |
The number of active tetramers of β-galactosidase enzyme was determined as described in Materials and Methods.
NCE medium was supplemented with MgCl2, NH4Cl, glycerol (as a carbon and energy source), methionine, and trace minerals. Also added were ethanolamine (EA; 50 mM), CN2Cbi (0.2 μM), 200 nM HOCbl (0.2 μM), AdoCbl (0.2 μM) and l-(+)-arabinose (200 μM). After 2 h of growth at 37°C, 1 ml of culture was centrifuged and cells were resuspended in 0.3 ml of sterile saline. CFU counts were determined by dilution plating on nutrient agar plates.
Complete genotypes of the strains are listed in Table 1. All strains carried a chromosomal eutE18::MudJ(lacZ+) element and mutations metE205 and ara-9.
pVOC, pBAD24 (17).
ND, not determined.
peutT+, pEUT7.
The eut operon encodes a corrinoid adenosyltransferase.
The existence of an alternative ATP:co(I)rrinoid adenosyltransferase was suggested by the slow but reproducible growth of the cobA strain on ethanolamine in medium supplemented with CNCbl. To investigate this possibility we performed NG mutagenesis (22) of strain JE2257 [Δ299(hisG-cobT) Δ902(cobA-trp)] searching for gain-of-function derivatives able to degrade ethanolamine. Strain JE2257 was used in this study to avoid dealing with the two known adenosyltransferases, i.e., CobA and PduO (19, 35). NG mutagenesis of strain JE2257 yielded a derivative (JE3193) with improved growth on ethanolamine as an N source in medium supplemented with CNCbl (data not shown). Hereafter we refer to this gain-of-function mutation as eut*-1149.
We used two nonbiased genetic approaches to locate the eut*-1149 mutation. First we used the Mud-P22 mapping kit of Benson and Goldman as previously described (3). Crude preparations of P22 tailspike protein were prepared (30, 39), sterilized, and spread onto selection plates to increase transduction efficiency (5, 21). By this method, the eut*-1149 mutation was located to the region of ∼50 centisomes, near the eut operon.
Second, we isolated a transposition-deficient Tn10d(tet+) element (37) near the eut* mutation. For this purpose we screened a pool of ∼100,000 strains carrying Tn10d(tet+) elements randomly inserted into the chromosome (11). The chromosomal location of the Tn10d(tet+) element was determined by sequencing the DNA flanking it. The latter was amplified by arbitrary PCR protocols (6, 23). The Tn10d(tet+) element near the eut* mutation was inserted into open reading frame STM2478 (unknown function; http://www.ncbi.nlm.nih.gov/genomes/altvik.cgi?gi=202&db=g&gene=stm2478), which is 7 genes from the 5′ end of the eut operon. Phage P22 grown on strain JE3193 carrying an insertion in open reading frame STM2478 was used to move the eut* mutation back into strain JE2257 (resulting in strain JE3604). The Tn10d(tet+) element and the eut*-1149 mutation were 80% cotransducible by phage P22, confirming that the eut*-1149 mutation was located in proximity to the eut operon.
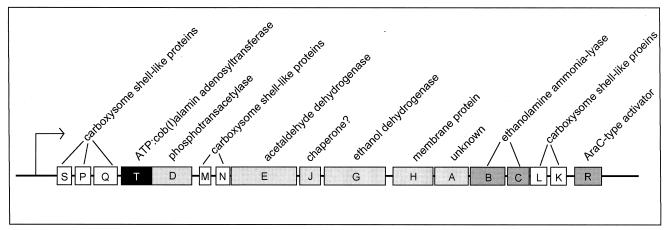
The nature of the eut*-1149 mutation was not established, but we hypothesized that it increased the expression of the eut operon, resulting in higher levels of a corrinoid adenosyltransferase encoded by the operon. Alternatively, the gain-of-function mutation could have improved the catalytic ability of the adenosyltransferase encoded by the operon. We focused our attention on the eutT gene (Fig. 2), which was annotated as encoding a putative cobalamin adenosyltransferase (http://www.ncbi.nlm.nih.gov/sutils/blink.cgi?pid=16765787&cut=95) on the basis of the fact that eutT function was not required for ethanolamine catabolism (20). As shown below, deleting the eutT gene had no effect on the ability of the strain to grow on ethanolamine as long as the cell had a functional cobA gene (Fig. 3, solid triangles).
FIG. 2.
The ethanolamine utilization (eut) operon of S. enterica.
