Abstract
The EzrA protein of Bacillus subtilis is a negative regulator for FtsZ (Z)-ring formation. It is able to modulate the frequency and position of Z-ring formation during cell division. The loss of this protein results in cells with multiple Z rings located at polar as well as medial sites; it also lowers the critical concentration of FtsZ required for ring formation (P. A. Levin, I. G. Kurster, and A. D. Grossman, Proc. Natl. Acad. Sci. USA 96:9642-9647, 1999). We have studied the regulation of ezrA expression during the growth of B. subtilis and its effects on the intracellular level of EzrA as well as the cell length of B. subtilis. With the aid of promoter probing, primer extension, in vitro transcription, and Western blotting analyses, two overlapping σA-type promoters, P1 and P2, located about 100 bp upstream of the initiation codon of ezrA, have been identified. P1, supposed to be an extended −10 promoter, was responsible for most of the ezrA expression during the growth of B. subtilis. Disruption of this promoter reduced the intracellular level of EzrA very significantly compared with disruption of P2. Moreover, deletion of both promoters completely abolished EzrA in B. subtilis. More importantly, the cell length and percentage of filamentous cells of B. subtilis were significantly increased by disruption of the promoter(s). Thus, EzrA is required for efficient cell division during the growth of B. subtilis, despite serving as a negative regulator for Z-ring formation.
Binary fission in rod-shaped bacteria entails the formation of a transverse septum that divides a progenitor cell into two daughter cells of equal size. In the initiation of cell division, the tubulin-like cell division protein FtsZ polymerizes at the mid-cell into a ring structure that is required for subsequent recruitment of other cell division proteins and assembly of the cell division machinery (7, 10, 29, 36, 40). Temporally, the division process is tightly coupled to chromosome replication, chromosome segregation, and cell growth to ensure that both daughter cells inherit complete genomes and are of appropriate size and shape (12, 16, 17, 35).
Two mechanisms, nucleoid occlusion and the Min system, are involved in selection of the correct mid-cell site for cell division (1, 12, 17, 30). Nucleoid occlusion, although poorly defined, was revealed by the observation that cell division is largely inhibited in the vicinity of the nucleoid of cells in which DNA replication and/or segregation is perturbed (33). In other words, Z-ring assembly and cell wall synthesis is inhibited in the immediate vicinity of the actively replicating nucleoid. Since the Z ring appears to form at a position where the DNA concentration is low compared to the wild-type situations (9, 15, 35, 38), it is assumed that it is not the presence of DNA per se but the concentration of DNA which determines the position of Z-ring formation. The Min system, which inhibits FtsZ polymerization and also division at cell poles, has been extensively characterized for both Escherichia coli and Bacillus subtilis. For B. subtilis, the MinCD complex is recruited to the pole by a cell pole-associated protein, DivIVA, probably through a direct interaction with MinD (5, 11, 23, 31). Moreover, the DivIVA-MinCD complex remains associated with the newly formed pole after division, thereby preventing future division at these polar sites (11, 31).
The B. subtilis EzrA protein is a negative regulator for Z-ring formation. It is able to modulate the frequency and position of Z-ring formation during cell division. The lack of this protein causes cells with multiple Z rings located at polar as well as medial sites and lowers the critical concentration of FtsZ required for ring formation (26). The EzrA protein is homogeneously distributed in the cell membrane and localized to the cell division site once the Z ring is assembled, presumably via an interaction with FtsZ (26). A null mutation of ezrA has been found to suppress the defects in FtsZ polymer stability associated with minCD overexpression (27). Moreover, the effect of the loss of EzrA on cell division is enhanced by ZapA (a Z-ring-associated protein). The absence of ZapA and EzrA, but not ZapA itself, causes a severe block in cytokinesis of B. subtilis (14), suggesting that EzrA may play a positive role during cell division. Furthermore, EzrA may also participate in asymmetric division, since it is detectable in the spiral-like structure in sporulating cells (3). Recently, it was reported that EzrA can be degraded by an ATP-dependent CodWX protease in vitro (22). However, it remains elusive whether the intracellular level of EzrA is regulated during the growth of B. subtilis. This work was aimed at studying the transcription regulation of ezrA and its effect on B. subtilis cell division.
