Abstract
McrA is one of three functions that restrict modified foreign DNA in Escherichia coli K-12, affecting both methylated and hydroxymethylated substrates. We present here the first systematic analysis of the functional organization of McrA by using the GPS-LS insertion scanning system. We collected in-frame insertions of five amino acids at 46 independent locations and C-terminal truncations at 20 independent locations in the McrA protein. Each mutant was assayed for in vivo restriction of both methylated and hydroxymethylated bacteriophage (M.HpaII-modified λ and T4gt, respectively) and for induction of the E. coli SOS response in the presence of M.HpaII methylation, indicative of DNA damage. Our findings suggest the presence of an N-terminal DNA-binding domain and a C-terminal catalytic nuclease domain connected by a linker region largely tolerant of amino acid insertions. DNA damage inflicted by a functional C-terminal domain is required for restriction of phage T4gt. Disruption of the N-terminal domain abolishes restriction of both substrates. Surprisingly, truncation mutations that spare the N-terminal domain do not mediate DNA damage, as measured by SOS induction, but nevertheless partially restrict M.HpaII-modified λ in vivo. We suggest a common explanation for this “restriction without damage” and a similar observation seen in vivo with McrB, a component of another of the modified-DNA restriction functions. Briefly, we propose that unproductive site-specific binding of the protein to a vulnerable position in the λ genome disrupts the phage development program at an early stage. We also identified a single mutant, carrying an insertion in the N-terminal domain, which could fully restrict λ but did not restrict T4gt at all. This mutant may have a selective impairment in substrate recognition, distinguishing methylated from hydroxymethylated substrates. The study shows that the technically easy insertion scanning method can provide a rich source of functional information when coupled with effective phenotype tests.
The McrA, McrBC, and Mrr endonucleases of Escherichia coli K-12 recognize and cleave specific DNA sequences only when those sequences are modified. This is a departure from the general paradigm of restriction-modification systems, in which endonucleases cleave unmodified DNA and accompanying methyltransferases modify DNA to prevent cleavage. When first discovered, McrA and McrBC were termed RglA and RglB, respectively (for restricts glucoseless phage) for their ability to restrict glucosyltransferase-deficient T-even bacteriophages such as T4gt and T6gt. These contain 5-hydroxymethylcytosine instead of cytosine residues, without the further modification by glucose present in the wild-type phage. Subsequently, the McrA and McrBC (modified cytosine restriction) functions, which restrict substrates bearing 5-methylcytosine (5mC) modification, were discovered and shown to be identical to RglA and RglB (26, 27). By convention, phenotypes describing restriction of hydroxymethylated substrates such as T-even bacteriophages are still denoted Rgl, whereas those describing restriction of methylated substrates are referred to as Mcr (24).
All three endonucleases exhibit sequence specificity, although for the well-characterized McrBC enzyme, the specificity is weak (34). The specificity of McrA is poorly known, since its in vitro action has not been studied. Furthermore, evidence that McrA actually cleaves DNA is indirect, consisting of the bacteriophage restriction phenomenon and the ability of a partially disabled mcrA gene to mediate induction of a DNA damage reporter in the presence of a sensitive methyltransferase gene (22). Substrates sensitive to McrA restriction in vivo include those modified by M.HpaII (C5mCGG), M.Eco1831I (C5mCSGG), and M.SssI (5mCG) (14, 16, 27); however, a comprehensive survey of 5-methylcytosine methyltransferases with regard to McrA restriction has not yet been undertaken, so the precise recognition sequence remains unclear.
Genetically, mcrA resides in the E. coli K-12 genome on e14 (26), a 14-kb defective prophage which self-excises following induction of the DNA damage-inducible (SOS) regulon; this property has proven useful in the construction of McrA− strains more tractable for cloning methylated foreign DNA than wild-type E. coli K-12. The mcrA gene has been cloned and found to encode a 31-kDa protein product with 277 residues (13, 28). It is normally expressed at low levels in the cell, and regulation at the translational level has been suggested (29).
McrA has no close homologues in the public sequence databases (3), but two conserved motifs have been noted in its carboxy-terminal domain. First, four cysteine residues define a zinc finger-like motif common to the reverse transcriptase/intron nuclease-like proteins encoded in many group II introns (8). Second, an H-N-H endonuclease motif, common to both group I and II intron-encoded proteins as well as phage proteins and bacteriocins, overlaps the zinc finger motif (10, 30). As the zinc finger motif is not conserved among all of the H-N-H endonucleases, its presence or absence is not thought to affect the fold of the endonuclease domain (10). In keeping with this, a recent sequence-threading model of McrA residues 159 to 272 shows the zinc finger stabilizing the arrangement of the presumptive catalytic histidine residues rather than participating directly in the active site of the protein (4). Outside the modeled region, proposed to be the catalytic core of the enzyme, McrA exhibits weak similarity only to the group II intron maturases, which have not been structurally characterized. Thus, there remains little insight into the structure and function of the majority of the McrA protein.
The relationship between the McrA and RglA phenotypes is also poorly understood. Of the mcrA mutants that have been described previously, several appeared to separate the McrA and RglA phenotypes, i.e., abolishing one activity but not the other (13, 29). However, the published results are inconsistent: two very similar mutants isolated by independent groups were characterized as McrA+ RglA− by one group and McrA− RglA+ by the other. This inconsistency aside, such mutants may ultimately help characterize the recognition elements responsible for distinguishing methylated from hydroxymethylated substrates.
The present study sought to characterize the functional domain architecture of McrA genetically by insertion scanning mutagenesis. These insertion mutants contain five additional contiguous amino acids interpolated at random locations within the polypeptide chain; the resulting structural perturbations will have different effects on the protein's activity depending on the location and composition of the inserted residues. Previous studies with this technique have yielded significant structure-function relationship information on proteins whose structure is known, including the XerD recombinase from Salmonella enterica serovar Typhimurium and β-lactamase from pBR322 (5, 11). As might be expected, insertions near a protein's active site as well as those that disrupt regions of secondary structure were in general shown to have more deleterious effects on activity than those occurring in linker regions and surface loops. In addition, some insertions affecting the substrate binding cleft of β-lactamase significantly altered the substrate specificity of that enzyme while sparing its catalytic activity (11). This study probed the domain structure of a protein of largely unknown structure by the generation of similar insertion mutants.