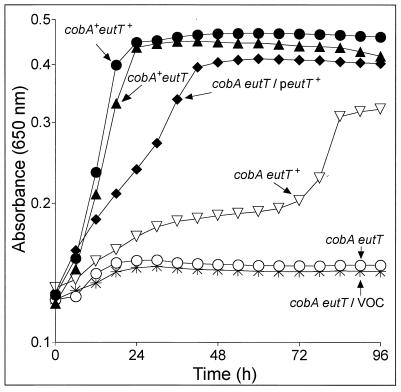
FIG. 3.
In vivo evidence that eutT encodes a cobalamin adenosyltransferase enzyme. Strains were grown in NCE minimal medium supplemented with ethanolamine as a carbon and energy source and CNCbl (200 nM) at 37°C. Cultures of cobA+ strains reached stationary phase approximately 22 h after inoculation. VOC, vector-only control; peutT+, pEUT7.
Phenotypic analysis of eutT strains.
The growth behavior of strain JE1293 [metE205 ara-9 cobA366::Tn10d(cat+)] on ethanolamine as an N source is shown in Fig. 3 (inverted open triangles). A nonpolar, in-frame deletion of eutT in strain JE1293 was constructed by using previously described protocols (9). The resulting strain, JE7180 [metE205 ara-9 cobA366::Tn10d(cat+) ΔeutT1145], failed to grow on ethanolamine even after 96 h of incubation (Fig. 3, open circles), suggesting that EutT was the alternative cobalamin adenosyltransferase. The eutT+ allele was placed under the control of the arabinose-inducible promoter (ParaBAD) in vector pBAD24, resulting in plasmid pEUT7 (ParaBAD-eutT+). Expression of eutT+ from plasmid pEUT7 in strain JE7205 [metE205 ara-9 cobA366::Tn10d(cat+) ΔeutT1145/pEUT7 ParaBAD-eutT+] abolished the lag and increased the cell density of the culture to almost wild-type levels (Fig. 3, solid diamonds). These results indicated that the chromosomal deletion of eutT in the strains did not have deleterious effects on the expression of genes downstream of it. The lack of eutT function had a strong negative effect on the ability of S. enterica to grow on ethanolamine only in a cell devoid of the ATP:co(I)rrinoid adenosyltransferase (CobA) enzyme (Fig. 3, compare solid triangles with open circles). This conditionality indicated functional redundancy, yet no evidence of a shared lineage was detectible by nucleotide or primary amino acid sequence comparisons of the EutT and CobA proteins or the eutT and cobA genes (data not shown). The observed lack of homology between CobA and EutT suggests that although these enzymes generate the same product, they probably do so through different mechanisms.
In vitro evidence supporting the synthesis of AdoCbl by EutT.
We obtained cell extracts of strains JE7204 [metE205 ara-9 cobA366::Tn10d(cat+) ΔeutT1145/pBAD24 ParaBAD] and JE7205 [metE205 ara-9 cobA366::Tn10d(cat+) ΔeutT1145/pEUT7 ParaBAD-eutT+] grown aerobically at 37°C for 22 h in NCE minimal medium supplemented with ethanolamine (as a carbon and energy source), MgSO4, NH4Cl, l-methionine, trace minerals, l-(+)-arabinose (as an inducer), and AdoCbl.
ATP:cob(I)alamin adenosyltransferase activity (6.1 ± 0.3 nmol of AdoCbl min−1 mg−1 of protein) was detected in strains carrying the complementation plasmid (eutT+) but not in the vector-only control strain (Fig. 4). Assay results were reproducible when the amount of KBH4 used to reduce cob(III)alamin to cob(II)alamin was 3 μmol. To eliminate the need for a reductant, subsequent reactions were performed with cob(II)alamin as the substrate. AdoCbl was detected when CobA or EutT extract was added (Fig. 5) but not when ATP was present alone in the reaction mixture. The yield of AdoCbl was half of the starting cob(II)alamin in the reaction mixture, and increasing the reaction time or the amount of protein did not improve the product yield. This result was not unexpected, since cob(II)alamin is known to be disproportionate to cob(I)alamin and cob(III)alamin (38). We posit that cob(I)alamin generated by cob(II)alamin disporportionation is used by CobA or EutT as a substrate. Taken together, the data indicate that EutT is an ATP:cob(I)alamin adenosyltransferase.
FIG. 4.