The putative promoter region(s) upstream of ezrA.
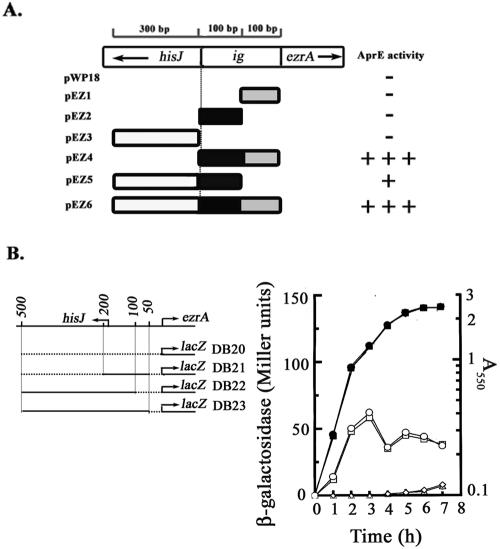
On the basis of the data of sequence analysis of the DNA encompassing ezrA and its upstream region, it was believed that ezrA constitutes a single-gene operon with the transcription direction opposite to that of hisJ (Fig. 1A). To search for the promoter(s) controlling ezrA expression, six different DNA fragments of up to 500 bp in length and upstream of the initiation codon of ezrA (Fig. 1A) were first synthesized by PCR using specific sets of primers (Table 1), digested with designed restriction enzymes, inserted into the promoter-probing vector, pWP18 (34), transformed into B. subtilis DB430, and analyzed for AprE activity on a skim-milk plate (Fig. 1A). Our data showed that both the DNA fragments containing the whole intergenic region (ig) between ezrA and hisJ, as shown for pEZ4 and pEZ6, were relatively high in producing the AprE activity (Fig. 1A). Dissection of this intergenic region into the two 100-bp halves, as shown for pEZ1 and pEZ2, completely eliminated the activity of each half, indicating that a putative promoter region is located in the junction of these two halves of DNA. However, only relatively weak AprE activity was observed for the DNA inserted into pEZ5. This weak activity was eliminated as the inserted DNA was further separated into two shorter halves, as shown for pEZ2 and pEZ3, suggesting that a second putative promoter region is located about 200 bp upstream of ezrA.
FIG. 1.
Localization of putative promoter region(s) upstream of ezrA and expression of putative promoter-lacZ fusions during B. subtilis growth. (A) Localization of the putative promoter region(s) upstream of ezrA. The DNA fragment containing the intergenic region (ig), hisJ coding region, or both regions was cloned into the promoter probing vector, pWP18, to generate pEZ1 through pEZ6. B. subtilis DB430 harboring each of the plasmids was then patched on the skim-milk plate (1% skim milk in 2× SG medium) and analyzed for the halo size. + to +++ indicates the relative size of the halo. − indicates no significant halo observed. (B) Expression of the putative promoter-lacZ fusions in B. subtilis. The solid lines indicate the DNA fragments inserted upstream of lacZ. Each of the B. subtilis strains was grown in 2× SG medium and analyzed for lacZ expression throughout the growth. The procedure used for β-galactosidase assay was derived from the work of Miller (32). The symbol ▴, ▪, ♦, or • indicates the growth curve of B. subtilis DB20, DB21, DB22, or DB23, respectively. The symbol, Δ, □, ⋄, or ○ indicates the expression of lacZ in the corresponding strain of B. subtilis.
TABLE 1.