MATERIALS AND METHODS
Bacterial strains and media.
pJEK8 plasmid construction was carried out in E. coli strains ER2683 [fhuA2 glnV44 e14− rfbD1? relA1? endA1 spoT1? thi-1 Δ(mcrC-mrr)114::IS10 Δ(lacI-lacA)200 (F′ proAB lacIq ΔlacZM15 [Kanr] mini-Tn10)] (14) and ER1793 [fhuA2 Δ(lacZ)r1 glnV44 e14− trp-31 his-1 rpsL104 xyl-7 mtl-2 metB1 Δ(mcrC-mrr)114::IS10] (31) grown in Luria (2) or Luria-Bertani (32) medium supplemented with ampicillin. Cultures for other plasmid preparations were carried out in ER1793 grown in Luria-Bertani medium supplemented with antibiotics as necessary. Phage restriction assays were also performed in ER1793 grown in lambda broth (15) supplemented with 20 μg of ampicillin per ml. Bacteriophages were diluted in a 9:1 mixture of λdil (10 mM Tris-HCl [pH 7.5], 10 mM MgSO4) and 100 mM MgSO4.
DNA damage assays were carried out in E. coli strain ER2171 [F− fhuA2 Δ(argF-lac)U169 glnV44 e14− trp-31 his-1 rpsL104 xyl-7 mtl-2 metB1 dinD2::MudI1734 (Kanr LacZ[Ts]) Δ(mcrC-mrr)114::IS10] containing pACYC-MHpaII grown on Luria medium supplemented with ampicillin, chloramphenicol, and 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml. ER2171 was constructed by introducing the dinD::lacZ reporter of AP1-200 (22) into ER1996 (20). The reporter allele in AP1-200 was found to be temperature sensitive for β-galactosidase activity, unlike the parental reporter (12). A nalidixic acid gradient will cause dose-dependent induction of blue color on X-Gal at 30 or 37°C, but at 42°C strains carrying this allele are white regardless of DNA damage. The β-galactosidase activity is low enough that uninduced colonies are white at low temperature, where the parental reporter gives a light blue color in the uninduced state that grows darker with age. This background reduction improves discrimination between colonies suffering DNA damage and those not when tested at 37°C or below. We presume that this allele carries a mutation resulting in lower specific activity of the β-galactosidase moiety even at permissive temperatures.
Enzymes and general techniques.
All enzymes were from New England Biolabs, Inc., and used according to the manufacturer's instructions. Plasmids were isolated with the QIAprep Spin miniprep kit (Qiagen, Inc.) or the Compass mini plasmid prep kit (American Bioanalytical, Inc.). Plasmid pNEB193 was from New England Biolabs, Inc.
Plasmid construction.
pJEK8 was constructed from pER137, which has been described previously (26), in several steps. The resulting construct (3,789 bp) consists of a roughly 1.1-kb HpaI-BsrGI E. coli genomic DNA fragment containing mcrA fused to the 2.7-kb BsgI-EcoRI fragment of pBR322.
pACYC-MHpaII was constructed by inserting the 2.0-kb HindIII fragment containing the hpaIIM gene (6) into the HindIII site of pACYC184. The orientation of the insert is unknown, but M.HpaII expression was confirmed by complete protection of the construct from cleavage by HpaII.
DNA sequence.
The nucleotide coordinates discussed in this work refer to the 1,040-bp DNA sequence of the E. coli genomic fragment containing mcrA from GenBank accession no. Z19104 (28). pJEK8 contains nucleotides 35 to 1037 of this sequence, and the mcrA coding region is nucleotides 124 to 957 of this sequence.
Bacteriophage restriction assays.
All bacteriophage assays were performed on phage medium plates supplemented with 100 μg of ampicillin per ml. Cross streak and spot tests were performed as described previously (26). For full plate titers, 100 μl of cells prepared as for the spot tests was combined with 100 μl of bacteriophage (T4gt for RglA assays or λ methylated by M.HpaII for McrA assays) and incubated for 20 min at room temperature; 2 ml of melted top agar was combined with the mixture and spread on phage plates supplemented with ampicillin.
Linker scanning mutagenesis.
Mutagenesis was performed with the GPS-LS linker scanning system (New England Biolabs, Inc., Beverly, Mass.) according to the manufacturer's instructions. ER1793 cells were transformed by electroporation or chemical means with between 1 and 10 μl of the reaction and selected with ampicillin and either kanamycin or chloramphenicol, depending on the donor plasmid used. Transformants were assayed by cross-streaking, and plasmids were isolated from RglA− clones; 20 μl of each plasmid was digested with PmeI for 2 h at 37°C in a 50-μl volume, and the enzyme was heat killed by incubation for 20 min at 65°C. Half of each reaction was examined by gel electrophoresis to determine the completeness of the reaction and the correctness of the fragment sizes. The remainder was ligated overnight at 16°C with 400 U of T4 DNA ligase in a total volume of 100 μl. ER1793 cells were transformed again with 4 μl of each ligation reaction and selected with ampicillin. Transformants were assayed for RglA and/or McrA activity by cross-streaking, spot test, or full plate titer. In addition, loss of the PmeI fragment containing the selectable marker was confirmed by restriction analysis. Insertions were located by DNA sequencing with the GPS-N and GPS-S primers.
RESULTS
Strategy for analysis.
Mutagenesis of mcrA was accomplished with the GPS-LS kit, which employs a Tn7-derived minitransposon carrying a selectable marker, with ends modified to encode PmeI sites at the penultimate positions (1). In an in vitro reaction including a transposon donor plasmid, a target DNA, and the transposase, the transposase excises the transposon from the donor and inserts it into the target. Five base pairs of target sequence are duplicated in the process, with one copy present at each side of the new insertion. The site of insertion is essentially random. The reaction is then transformed into a strain nonpermissive for donor plasmid replication. When a marker in the transposon is selected, only transposition reaction products are recovered. In this case, plasmid pJEK8, carrying mcrA, was used as a target, resulting in a collection of derivatives each with a single transposon insertion at a random location. Digestion of these derivatives with PmeI removes nearly the entire transposon, and recircularization leaves behind, at the original site of transposition, 15 bp not present in pJEK8: 10 bp are contributed by the residual transposon itself, and 5 bp by the target site duplication (1). When inserted into a coding sequence, these 15 bp are translated to five new amino acids in two of three reading frames; the third frame is not open and results in a truncated protein product (Fig. 1).