Corrinoid adenosyltransferase activity is detectable in cobA cell lysates. Strains JE7204 (cobA eutT/pBAD24) and JE7205 (cobA eutT/pEUT7) were grown on ethanolamine as the sole C source in minimal medium supplemented with AdoCbl. Cells were harvested and lysed anoxically with 0.2 M Tris Cl (pH 8.0) in 1× BugBuster reagent (Novagen) containing protease inhibitor. Protein content was normalized with fresh buffer. The KBH4 corrinoid adenosylation procedure was followed (35), except that the amount of reductant was reduced to 3 μmol per reaction mixture. Reactions were allowed to proceed for 1 h at 37°C, and the AdoCbl concentration was determined by photolysis. Lysate containing EutT by overexpression from the pEUT7 plasmid produced AdoCbl at levels dependent on the amount of protein added per reaction mixture. Only a background level decrease in absorbance was detected from the vector-only negative control. Each data point represents the average of at least three reactions.
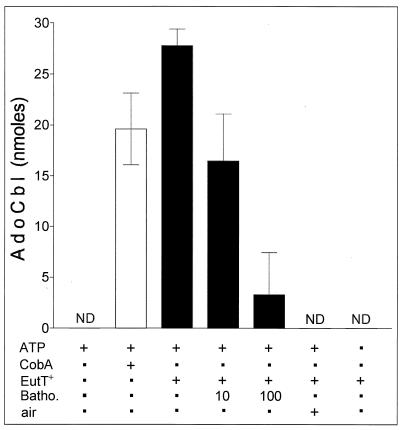
FIG. 5.
EutT adenosyltransferase activity is sensitive to air and metal chelation. Substrates and protein were added, and the reaction was initiated by a shift to 37°C. EutT-dialyzed extract was prepared as described in Materials and Methods. ATP, 500 μM; CobA, 40 μg of protein; EutT+, 100 μg of protein (EutT-enriched extract); Batho., 10 or 100 μM bathophenanthroline; air, EutT-enriched extract exposed to air for 20 min and then flushed with O2-free N2 gas before addition to the reaction mixture. AdoCbl production was determined by comparing the A525 after 30 min with that after 10 min of photolysis. Each data point represents the average of at least two reactions. ND, not detected.
In vivo evidence that AdoCbl is the product of the EutT reaction.
To confirm that the product of the EutT reaction was AdoCbl, we used a sensitive fluorimetric method to measure expression of the eut-lacZ operon fusion as a function of EutT activity. Expression of the reporter was measured in strains JE7202 [metE205 ara-9 cobA366::Tn10d(cat+) eutE18::MudJ/pBAD24] and JE7203 [metE205 ara-9 cobA366::Tn10d(cat+) eutE18::MudJ/pEUT7 ParaBAD-eutT+]. These strains were grown in medium containing ethanolamine and either (CN)2Cbi or HOCbl. Expression of the fusion in strain JE7203 was about sixfold higher than in control strain JE7202 when either (CN)2Cbi or HOCbl was added. However, in strain JE7203 the level of expression of the fusion was much higher when HOCbl was added to the medium (Table 2). These results confirmed that AdoCbl was the product of the EutT reaction and suggested that EutT may have higher affinity for a complete corrinoid substrate. It is unclear why the presence of CobA resulted in an increase in transcription of the eut operon even when no ethanolamine or corrin was added to the medium. The increase in eut expression in a cobA/peutT+ strain versus a cobA+ strain when ethanolamine and hydroxycobalamin are added to the medium likely reflects the effect of arabinose on eutT+ expression and the fact that eutT+ was present in a multicopy plasmid.
Evidence suggesting the involvement of Fe.
Like CobA, EutT adenosylates cob(I)alamin with ATP as a cosubstrate. Unlike CobA, EutT does not have an ATP-binding P-loop motif (2, 31), and the amino acid sequence of EutT is distinct from that of CobA and PduO cobalamin adenosyltransferases. Close examination of the primary amino acid sequence of EutT revealed a cysteine-rich region reminiscent of the conserved S-adenosylmethionine Fe-S cluster motif identified by Sofia et al. (32).
Two results suggested that Fe-S centers might be present in the S. enterica EutT protein. First, EutT activity was lost upon exposure to oxygen. Second, EutT activity was lost as a function of the concentration of bathophenanthroline (an Fe2+ chelator) (16) in the reaction mixture (Fig. 5). Addition of 10 μM bathophenanthroline to the reaction mixture resulted in a 40% loss of EutT-dependent activity despite the presence of 0.8 mM MnCl2 in the reaction mixture. Attempts to substitute S-adenosylmethionine for ATP as a substrate for EutT did not yield any product (data not shown). Electron paramagnetic resonance experiments aimed at confirming the presence and determining the type of Fe-S centers in EutT await isolation of the enzyme.