Primers used in this study
| Description of primer | Direction and sequencea |
|---|---|
| Overexpression of EzrA | Forward, 5′-AGACTACCATGGAGTTTGGATTA-3′ |
| Reverse, 5′-CGTTACCTCGAGAGCGGATATGTCAGCTTTG-3′ | |
| Construction of pWP18 derivatives | |
| pEZ1 | Forward, 5′-AGACTAGAATTCTAACAATGAAAACGACAGTTTTTC-3′ |
| Reverse, 5′-CGTTACGGATCCAATGAGCCCCCTTGCTGTT-3′ | |
| pEZ2 | Forward, 5′-AGACTAGAATTCGCATTGATGTCACCCCATG-3′ |
| Reverse, 5′-CGTTACGGATCCTTATGATACCATGTAACAAAC-3′ | |
| pEZ3 | Forward, 5′-AGACTAGAATTCATGTAATTTCGTCTTCAAATTC-3′ |
| Reverse, 5′-CGTTACGGATCCAAAAGCGAGACGGACATATTC-3′ | |
| pEZ4 | Forward, 5′-AGACTAGAATTCGCATTGATGTCACCCCATG-3′ |
| Reverse, 5′-CGTTACGGATCCAATGAGCCCCCTTGCTGTT-3′ | |
| pEZ5 | Forward, 5′-AGACTAGAATTCATGTAATTTCGTCTTCAAATTC-3′ |
| Reverse, 5′-CGTTACGGATCCTTATGATACCATGTAACAAAC-3′ | |
| pEZ6 | Forward, 5′-AGACTAGAATTCATGTAATTTCGTCTTCAAATTC-3′ |
| Reverse, 5′-CGTTACGGATCCAATGAGCCCCCTTGCTGTT-3′ | |
| Construction of pCoiZA derivatives | |
| pEZ8 | Forward, 5′-AGACTACTGCAGATGTAATTTCGTCTTCAAATTC-3′ |
| Reverse, 5′-CGTTACGGATCCTTATGATACCATGTAACAAAC-3′ | |
| pEZ10 | Forward, 5′-AGACTACTGCAGATGTAATTTCGTCTTCAAATTC-3′ |
| Reverse, 5′-CGTTACGGATCCAATGAGCCCCCTTGCTGTT-3′ | |
| pEZ11 | Forward, 5′-AGACTACTGCAGATGTAATTTCGTCTTCAAATTC-3′ |
| Reverse, 5′-CGTTACGGATCCTATTCAGAAAGATTTCAGG-3′ | |
| pEZ12 | Forward, 5′-AGACTACTGCAGGTTTGTTACATGGTATCATAATAAC-3′ |
| Reverse, 5′-CGTTACGGATCCTATTCAGAAAGATTTCAGG-3′ | |
| pEZ13 | Forward a, 5′-AGACTACTGCAGGCATTGATGTCACCCCATG-3′ |
| Reverse b, 5′-CAGAAAGATTTCATTGTTATTATGATACCATGTAAC-3′ | |
| Forward c, 5′-ATAACAATGAAATCTTTCTGAATAAATAAAAC-3′ | |
| Reverse d, 5′-CGTTACGGATCCTATTCAGAAAGATTTCAGG-3′ | |
| Deletion of ezrA promoters | |
| P1 deletion strain (DB2001) | Forward, 5′-AGACTAGAATTCGTTTGTTACATGGTATCATAATAAC-3′ |
| Reverse, 5′-CGTTACCTGCAGTCCGCTCGTTTCAGCTGG-3′ | |
| P2 deletion strain (DB2002) | Forward a, 5′-AGACTAGAATTCGCATTGATGTCACCCCATG-3′ |
| Reverse b, 5′-CAGAAAGATTTCATTGTTATTATGATACCATGTAAC-3′ | |
| Forward c, 5′-ATAACAATGAAATCTTTCTGAATAAATAAAAC-3′ | |
| Reverse d, 5′-CGTTACCTGCAGTCCGCTCGTTTCAGCTGG-3′ | |
| P1P2 deletion strain (DB2003) | Forward, 5′-AGACTAGAATTCCGTGGAAGATTGAAATTCTG-3′ |
| Reverse, 5′-CGTTACCTGCAGTCCGCTCGTTTCAGCTGG-3′ | |
| Primer extension | 5′-AATGAGCCCCCTTGCYGTTTAC-3′ |
The underlined, doubly underlined, bold, italic, and bold italic bases indicate the NcoI, XhoI, EcoRI, BamHI, and PstI restriction sites, respectively. Forward a, Reverse b, Forward c, and Reverse d primers were used for construction and amplification of P2-disrupted promoter DNA fragments by overlapping extension PCR. Reverse b and Forward c are primers containing overlapping sequences and mutation sites.