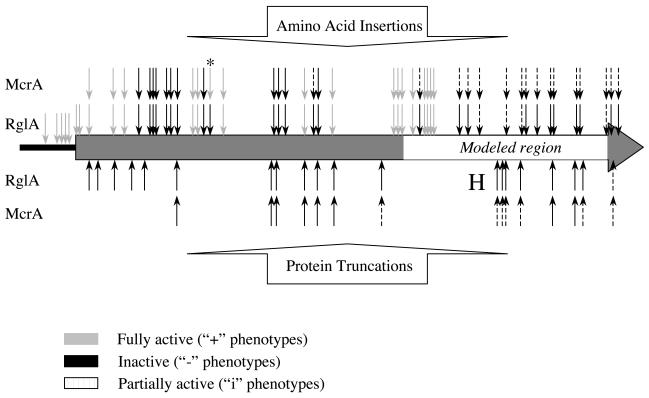
FIG. 1.
Nucleotides and amino acids added following transposon insertion and subsequent removal with PmeI. Three codons of the target sequence are shown, with numerals representing specific bases. x1, x2, and x3 represent the amino acids encoded by codons 123, 456, and 789, respectively. Insertion occurs in one of three reading frames: frame 1 (following base 5), frame 2 (following base 6), or frame 3 (following base 7). The 5-base target site duplication that results from the transposon insertion is underlined, and the amino acids added relative to the original sequence are boxed in gray. Some added amino acids are invariant, whereas others (represented by α, β, γ, and δ) are variable and dependent on the target sequence. Insertions in frame 1 almost always result in premature truncation of the expressed protein. However, in the rare case when codon 456 is itself a stop codon, an insertion in frame 1 will actually add two residues (ɛV, where ɛ = Y or C) to the C terminus of the expressed protein rather than truncate prematurely. α = any amino acid except M, E, K, Q, or W (in approximately 75% of cases it will be x2); β = I, K, M, N, R, S, or T; γ = L, M, or V; δ = H or Q.
The overall strategy for generating and analyzing the McrA mutants was as follows. Transposon insertions in pJEK8 were generated and categorized with the rapid RglA phenotype classification. Only plasmids conferring the RglA− phenotype were analyzed further. The precise location of the transposon insertion in each of these plasmids was determined by bidirectional sequencing from universal priming sites within the transposon. Next, a corresponding linker insertion was obtained from each transposon insertion by PmeI digestion and religation as described above. The RglA and McrA phenotypes were then determined for each linker mutant. Although all of the corresponding transposon insertions had been RglA−, the 15-bp insertions were expected to be far less disruptive than the full-length transposons. Indeed, RglA activity was restored for some but not all linker mutants. Finally, for a selected set of linker mutants, the ability to mediate DNA damage as revealed by SOS induction in the presence of M.HpaII was determined with a lacZ-based reporter system. Each step in the process will now be discussed in more detail.
Transposon mutagenesis.
The mcrA-containing plasmid pJEK8 was mutagenized in vitro with minitransposons in six separate transposition reactions: five reactions employed the transposon Transprimer-5, encoding resistance to kanamycin, and one used Transprimer-4, encoding chloramphenicol resistance. A total of 470 kanamycin- or chloramphenicol-resistant colonies were examined by cross-streaking analysis, and 129 (27%) were scored as RglA− (Table 1). We assumed that all mutants carrying insertions in the ampicillin resistance marker and origin of replication would be inviable, all mutants carrying transposon insertions in the mcrA coding region would be RglA−, and all mutants carrying transposon insertions outside the mcrA coding region would be RglA+. Given these assumptions, about 36% of transformants were expected to be RglA−, given a random distribution of insertions, in good agreement with the 27% obtained. The small discrepancy between the expected and observed values may be attributed to the failure of any of the above assumptions in certain cases, misinterpretation of cross-streaking results, or stochastic variation. None of the RglA+ clones were sequenced.
TABLE 1.
Summary of linker mutants generated
| Expt | Transposon | No. of transformantsa | No. RglA− | No. used in final analysis |
|---|---|---|---|---|
| 1 | Transprimer-5 | 100 | 21 | 13 |
| 2 | Transprimer-5 | 100 | 25 | 13 |
| 3 | Transprimer-5 | 99 | 35 | 24 |
| 4 | Transprimer-4 | 27 | 15 | 12 |
| 5 | Transprimer-5 | 45 | 12 | 10 |
| 6 | Transprimer-5 | 99 | 21 | 12 |
| Total | 470 | 129 | 84 |
Number of transformants examined by cross-streaking for loss of RglA activity.
The transposon insertion sites in those clones for which linker insertions were successfully obtained (a total of 84; see below) were located by DNA sequencing. In a large majority of cases, sequence data was obtained with two primers, GPS-N and GPS-S, reading out from the right and left ends of the transposon, respectively. In each of these cases, the expected 5-bp target site duplication was present. In the few cases in which sequence was obtained from only one of the two primers, the duplication was assumed to be present. In several cases in which sequence data from GPS-N and GPS-S was ambiguous or conflicting, sequence was obtained from the corresponding linker mutant with independent primers hybridizing to the mcrA sequence. In all of these cases, the 15-bp insertion was present as expected.
Generation and analysis of linker mutants.
The target plasmid pJEK8 contained no PmeI sites, so digestion of the transposon insertion plasmids with PmeI generated two fragments, 3.8 kb (including all of pJEK8 plus 15 bp of new DNA at the site of transposon insertion; see Fig. 1) and either 1.7 or 1.4 kb (including the bulk of Transprimer-5 or Transprimer-4, respectively). Those of the PmeI-digested RglA− clones which yielded the expected digestion pattern were religated, yielding 3,804-bp plasmids containing a functional origin of replication and encoding the mcrA linker mutants. Of the 129 RglA− clones, 84 were used in the final analysis (Table 1). The remaining 45 were eliminated for a variety of reasons, specifically: aberrant PmeI digest patterns either before or after loss of the transposon fragment (28), poor sequencing results, possibly due to double transposon insertion (3), location of the insertion outside the mcrA fragment (2), clerical errors specific to one experiment (8), and miscellaneous problems with strain growth, plasmid copy number, or spot test results (4).