In summary, we have provided in vivo and in vitro evidence to support the previous proposal by Kofoid et al. (20) that EutT is a corrinoid adenosyltransferase. We have also shown that EutT uses ATP as a donor of the adenosyl moiety and that it contains a metal ion that can be chelated by bathophenanthroline. We suggest that oxidation of the latter may cause the loss of activity observed upon exposure of the enzyme to air. The sensitivity of EutT to air and metal chelation set it apart from the other two known ATP:corrinoid adenosyltransferases described in S. enterica, CobA and PduO. These observations further support the idea that EutT catalyzes the synthesis of AdoCbl via a mechanism different from the one used by CobA and probably PduO. The isolation and characterization of the EutT enzyme are in progress.
EutT homologs.
The eutT gene from S. enterica is 88% identical and 93% similar to its homolog in E. coli. The predicted sequences of these EutT proteins contain the same cysteine-rich sequence we think may play a role in metal ligation. Other members of the COG4812 family (http://www.ncbi.nlm.nih.gov/COG/new/release/cow.cgi?cog=COG4812) are 30 to 27% identical and 48 to 51% similar to the S. enterica EutT protein, but a cysteine-rich sequence is not obvious in these orthologs.
Why is a corrinoid adenosyltransferase encoded by an operon requiring AdoCbl for expression? There is no clear answer to this question. Cells are able to grow on ethanolamine in the absence of the housekeeping adenosyltransferase (CobA) enzyme after a long lag (Fig. 3, open triangles), suggesting that there is a low basal level of transcription of the eut operon. We propose that the lag is a period of time during which sufficient levels of EutT accumulate to synthesize enough AdoCbl for EutR to fully activate expression of the operon. It is possible that EutT is encoded by the eut operon because EutT needs to be localized to the carboxysome-like structure, where ethanolamine degradation is thought to occur (20, 33). Whether EutT is localized within the carboxysome-like structure remains an open question.
Acknowledgments
This work was supported in part by NIH grant GM40313 to J.C.E.-S. N.R.B. was a Howard Hughes Medical Institute predoctoral fellow.
REFERENCES
- 1.Balch, W. E., and R. S. Wolfe. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 32:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer, C. B., M. V. Fonseca, H. M. Holden, J. B. Thoden, T. B. Thompson, J. C. Escalante-Semerena, and I. Rayment. 2001. Three-dimensional structure of ATP:corrinoid adenosyltransferase from Salmonella typhimurium in its free state, complexed with MgATP, or complexed with hydroxycobalamin and MgATP. Biochemistry 40:361-374. [DOI] [PubMed] [Google Scholar]
- 3.Benson, N. R., and B. S. Goldman. 1992. Rapid mapping in Salmonella typhimurium with Mud-P22 prophages. J. Bacteriol. 174:1673-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkowitz, D., J. M. Hushon, H. J. Whitfield, J. Roth, and B. N. Ames. 1968. Procedure for identifying nonsense mutations. J. Bacteriol. 96:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochner, B. R., H.-C. Huang, G. L. Schieven, and B. N. Ames. 1980. Positive selection for loss of tetracycline resistance. J. Bacteriol. 143:926-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caetano-Annoles, G. 1993. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 3:85-92. [DOI] [PubMed] [Google Scholar]
- 7.Castilho, B. A., P. Olfson, and M. J. Casadaban. 1984. Plasmid insertion mutagenesis and lac gene fusions with mini-Mu bacteriophage transposons. J. Bacteriol. 158:488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, R. K., D. Botstein, T. Watanabe, and Y. Ogata. 1972. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high transducing lysate. Virology 50:883-898. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, R. W., D. Botstein, and J. R. Roth. 1980. A manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 11.Elliott, T., and J. R. Roth. 1988. Characterization of Tn10d-Cam: a transposition-defective Tn10 specifying chloramphenicol resistance. Mol. Gen. Genet. 213:332-338. [DOI] [PubMed] [Google Scholar]
- 12.Escalante-Semerena, J. C., S.-J. Suh, and J. R. Roth. 1990. cobA function is required for both de novo cobalamin biosynthesis and assimilation of exogenous corrinoids in Salmonella typhimurium. J. Bacteriol. 172:273-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fonseca, M. V., N. R. Buan, A. R. Horswill, I. Rayment, and J. C. Escalante-Semerena. 2002. The ATP:co(I)rrinoid adenosyltransferase (CobA) enzyme of Salmonella enterica requires the 2′-OH group of ATP for function and yields inorganic triphosphate as its reaction byproduct. J. Biol. Chem. 277:33127-33131. [DOI] [PubMed] [Google Scholar]