The promoter region 100 bp upstream of ezrA is responsible for most of the expression of ezrA.
To analyze whether both of the putative promoter regions were critical for ezrA expression, transcriptional fusions of promoter-lacZ, as shown in the left part of Fig. 1B, were constructed. To make these constructs, each of the promoter regions was amplified by PCR, using the chromosomal DNA of B. subtilis DB2 as the template. The forward, reverse, and overlapping primers are shown in Table 1. The synthesized putative promoter DNA fragments were digested with PstI and BamHI and cloned into the integration vector, pCoiZA (34). The resultant plasmids, pEZ10, pEZ8, and pEZ11 (Table 2), as well as the vector, pCoiZA, were integrated, respectively, into the B. subtilis DB2 chromosome through homologous recombination at the aprE locus to generate B. subtilis DB21, DB22, DB23, and DB20. The expression of lacZ in each of the strains was then measured throughout the growth in 2× SG medium (25). Similar to that observed for the negative control strain, B. subtilis DB20, no significant expression of lacZ was detected for B. subtilis DB22 in which the lacZ gene was under the control of the putative promoter region located at about 200 bp upstream of ezrA (Fig. 1B). These results suggested that this putative promoter region is of no significance to the expression of ezrA. In contrast, a significant two-peak expression pattern of lacZ was observed when DNA fragments containing the putative promoter region located at about 100 bp upstream of ezrA were cloned in front of lacZ as shown for B. subtilis DB21 and DB23 (Fig. 1B), indicating that this putative promoter region is responsible for most of the expression of ezrA.
TABLE 2.
B. subtilis strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| B. subtilis strains | ||
| DB2 | trpC2 ezrA (wild type) | B. subtilis 168 |
| DB430 | trpC2 npr apr epr bfp isp1 | 18 |
| DB2001 | trpC2 ezrA (ΔP1) Cmr | This study |
| DB2002 | trpC2 ezrA (ΔP2) Cmr | This study |
| DB2003 | trpC2 ezrA (ΔP1P2) Cmr | This study |
| Plasmids | ||
| pET21d | E. coli protein overexpression vector, Ampr | Novagen Inc. |
| pKM1 | Derivative of pET21d, containing an expressible ezrA from B. subtilis DB2 | This study |
| pCT20 | pCT18-derived plasmid which contains P1P2, P7, and P8 promoters of sigA operon, followed by the T1T2 terminators of E. coli rrnB operon | 28 |
| pKM3 | pCT20-derived plasmid containing DNA fragment encompassing the 500-bp of DNA upstream of ezrA followed by T1T2 terminators of E. coli rrnB operon | This study |
| pWP18 | Promoter probing vector with aprE reporter | 34 |
| pEZ1 to -6 | pWP18 derivative containing putative promoter DNA fragments with different length | This study |
| pCioZA | Integration vector with lacZ as reporter; Ampr Ermr | 34 |
| pEZ8, -10 to -13 | pCoiZA derivative containing different putative promoter DNA fragments and used for construction of B. subtilis DB22, DB21, DB23, DB24, DB25, respectively, by integration into the aprE locus on B. subtilis DB2 chromosome | This study |
| pDP1 | Integration vector used for construction of B. subtilis DB2001 | This study |
| pDP2 | Integration vector used for construction of B. subtilis DB2002 | This study |
| pDP12 | Integration vector used for construction of B. subtilis DB2003 | This study |
Identification and characterization of promoters critical for ezrA expression.