The linker mutants were tested for RglA activity by spot test with phage T4gt. These showed a range of restriction of greater than 106-fold between the RglA+ phenotype from ER1793(pJEK8) cells expressing wild-type McrA and the RglA− phenotype from ER1793(pBR322) cells not expressing McrA. Tables 2 and 3 show the degree of restriction and corresponding phenotypes of the 84 McrA mutants generated in this study with amino acid insertions and C-terminal truncations, respectively. The phenotypes of most mutants resembled either the archetypal RglA+ or RglA− phenotype, but several exhibited clearly intermediate degrees of T4gt restriction, and these were collectively termed RglAi phenotypes.
TABLE 2.
Linker mutation locations and phenotypes (amino acid insertions)
| Nucleo- tide | Amino acid no. | Residues inserted | Isolate(s) | Phenotypea
|
||
|---|---|---|---|---|---|---|
| RglA | McrA | SOS | ||||
| pBR322 | − | − | White | |||
| pJEK8 | + | + | X | |||
| 85 | 3-58 | + | ||||
| 108 | 1-2 | + | ||||
| 112 | 1-39 | + | ||||
| 115 | 3-27 | + | ||||
| 117 | 5-32 | + | ||||
| 124 | 0 | MFKHI | 1-66 | + | ||
| 125b | 4-16 | + | ||||
| 133 | 3 | LFKHV | 6-19 | + | + | |
| 168 | 15 | CLNKC | 1-20, 1-68, 3-42, 6-97 | + | + | |
| 184 | 20 | VFKQE | 4-11, 4-21 | + | + | |
| 208 | 28 | VFKHL | 4-22 | − | − | White |
| 235 | 37 | MFKQN | 3-29 | − | − | White |
| 237 | 38 | CLNNK | 5-45 | − | − | White |
| 238 | 38 | VFKHK | 3-89 | − | − | White |
| 256 | 44 | VFKHL | 5-30 | − | − | White |
| 261 | 46 | CLNNY | 2-85, 3-34, 5-49 | − | − | White |
| 271 | 49 | LFKHE | 3-21 | − | − | Pale blue |
| 298 | 58 | VFKHQ | 5-34 | + | + | X |
| 303 | 60 | CLNIV | 5-57 | + | + | X |
| 313 | 63 | VFKHL | 3-55 | − | − | Pale blue |
| 324 | 67 | CLNTS | 1-75 | − | + | Mixedc |
| 345 | 74 | CLNNS | 3-17 | + | + | X |
| 418 | 98 | MFKHL | 1-24, 2-47 | − | − | White |
| 423 | 100 | CLNKM | 3-33 | − | − | Pale blue |
| 432 | 103 | CLNSY | 3-36 | − | − | White |
| 477 | 118 | CLNTG | 4-3 | + | + | X |
| 490 | 122 | LFKQK | 3-8, 3-22 | − | i | Blue |
| 495 | 124 | CLNRI | 3-66 | − | − | White |
| 508 | 128 | LFKHV | 1-54 | + | + | X |
| 594 | 157 | CLNTL | 1-77 | + | + | |
| 597 | 158 | CLNIL | 6-16 | + | + | |
| 600 | 159 | CLNMN | 2-66 | + | + | |
| 625 | 167 | MFKHK | 6-98 | + | + | X |
| 640 | 172 | LFKHQ | 3-57 | − | i | White |
| 645 | 174 | CLNTE | 4-12 | + | + | |
| 646 | 174 | VFKHE | 3-30 | + | + | |
| 654 | 177 | CLNTR | 4-6 | + | + | |
| 655 | 177 | MFKHR | 6-6 | + | + | Xd |
| 696 | 191 | CLNIR | 3-24 | − | i | White |
| 715 | 197 | LFKQA | 6-58 | − | i | White |
| 732 | 203 | CLNKS | 1-33 | i | i | Blue |
| 774 | 217 | CLNIY | 6-28 | i | i | Blue |
| 795 | 224 | CLNTY | 3-18 | i | i | Pale blue |
| 798 | 225 | CLNNL | 1-42 | − | i | White |
| 810 | 229 | CLNNH | 2-6 | − | i | White |
| 828 | 235 | CLNTS | 1-84 | − | i | White |
| 829 | 235 | VFKHS | 5-66 | − | i | White |
| 874 | 250 | LFKQN | 6-18 | − | i | White |
| 876 | 251 | CLNNC | 6-12 | − | i | White |
| 921 | 266 | CLNIE | 2-5, 4-27 | i | i | Blue |
| 925 | 267 | LFKQM | 1-57, 3-48 | i | i | Blue |
| 937 | 271 | MFKHN | 3-46 | i | i | Blue |
Both RglA and McrA phenotypes were defined as follows: −, <10-fold restriction; i, intermediate, 10- to 1,000-fold restriction; +, >1,000-fold restriction. SOS phenotyes are described by the color of the colony on X-Gal. X, no colonies were obtained.
The insertion here is in frame 1 but results only in additional nucleotides upstream of the start codon, not in truncation of the expressed protein.
Mutant 1-75 yielded a mixture of white and blue colonies on X-Gal.
Mutant 6-6 yielded several white colonies when transformed at high concentration.
TABLE 3.
Linker mutation locations and phenotypes (protein truncations)
| Nucleo- tide no. | Amino acid no. | Residue(s) inserteda | Isolate(s) | Phenotypeb
|
||
|---|---|---|---|---|---|---|
| RglA | McrA | SOS | ||||
| 134 | 4 | V* | 3-45 | − | ||
| 143 | 7 | V* | 3-56 | − | ||
| 170 | 16 | V* | 4-23 | − | ||
| 194 | 24 | V* | 2-80 | − | ||
| 224 | 34 | V* | 5-55, 5-61 | − | ||
| 269 | 48 | DV* | 4-10, 6-56 | − | − | White |
| 413 | 97 | V* | 5-38 | − | − | |
| 419 | 98 | NV* | 2-56 | − | − | |
| 476 | 118 | V* | 6-77 | − | − | |
| 497 | 124 | FV* | 2-49 | − | − | |
| 509 | 129 | V* | 4-13 | − | − | |
| 569 | 149 | V* | 2-24 | − | i | White |
| 761 | 213 | V* | 2-55 | − | i | White |
| 770 | 216 | V* | 3-26 | − | i | White |
| 773 | 217 | V* | 4-17 | − | i | White |
| 788 | 222 | V* | 2-42 | − | i | White |
| 830 | 236 | V* | 6-83 | − | − | White |
| 869 | 249 | V* | 2-46 | − | − | White |
| 881 | 252 | SV* | 2-81 | − | i | White |
| 926 | 268 | V* | 3-7 | i | i | Blue |
Most linker mutants were also tested for McrA activity by full plate titers with a stock of M.HpaII-methylated λ phage that had been methylated and packaged in vitro [λ.HpaII, 1.6 × 106 PFU/ml titered on ER1793(pBR322), with a roughly 5 × 104-fold range of restriction] (14). As in the RglA assays, McrA+ and McrA− phenotypes were determined on ER1793(pJEK8) and ER1793(pBR322) cells, respectively. Tables 2 and 3 show the degree of restriction and corresponding McrA phenotypes of linker mutants with amino acid insertions and truncations, respectively. Mutants exhibiting clearly intermediate degrees of λ.HpaII restriction were designated as having McrAi phenotypes.