- 14.Fonseca, M. V., and J. C. Escalante-Semerena. 2001. An in vitro reducing system for the enzymic conversion of cobalamin to adenosylcobalamin. J. Biol. Chem. 276:32101-32108. [DOI] [PubMed] [Google Scholar]
- 15.Fonseca, M. V., and J. C. Escalante-Semerena. 2000. Reduction of cob(III)alamin to cob(II)alamin in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 182:4304-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham, G., R. S. Nairn, and G. W. Bates. 1978. Polyacrylamide gel staining with Fe2+-bathophenanthroline sulfonate. Anal. Biochem. 88:434-441. [DOI] [PubMed] [Google Scholar]
- 17.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Havemann, G. D., E. M. Sampson, and T. A. Bobik. 2002. PduA is a shell protein of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 184:1253-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, C. L., E. Pechonick, S. D. Park, G. D. Havemann, N. A. Leal, and T. A. Bobik. 2001. Functional genomic, biochemical, and genetic characterization of the Salmonella pduO gene, an ATP:cob(I)alamin adenosyltransferase gene. J. Bacteriol. 183:1577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kofoid, E., C. Rappleye, I. Stojiljkovic, and J. Roth. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 181:5317-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maloy, S. R., and W. D. Nunn. 1981. Selection for loss of tetracycline resistance by Escherichia coli. J. Bacteriol. 145:1110-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 24.Ratzkin, P., and J. R. Roth. 1978. Cluster of genes controlling proline degradation in Salmonella typhimurium. J. Bacteriol. 133:744-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roof, D. M., and J. R. Roth. 1992. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J. Bacteriol. 174:6634-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roof, D. M., and J. R. Roth. 1988. Ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 170:3855-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roth, J. R., J. G. Lawrence, M. Rubenfield, S. Kieffer-Higgins, and G. M. Church. 1993. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J. Bacteriol. 175:3303-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmieger, H. 1971. A method for detection of phage mutants with altered transduction ability. Mol. Gen. Genet. 100:378-381. [DOI] [PubMed] [Google Scholar]
- 29.Schmieger, H., and H. Bakhaus. 1973. The origin of DNA in transducing particles of P22 mutants with increased transduction frequencies (HT-mutants). Mol. Gen. Genet. 120:181-190. [DOI] [PubMed] [Google Scholar]
- 30.Schwarz, J. J., and P. B. Berget. 1989. The isolation and sequence of missense and nonsense mutations in the cloned bacteriophage P22 tailspike protein gene. Genetics 121:635-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, C. A., and I. Rayment. 1996. Active site comparisons highlight structural similarities between myosin and other P-loop proteins. Biophys. J. 70:1590-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sofia, H. J., G. Chen, B. G. Hetzler, J. F. Reyes-Spindola, and N. E. Miller. 2001. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29:1097-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stojiljkovic, I., A. J. Bäumler, and F. Heffron. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutj eutH gene cluster. J. Bacteriol. 177:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suh, S.-J. 1994. The role of CobA in adenosylation of corrinoids in Salmonella typhimurium. Ph.D. University of Wisconsin, Madison.
- 35.Suh, S.-J., and J. C. Escalante-Semerena. 1995. Purification and initial characterization of the ATP:corrinoid adenosyltransferase encoded by the cobA gene of Salmonella typhimurium. J. Bacteriol. 177:921-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warren, M. J., E. Raux, H. L. Schubert, and J. C. Escalante-Semerena. 2002. The biosynthesis of adenosylcobalamin (vitamin B12). Nat. Prod. Rep. 19:390-412. [DOI] [PubMed] [Google Scholar]
- 37.Way, J. C., M. A. Davis, D. Morisato, D. E. Roberts, and N. Kleckner. 1984. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32:369-379. [DOI] [PubMed] [Google Scholar]
- 38.Yamada, R.-H., S. Schimizu, and S. Fukui. 1968. Disproportionation of vitamin B12r under various mild conditions. Biochemistry 7:1713-1719. [DOI] [PubMed] [Google Scholar]
- 39.Youderian, P., P. Sugiono, K. L. Brewer, N. P. Higgins, and T. Elliott. 1988. Packaging specific segments of the Salmonella chromosome with locked-in Mud-P22 prophages. Genetics 118:581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]