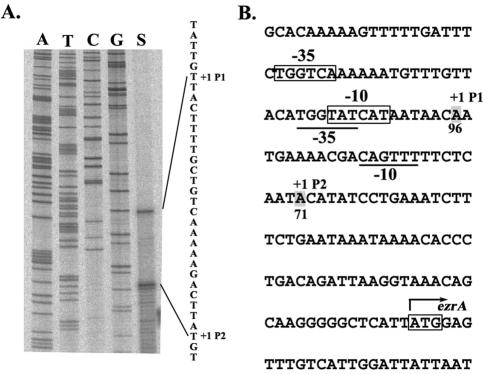
To locate exactly the position of the promoter(s) about 100 bp upstream of ezrA, primer extension analysis of total RNA extracted from B. subtilis DB430 harboring pEZ6 was performed. The protocol used for primer extension was similar to that reported previously (28). The primer used is 22 bases long, starting from the base immediately upstream of the initiation codon (Table 1). As shown in Fig. 2A, two major transcription initiation sites, A-71 and A-96, upstream of the initiation codon (ATG) of ezrA were found. The promoter identified by A-96 was designated as P1; it had nice fits (four of six and five of six, respectively) to the −35 (TTGACA) and −10 (TATAAT) consensus of the B. subtilis σA-type promoter besides the possession of a TG motif (TGnTATAAT) immediate upstream of the −10 consensus, indicative of an extended −10 promoter (Fig. 2B) (2, 4, 24). Initiation at A-71 identified another promoter, designated P2. This promoter had poor fits (three of six and two of six, respectively) to the −35 and −10 consensus of the σA-type promoters. No consensus for other σ-type promoters was found at this region. Thus, the expression of ezrA is controlled by two overlapping σA-type promoters, with P1 probably playing the major role (Fig. 2B).
FIG. 2.
Determination of the transcription initiation sites of B. subtilis ezrA. (A) Primer extension analysis of the transcription initiation sites of ezrA. The letter A, T, C, or G above each lane indicates the dideoxynucleotide used to terminate the sequencing reaction. The transcription initiation sites are indicated as +1. The DNA sequence shown in the right margin is read directly from the gel and represents the sequence of noncoding strand DNA. (B) Coding-strand DNA sequence of the two overlapping promoters of B. subtilis ezrA. The nucleotide sequence is given in the 5′-to-3′ direction. The −10 and −35 regions of P1 and P2 are boxed and underlined, respectively. The transcription +1 sites of the two overlapping promoters are indicated by the shadowed letter, A. The translation initiation codon (ATG) for ezrA is boxed and indicated by the rightward arrow.
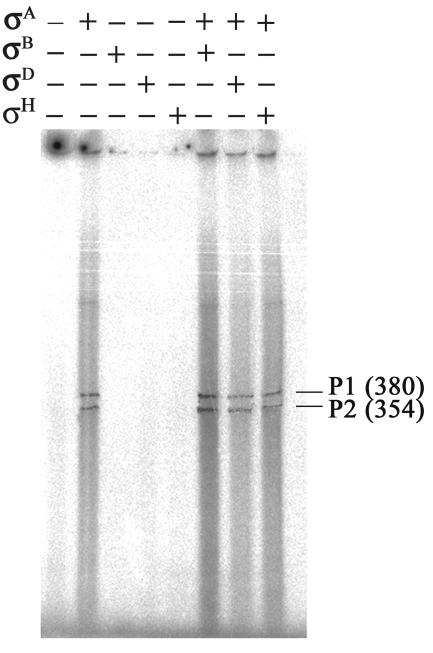
To confirm that P1 and P2 promoters are of the σA type, in vitro transcription assays were performed. The procedure used for the in vitro transcription assay was similar to that reported previously (28). Four different B. subtilis σ factors, including σA, σB, σD, and σH, were overexpressed, purified from E. coli (6, 8, 28, 39), and reconstituted with core RNA polymerase (20). Subsequently, each RNA polymerase holoenzyme or a mixture of two different RNA polymerase holoenzymes was examined for activity on transcribing the pKM3 plasmid containing the DNA sequence encompassing both P1 and P2 (Table 2). As shown in Fig. 3, only in the presence of σA-RNA polymerase holoenzyme was the promoter activity observed. Two mRNA transcripts that were supposed to be initiated from P1 and P2, respectively, and terminated at the T1 site of the T1T2 terminators (21) were produced. Thus, the two promoters controlling ezrA expression are both of the σA type.
FIG. 3.