RglA classification of linker mutants.
Linker insertions generated by this method result in either in-frame amino acid insertions or premature stops, depending on the reading frame. The amino acid insertions will have various composition, dependent partly on the target sequence at the site of insertion (Fig. 1). Fifty-six such mutants were isolated, representing 46 independent locations within the gene and 42 independent locations within the protein. Of the 46 distinct mutants, 17 restricted T4gt to a degree approximating that of wild-type McrA (RglA+), 23 exhibited no detectable restriction (RglA−), and 6 restricted the phage but to a degree significantly less than the wild type (RglAi). Figure 2 shows the distributions of these phenotypes across the gene graphically. Much of the protein is divided into tolerant and intolerant regions, judged by the clustering of black and gray RglA arrows in the upper half of the figure. A segment within the modeled region contained a cluster of mutations each giving partial activity.
FIG. 2.
Schematic location of GPS-LS insertions in the mcrA gene. The large horizontal arrow represents mcrA; some of the immediate upstream region is also shown. Vertical arrows above the gene represent amino acid insertion mutants (frames 2 and 3), and arrows below the gene represent truncation mutants (frame 1). Black arrows, RglA− or McrA−; dashed arrows, RglAi or McrAi; and gray arrows, RglA+ or McrA+. The region modeled by Bujnicki and coworkers (5) (residues 159 to 272 of the gene product) is indicated. The location of the HindIII site is indicated by the H, and the insertion in mutant 1-75 is marked with an asterisk (*).
The truncation mutants had linker insertions in frame 1 (Fig. 1). In this frame, the TAA sequence in the PmeI site is read as a stop codon, resulting in expressed proteins that are truncated at the point of insertion. Twenty-two such mutants were isolated, representing 20 distinct truncations. All of these mutants were RglA− with the exception of the C-terminal-most truncation, in which the last 9 residues of McrA (YVNINRLQK) are replaced with a single valine residue. This mutant exhibited partial RglA activity (RglAi). The amino acid insertions in this region were also RglAi (see Tables 2 and 3 and Fig. 2).
A final group of mutants, of which six were isolated, have linker insertions immediately upstream of the mcrA coding sequence. Although the expressed proteins should all be identical to wild-type McrA, the original transposon insertions were no doubt RglA− due to separation of the coding sequence from its promoter. In agreement with this, all corresponding linker mutants were RglA+. Even clone 4-16, in which the distance between the putative ribosome-binding site and the mcrA coding sequence is increased by 15 bp, appeared to be fully RglA+.
McrA classification of linker mutants.
In a surprisingly large number of cases (about one-third of all mutants tested), the McrA phenotype did not match the RglA phenotype, confirming the separation-of-phenotype phenomenon observed in earlier studies (13, 29) (see Discussion). The relationship between the two phenotypes can be described by the following rules, with one exception: (i) mutants that are RglA+ are also McrA+, and vice versa; (ii) mutants that are RglAi are also McrAi; and (iii) mutants that are RglA− can be either McrA− or McrAi. In most cases, RglA− mutants with insertions or truncations in the N-terminal half were McrA−, and RglA− mutants with insertions or truncations in the C-terminal half were McrAi.
To confirm that the McrAi phenotype observed in the truncation mutants did not result from spurious translational readthrough of the inserted stop codon, the AfeI-PmeI mcrA fragments from mutants 2-24 (McrAi) and 4-13 (McrA−) were subcloned into the PmeI site of pNEB193. These subclones did not contain any mcrA-derived sequence downstream of the stop codon but still conferred the same McrA phenotypes as their respective parental plasmids, indicating that the McrAi phenotype is intrinsic to the mutant protein (data not shown).
The single exception to the rules above was mutant 1-75, which had a 5-amino-acid insertion after residue 67 and was RglA− but fully McrA+. All cases demonstrating separation of phenotype indicated that RglA activity is more easily disrupted by mutation than is McrA activity, or conversely, λ.HpaII is more easily restricted than T4gt.
Assessment of DNA damage.
Piekarowicz and colleagues suggested that wild-type mcrA can mediate induction of the LexA regulon (the SOS response to DNA damage) in the presence of M.HpaII methylation (22). We exploited this property to determine which of the McrA mutants could inflict DNA damage in the presence of M.HpaII methylation. Each of the linker mutant plasmids was introduced into ER2171(pACYC-MHpaII), an E. coli reporter strain with lacZ fused to the damage-inducible dinD locus and expressing M.HpaII from a compatible plasmid. This strain yields blue colonies on X-Gal plates at 37°C and below when the SOS response has been induced. Similar reporter strains have been used to screen for restriction endonucleases and methyltransferases (9, 12, 22).
The final columns of Tables 2 and 3 show the colony colors obtained in this test. As expected, pBR322 yielded white colonies, as did 12 of 15 derivatives with RglA− McrA− phenotypes. No colonies were obtained from strains transformed with the wild type or any derivatives with the RglA+ McrA+ phenotypes. This was anticipated also; strains containing mcrA restrict plasmids containing hpaIIM (27), so it is reasonable to expect that the opposite configuration of genes would also be incompatible. All RglAi McrAi mutants yielded blue colonies, suggesting the presence of McrA-dependent DNA cleavage activity in these cases; 15 of 16 RglA− McrAi mutants yielded white colonies, suggesting the absence of DNA cleavage activity. The partial restriction of λ.HpaII by these mutants is therefore independent of apparent McrA nucleolytic activity and must result from some other mechanism.