Determination of the σ specificity of ezrA promoters by in vitro transcription assays. The pKM3 plasmid bearing the DNA sequence encompassing both P1 and P2 promoters upstream of ezrA was used as a template for in vitro transcription. A single RNA polymerase holoenzyme or a mixture of two RNA polymerase holoenzymes, as indicated by + and − signs, was used to identify the σ type of the promoters. The transcripts initiated from P1 and P2 and terminated at the first termination site (T1) of the T1T2 terminators are indicated by P1 and P2 with transcript length (in bases) shown in the right margin.
P1 plays the major role for ezrA expression during the growth of B. subtilis.
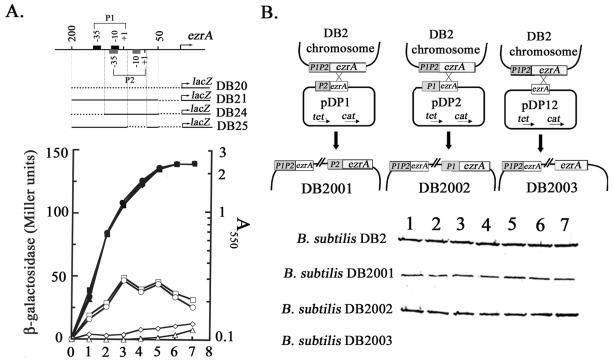
To examine the relative importance of each of the two promoters for ezrA expression, two more strains of B. subtilis, DB24 and DB25, with a mutant ezrA promoter (P1 or P2 disruption) transcriptionally fused to lacZ (Fig. 4A, upper part) were constructed using the same strategy as described for B. subtilis DB21, DB22, and DB23. Then, lacZ expression was measured for these strains throughout the growth in 2× SG medium. As shown in the lower part of Fig. 4A, B. subtilis DB25 with P2 being disrupted by removing the −10 region had a high expression pattern for lacZ similar to that of the wild-type counterpart, B. subtilis DB21, indicating that P2 plays only a minor role in transcription of ezrA. In contrast, disruption of P1 by removing its −35 region drastically reduced the expression of lacZ in B. subtilis DB24, albeit with a gradual increase in lacZ expression during growth. Thus, P1 is the major promoter for transcriptional control of ezrA in B. subtilis, while P2 is a supplemental promoter for the control, especially during the later stage of B. subtilis growth.
FIG. 4.
Effects of promoter mutations on transcription and intracellular level of EzrA. (A) The effect of promoter mutation on transcription of ezrA in B. subtilis. The transcriptional promoter-lacZ fusions are shown in the upper part of the figure. The horizontal dotted lines indicate the DNA sequences removed in the mutant promoters. Each of them was synthesized directly by PCR or indirectly by overlapping extension PCR (19). Each of the B. subtilis strains was grown in 2× SG medium and measured for lacZ expression. ▴, ▪, ♦, or • indicates the growth curve of B. subtilis DB20, DB21, DB24 or DB25, respectively, while ▵, □, ⋄, or ○ indicates the expression of lacZ throughout growth. (B) Effect of promoter mutation on intracellular level of EzrA during the growth of B. subtilis. The designs for constructing ezrA promoter mutants by homologous recombination are shown in the upper part of the figure. The boxed ezrA in pDPx (where x = 1, 2, or 12) indicate a 3′-truncated ezrA gene which is about 700 bp in length. B. subtilis DB2001, DB2002, and DB2003 are strains in which the P1, P2, and P1P2 promoters of ezrA, respectively, are disrupted. The promoter designs are the same as those shown at the upper part of panel A. The lower part of the figure shows the intracellular levels of EzrA during growth of B. subtilis. The cell samples analyzed were collected every hour throughout growth in 2× SG medium with an initial cell density (A550) of 0.1. Equal amounts of total proteins were loaded for Western blot analysis of EzrA. To prepare anti-EzrA, His-tagged EzrA was overexpressed in E. coli BL21(DE3)/pKM1, purified with TALON resin (CLONTECH), concentrated, mixed with adjuvant, and injected into rabbit.