There were a few exceptions to the above generalizations among the mutants with 5-amino-acid insertions. One RglA− McrAi mutant with an insertion in the middle of the protein yielded blue colonies. A more sensitive RglA assay indicated no more than threefold restriction of T4gt by this mutant. Although we consider restriction in this range to be RglA−, the blue phenotype on X-Gal suggests some level of DNA cleavage competence. Finally, three RglA− McrA− mutants carrying insertions in the middle of the protein yielded pale blue colonies on X-Gal. This color did not appear until about 24 h after plating and was significantly less intense than that scored as blue. We grouped these with whites when considering them below (Discussion).
Distribution of insertions.
The distribution of transposon insertions is shown in Fig. 2. Not shown are duplicate insertions. Nine of 72 sites yielded multiple insertions: one site had four insertions, one had three insertions, and seven sites had two insertions. In 12 of the 16 possible pairs of duplicate insertions, the two members were generated by different transposition reactions and thus were independent.
The distribution of transposons appears random and therefore is compatible with previous reports (1). The longest region that did not receive an insertion was between nucleotides 345 and 413 (68 bp long). Given a completely random distribution of 84 insertions in 872 bp (nucleotides 85 to 957 in our sequence), the longest run of sequence that we should expect to find without an insertion is approximately 67 bp, in excellent agreement with our observed value. (Expected run length was calculated as K = log1/[1−P] N, where K is the longest expected run, P = 84/872 is the probability of an insertion at any given location, and N is the number of windows of size K, which we approximate as the sequence length 872.) Furthermore, no obvious biases were observed specific to either Transprimer-4 or Transprimer-5 (Tables 2 and 3).
DISCUSSION
McrA structural predictions.
The phenotypes of the mutants obtained here are broadly compatible with the structural predictions of Bujnicki and coworkers (4), as far as they go. Endonucleases with the H-N-H catalytic motif suggested for McrA belong to the ββα-Me superfamily of nucleases (18). The core ββα-Me catalytic domain of McrA (residues 159 to 272) was modeled by Bujnicki and coworkers with a sequence-threading approach. This region contains three histidine residues (H-228, H-252, and H-256) predicted to coordinate the Mg2+ ion, as well as four cysteine residues (C-207, C-210, C-248, and C-251) which form a putative zinc finger, most likely involved in stabilizing the structure.
As expected from this model, all C-terminal truncations within this region abolished RglA activity and SOS induction with the exception of isolate 3-7 (Table 3), which lacks only the extreme C-terminal 9 residues. This mutant shows an intermediate phenotype (RglAi McrAi). Furthermore, the phenotypes of amino acid insertion mutants are consistent with the proposed structure. Within the modeled region, only amino acid insertions after residues 159, 167, 174, and 177 resulted in full activity (RglA+ McrA+). The structural elements proposed for these locations are loops or regions connecting secondary structural elements at the periphery of the structure. The region may represent the C-terminal end of a larger connector region extending outside of the modeled area. Six other amino acid insertion mutants retained partial restriction activity (after residues 203, 217, 224, 266, 267, and 271) and also induced the SOS response. While these insertions were also in proposed surface loop regions, they were more centrally located in the proposed tertiary structure. This could cause enough structural distortion to affect catalytic activity. The remaining nine insertions in this region, many of which were located within putative helices or proximal to metal ion-coordinating residues, abolished both RglA restriction and SOS induction completely.
Outside of the modeled region, we found additional segments critical for restriction activity in the N-terminal domain of the protein. Between residues 28 and 124, a majority of insertions (12 of 18) were inactive in all tests. Since the active site likely resides in the modeled C-terminal region, DNA binding and sequence specificity are probable roles for these N-terminal intolerant segments. The extreme N terminus (residues 1 to 20) and the putative interdomain connector region (residues 128 to 177) were highly tolerant of the five-residue insertions generated in this study. Mutants with insertions in the extreme C terminus, on the other hand, suffered partial loss of both restriction activities. This may result from spatial proximity of this region to the cysteine residues coordinating the Zn2+ ion as per the threading model (4); distortion of this region by insertion may cause loss of or reduced affinity for the metal ion.
Effect of amino acid identity in insertions.
The insertion of five amino acid residues within a protein will affect the overall structure both by distorting the backbone and by introducing new noncovalent, and potentially covalent, interactions with the remainder of the protein via the new side chains. Among the mutants generated here, there were four cases in which different sets of amino acids were introduced at the same location, after residues 38, 174, 177, and 235. In all four cases, the phenotypes of both insertions were identical. Based on this evidence, it seems that the backbone distortion is the more influential of the two structural effects, but side chain effects may predominate in other cases.
Comparison with published mutants.
The small number of nondeletion mutants of McrA which have been described previously are summarized in Table 4. Entries include both characterized mutants, for which the precise changes relative to the wild-type sequence are known, and mutagenized E. coli strains, for which the sequence change(s) underlying the restriction phenotypes is unknown. The insertion mutation mcrA1272::Tn10 abolished RglA restriction activity, as did all of our full-length transposon insertions.
TABLE 4.
Summary of previously characterized mcrA mutants
| Mutant | Phenotypea
|
Description | Reference | |
|---|---|---|---|---|
| McrA | RglA | |||
| mcrA1272::Tn10 | − | − | Tn10 insertion approx. 100 bp upstream of HindIII site | 25 |
| AP1-100-1 | +(Ts) | +(Ts) | Uncharacterized; strain derived from AP1-100 | 21 |
| AP1-100-3 | +(Ts) | i | Uncharacterized; strain derived from AP1-100 | 21 |
| MM294 ES1 | +(Ts) | i | Uncharacterized; strain derived from MM294 | 21 |
| AP1-100-9 | − | − | Uncharacterized; strain derived from AP1-100 | 21 |
| pMH611 | − | + | McrA 1-197 + 21 vector-derived residues at C terminus | 13 |
| pDRR551.6 | + | − | McrA 1-197 + 7 residues after frameshift at C terminus | 29 |
| pPA2 | + | − | 7 vector-derived residues at N terminus + McrA 14-277 | 29 |
Ts, temperature sensitive. These phenotypes were obtained at 30°C; restriction activity in all cases was lower at 42°C.