The difference in the importance of P1 and P2 to ezrA expression indicated that the intracellular level of EzrA might be also differentially affected by disruption of each of the promoters. To check this idea, B. subtilis DB2001, DB2002, and DB2003, with P1, P2, or both P1 and P2 being disrupted, respectively, were constructed (Fig. 4B, upper part) and analyzed for relative intracellular levels of EzrA by Western blotting (Fig. 4B, lower part). As expected, P1 disruption led to a substantial reduction in the content of EzrA in B. subtilis DB2001; P2 disruption caused only a minor reduction in EzrA (see B. subtilis DB2002). Deletion of both P1 and P2 completely abolished EzrA in B. subtilis DB2003. These results were consistent with those obtained for lacZ expression (Fig. 4A), strongly supporting that the expression of ezrA is exclusively controlled by the two promoters. Furthermore, a rather constant level of EzrA was observed for each of the B. subtilis strains throughout growth, indicating that EzrA is constitutively expressed and that the synthesis and degradation of EzrA are maintained at equal rates during the growth. The constitutive expression of EzrA is consistent with the participation of EzrA in Z-ring formation in both vegetative and sporulation phases of B. subtilis.
The intracellular level of EzrA affects the cell length and percentage of filamentous cells of B. subtilis during growth.
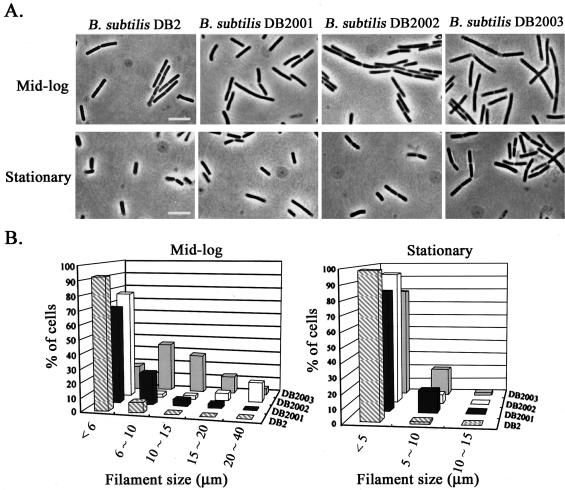
It has been reported that the ezrA null mutation appears to cause a delay in cell division, resulting in cells 20% longer on average than the wild-type counterpart grown in minimal medium (26); however, it remains unclear whether the difference in the intracellular level of EzrA would have differential effects on cell division of B. subtilis during growth. To answer this question, the overnight culture of B. subtilis DB2, DB2001, DB2002, or DB2003 was inoculated into 2× SG medium to an initial cell density (A550) of 0.1. The cell length and filament size of each strain of B. subtilis were then measured at mid-log (A550 = 0.9) and stationary (A550 = 2.4) phases after fixation with 70% ethanol. The phase-contrast micrographs for each strain of B. subtilis are shown in Fig. 5A. During mid-log phase, the cells of B. subtilis DB2001, DB2002, and DB2003 were 21, 5, and 25%, respectively, longer on average than the wild-type counterpart (DB2). During stationary phase, both B. subtilis DB2001 and DB2003, in which the intracellular level of EzrA was either drastically reduced or completely abolished, remained 17% longer on average than DB2. However, B. subtilis DB2002, with only a minor reduction in the intracellular level of EzrA, was of about the same size as DB2. Moreover, higher percentages of septate filaments were observed for all of the three mutant strains of B. subtilis than with the wild-type at both growth stages (Fig. 5), and nonseptate filaments were clearly observed only for B. subtilis DB2001 and DB2003. The increase in the percentage of filamentous cells of B. subtilis correlated with the decrease in the intracellular level of EzrA, regardless of the size distribution of filaments (Fig. 5B). The increase in both cell length and percentage of filamentous cells for B. subtilis containing reduced levels of EzrA indicates that EzrA is required for efficient cell division.
FIG. 5.