The remaining three characterized mutants in Table 4 as well as two of the four uncharacterized mutant strains were reported to have selectively lost either RglA or McrA activity. Curiously, two independently isolated mutations involving frameshifts at the same HindIII site were described as giving opposite phenotypes: pMH611 was RglA+ McrA− (29) and pDRR551.6 was RglA− McrA+ (13). Both mutant proteins have lost the C-terminal 80 residues of the wild-type protein (i.e., were truncated after residue 197), and each has substituted a different vector-derived residue sequence. It seems unlikely from our results (discussed below) that the different C-terminal appendages have different effects. Our results are compatible with the report of Shivapriya et al. (29) and contradict those of Hiom and Sedgwick (13).
Separation of restriction phenotypes.
All 17 mutants exhibiting phenotype separation were RglA−; of these, only one retained full McrA+ activity, and this one will be discussed later. The remaining 16 showed partial activity (McrAi) and all carried mutations located in the C-terminal half of the protein. Six of these were truncation mutants, most comparable to the HindIII fill-in mutants reported earlier; 15 of these 16 McrAi mutants (including all of the truncations) were also white in the SOS-reporting strain, and therefore we conclude that they are not able to inflict DNA damage in the presence of HpaII-methylated DNA, the presumed target of restriction.
We propose that for these 15 mutants, the separation of phenotype reflects loss of cleavage activity but not DNA binding activity for both targets. The residual restriction phenotype, we suggest, is due to interference with phage development by binding rather than cleavage. The RglA and McrA phenotype assays differ in two fundamental ways, the type of bacteriophage (T4 or λ) used for the assay and the type of DNA modification present (5-hydroxymethylcytosine or 5-methylcytosine). λ is a temperate phage dependent on the host RNA and DNA polymerases, all replication functions except initiator proteins O and P, and all translation functions. T4 is a virulent phage that modifies the host's RNA polymerase, elaborates all its own replication functions, and brings its own tRNA to the translation process (19). A DNA binding protein acting at a critical site on an infecting λ genome might disrupt phage development efficiently, while the same protein bound to an infecting T4 genome might be without effect. The boundaries of the DNA binding domain are delineated by truncation mutants 4-13 (residue 129, null phenotype) and 2-24 (residue 149, RglA− McrAi/white; Table 3); the first 130 to 149 residues are required to confer McrAi and, we propose, for functional DNA binding activity.
This explanation for residual restriction of λ but not T4 might also be extended to the properties of certain mutants of the unrelated McrBC system. Mutants defective in McrC do not restrict T4gt but still restrict MspI-modified λ (λ.MspI) partially (7). M.MspI methylates the same sequence as does M.HpaII (CCGG) but at the first cytosine rather than the second. Studies subsequent to the original report have shown that McrC is absolutely required for cleavage of all substrates tested in vitro but that McrB alone retains DNA binding activity (17, 23, 33, 34). Since site-specific DNA binding of truncated McrA and of McrB would be directed to a similar set of sites (CCGG, differently modified) in the phage genomes, this proposal economically resolves the paradoxical behavior in both systems.
A further inference can be drawn; there may be a privileged location in λ that contains CCGG and is particularly sensitive to interference by DNA binding proteins. This inference rests on the observation that not all McrBC-sensitive methyltransferases give λ substrates that behave like λ.MspI (7, 14). Thus, binding of McrB at different locations, as dictated by a different methylation patterns, yields different behaviors, but binding of a wholly different protein to a similar set (or subset) of sites yields similar behavior.
Other partial-restriction phenotypes.
Mutants carrying mutations that only partially disrupt the catalytic domain should be capable of partially restricting both T4gt and λ.HpaII as well as inducing SOS. This combination of properties was observed in the RglAi McrAi mutants, all with insertions in the proposed catalytic domain (Table 2 and Fig. 2).
One RglA− McrAi mutant carrying an insertion near the border of the proposed binding domain (residue 122) retained blue color in the SOS reporter strain, and three mutants carrying insertions within the proposed boundaries of the DNA binding domain were RglA− McrA− but gave a pale blue color on X-Gal. These mutants may retain limited cleavage activity but represent various degrees of debilitation of enzymatic activity or protein stability relative to the RglAi McrAi class.
The last mutant showing phenotype separation is mutant 1-75. This mutant did not display partial disability in any assay; it was fully RglA− but also fully McrA+. Moreover, it could not be stably maintained in the presence of M.HpaII expression; both blue and white colonies were observed on X-Gal in the SOS induction assay, presumably resulting from strong selection for further mutations that inactivate the enzyme completely. This mutation is a five-residue insertion in the proposed DNA binding domain. A reasonable proposal is that this mutation does indeed disable the recognition of hydroxymethylated but not methylated substrates.
Utility of insertion scanning mutagenesis.
The ease with which many mutants can be generated, coupled with a variety of phenotypic assays, has enabled us to propose a model for the functional domains of the McrA protein, an N-terminal DNA binding domain and a C-terminal cleavage domain. In addition, the convenient restriction sites associated with the mutations provide ready tools for further manipulation. A straightforward test of the model will be isolation of protein from truncation mutants that spare McrA activity partially; these should enable isolation of a DNA-binding protein. Characterization of the sole mutant that is fully McrA+ and fully RglA− may enable better understanding of sequence recognition elements in this protein.
Acknowledgments
We thank Julie Kaminski and Rebecca Leary for excellent technical assistance and Marion Sibley for assistance with plasmid construction. We also thank Julia Kelleher Thompson for preparation of methylated λ phage.
REFERENCES
- 1.Biery, M. C., F. J. Stewart, A. E. Stellwagen, E. A. Raleigh, and N. L. Craig. 2000. A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res. 28:1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooks, J. E., J. S. Benner, D. F. Heiter, K. R. Silber, L. A. Sznyter, T. Jager-Quinton, L. S. Moran, B. E. Slatko, G. G. Wilson, and D. O. Nwankwo. 1989. Cloning the BamHI restriction modification system. Nucleic Acids Res. 17:979-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bujnicki, J. M. 2000. Phylogeny of the restriction endonuclease-like superfamily inferred from comparison of protein structures. J. Mol. Evol. 50:39-44. [DOI] [PubMed] [Google Scholar]
- 4.Bujnicki, J. M., M. Radlinska, and L. Rychlewski. 2000. Atomic model of the 5-methylcytosine-specific restriction enzyme McrA reveals an atypical zinc finger and structural similarity to ββαMe endonucleases. Mol. Microbiol. 37:1280-1281. [DOI] [PubMed] [Google Scholar]
- 5.Cao, Y., B. Hallet, D. J. Sherratt, and F. Hayes. 1997. Structure-function correlations in the XerD site-specific recombinase revealed by pentapeptide scanning mutagenesis. J. Mol. Biol. 274:39-53. [DOI] [PubMed] [Google Scholar]
- 6.Card, C. O., G. G. Wilson, K. Weule, J. Hasapes, A. Kiss, and R. J. Roberts. 1990. Cloning and characterization of the HpaII methylase gene. Nucleic Acids Res. 18:1377-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dila, D., E. Sutherland, L. Moran, B. Slatko, and E. A. Raleigh. 1990. Genetic and sequence organization of the mcrBC locus of Escherichia coli K-12. J. Bacteriol. 172:4888-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferat, J.-L., and F. Michel. 1993. Group II self-splicing introns in bacteria. Nature 364:358-361. [DOI] [PubMed] [Google Scholar]
- 9.Fomenkov, A., J. P. Xiao, D. Dila, E. Raleigh, and S. Y. Xu. 1994. The ‘endo-blue method’ for direct cloning of restriction endonuclease genes in E. coli. Nucleic Acids Res. 22:2399-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorbalenya, A. E. 1994. Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci. 3:1117-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallet, B., D. J. Sherratt, and F. Hayes. 1997. Pentapeptide scanning mutagenesis: random insertion of a variable five amino acid cassette in a target protein. Nucleic Acids Res. 25:1866-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heitman, J., and P. Model. 1987. Site-specific methylases induce the SOS DNA repair response in Escherichia coli. J. Bacteriol. 169:3243-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiom, K., and S. G. Sedgwick. 1991. Cloning and structural characterization of the mcrA locus of Escherichia coli. J. Bacteriol. 173:7368-7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelleher, J., and E. A. Raleigh. 1991. A novel activity in Escherichia coli K-12 that directs restriction of DNA modified at CG dinucleotides. J. Bacteriol. 173:5220-25223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleckner, N., D. F. Barker, D. G. Ross, and D. Botstein. 1978. Properties of the translocatable tetracycline-resistance element Tn10 in Escherichia coli and bacteriophage lambda. Genetics 90:427-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kravetz, A. N., M. V. Zakharova, I. V. Beletskaya, E. V. Sineva, M. M. Denjmuchametov, S. I. Petrov, L. I. Glatman, and A. S. Solonin. 1993. The cleavage sites and localization of genes encoding the restriction endonucleases Eco1831I and EcoHI. Gene 129:15304. [DOI] [PubMed] [Google Scholar]
- 17.Kruger, T., C. Wild, and M. Noyer-Weidner. 1995. McrB: a prokaryotic protein specifically recognizing DNA containing modified cytosine residues. EMBO J. 14:2661-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhlmann, U. C., G. R. Moore, R. James, C. Kleanthous, and A. M. Hemmings. 1999. Structural parsimony in endonuclease active sites: should the number of homing endonuclease families be redefined? FEBS Lett. 463:1-2. [DOI] [PubMed] [Google Scholar]
- 19.Mathews, C. K. 1994. An overview of the T4 developmental program, p. 1-8. In J. D. Karam (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 20.Mi, S., and R. J. Roberts. 1992. How M.MspI and M.HpaII decide which base to methylate. Nucleic Acids Res. 20:4811-4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piekarowicz, A., R. Yuan, and D. C. Stein. 1991. Isolation of temperature-sensitive McrA and McrB mutations and complementation analysis of the McrBC region of Escherichia coli K-12. J. Bacteriol. 173:150-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piekarowicz, A., R. Yuan, and D. C. Stein. 1991. A new method for the rapid identification of genes encoding restriction and modification enzymes. Nucleic Acids Res. 19:1831-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pieper, U., and A. Pingoud. 2002. A mutational analysis of the PD.D/EXK motif suggests that McrC harbors the catalytic center for DNA cleavage by the GTP-dependent restriction enzyme McrBC from Escherichia coli. Biochemistry 41:5236-5244. [DOI] [PubMed] [Google Scholar]
- 24.Raleigh, E. A., J. Benner, F. Bloom, H. D. Braymer, E. DeCruz, K. Dharmalingam, J. Heitman, M. Noyer-Weidner, A. Piekarowicz, P. L. Kretz, J. M. Short, and D. Woodcock. 1991. Nomenclature relating to restriction of modified DNA in Escherichia coli. J. Bacteriol. 173:2707-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raleigh, E. A., N. E. Murray, H. Revel, R. M. Blumenthal, D. Westaway, A. D. Reith, P. W. J. Rigby, J. Elhai, and D. Hanahan. 1988. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 15:1563-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raleigh, E. A., R. Trimarchi, and H. Revel. 1989. Genetic and physical mapping of the mcrA (rglA) and mcrB (rglB) loci of Escherichia coli K-12. Genetics 122:279-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raleigh, E. A., and G. Wilson. 1986. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc. Natl. Acad. Sci. USA 83:9070-9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramalingam, R., R. Prasad, R. Shivapriya, and K. Dharmalingam. 1992. Molecular cloning and sequencing of mcrA locus and identification of McrA protein in Escherichia coli. J. Biosci. 17:217-232. [Google Scholar]
- 29.Shivapriya, R., R. Prasad, I. L. Narayanan, S. Krishnaswamy, and K. Dharmalingam. 1995. Expression of the mcrA gene of Escherichia coli is regulated posttranscriptionally, possibly by sequestration of the Shine-Dalgarno region. Gene 157:201-207. [DOI] [PubMed] [Google Scholar]
- 30.Shub, D. A., H. Goodrich-Blair, and S. R. Eddy. 1994. Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem. Sci. 19:402-404. [DOI] [PubMed] [Google Scholar]
- 31.Sibley, M. H., and E. A. Raleigh. 2004. Cassette-like variation of restriction enzyme genes in Escherichia coli C and relatives. Nucleic Acids Res. 32:522-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Stewart, F. J., D. Panne, T. A. Bickle, and E. A. Raleigh. 2000. Methyl-specific DNA binding by McrBC, a modification-dependent restriction enzyme. J. Mol. Biol. 298:611-622. [DOI] [PubMed] [Google Scholar]
- 34.Sutherland, E., L. Coe, and E. A. Raleigh. 1992. McrBC: a multisubunit GTP-dependent restriction endonuclease. J. Mol. Biol. 225:327-348. [DOI] [PubMed] [Google Scholar]