The effects of ezrA promoter disruption on cell length and filament size of B. subtilis. (A) Phase-contrast microscopy of B. subtilis at ×1,000 magnification. Bar, 10 μm. (B) The filament sizes for each strain of B. subtilis. The overnight culture of B. subtilis was diluted with 2× SG medium to an initial cell density (A550 = 0.1), grown at 37°C, harvested at mid-log (A550 = 0.9) or stationary (A550 = 2.4) phases, fixed in 70% ethanol for 2 h, and resuspended with PBS buffer (5.4 mM Na2HPO4, 1.7 mM NaH2PO4, 137 mM NaCl, and 3 mM KCl) before measuring the cell length (×1,000) and filament size (×400) with a phase-contrast microscope. The measurement was repeated at least five times, and the data are reproducible.
The possible roles of EzrA during cell division.
It has been reported that EzrA recruited to the division site, partially dependent on FtsA, is a negative regulator for Z-ring formation (13, 26). However, EzrA was shown in this study to be required for efficient cell division. How does EzrA work to fulfill the two seemingly contradictory functions? The paradox can be partly solved if the effect of EzrA on Z-ring formation is not restricted to a single round of cell division: more precisely, if the negative effects of EzrA on a round of Z-ring formation become positive for the next. It was proposed that EzrA contributes to Z-ring remodeling by accelerating the disassembly of Z ring (14, 37). Thus, in the absence or deficiency of EzrA, the slow disassembly of Z ring in a round of cell division may reduce the concentration of free FtsZ monomers or protofilaments in the cytoplasmic pool and therefore the efficiency of the next round of Z-ring formation. However, Z-ring remodeling seems unable to explain why the ezrA null mutation is capable of suppressing the inhibition of cell division caused by overexpression of MinCD (27). Probably, EzrA is also able to interact either directly or indirectly with MinC or MinCD complex to affect their functions before being recruited to the division site. Actually, EzrA may also serve as a membrane anchor for coordinate membrane invagination at the later stage of cell division, since it shares similar membrane topologies with the homologous E. coli ZipA (12). Thus, EzrA may be multifunctional during cell division and present in B. subtilis all the time. The presence of a constant level of EzrA in B. subtilis during growth (Fig. 4B) and the requirement of EzrA for efficient cell division are consistent with this idea.
Acknowledgments
This research was supported by the National Science Council, Taiwan, Republic of China.
REFERENCES
- 1.Autret, S., R. Nair, and J. Errington. 2001. Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein. Mol. Microbiol. 41:743-755. [DOI] [PubMed] [Google Scholar]
- 2.Barne, K. A., J. A. Bown, S. J. Busby, and S. D. Minchin. 1997. Region 2.5 of the Escherichia coli RNA polymerase sigma 70 subunit is responsible for the recognition of the “extended −10” motif at promoters. EMBO J. 16:4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Yehuda, S., and R. Losick. 2002. Asymmetric cell division in B. subtilis involved a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257-266. [DOI] [PubMed] [Google Scholar]
- 4.Camacho, A., and M. Salas. 1999. Effect of mutations in the “extended −10” motif of three Bacillus subtilis σA-RNA polymerase-dependent promoters. J. Mol. Biol. 286:683-693. [DOI] [PubMed] [Google Scholar]
- 5.Cha, J. H., and G. C. Stewart. 1997. The divIVA minicell locus of Bacillus subtilis. J. Bacteriol. 179:1671-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, B. Y., and R. H. Doi. 1990. Overexpression, purification, and characterization of B. subtilis sigma-A factor. J. Bacteriol. 172:3257-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J. C., D. S. Weiss, J. M. Ghigo, and J. Beckwith. 1999. Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J. Bacteriol. 181:521-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu, C. H. 2003. Characterization of a Bacillus subtilis sigA mutant with filamentous phenotype. Master's thesis. National Chung-Hsing University, Taiwan, Republic of China.
- 9.Cook, W. R., and L. I. Rothfield. 1999. Nucleoid-independent identification of cell division sites in Escherichia coli. J. Bacteriol. 181:1900-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel, R. A., E. J. Harry, V. L. Katis, R. G. Wake, and J. Errington. 1998. Characterization of the essential cell division gene ftsL (ylld) of Bacillus subtilis and its role in the assembly of division apparatus. Mol. Microbiol. 29:593-604. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, D. H., and J. Errington. 1997. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol. 24:905-915. [DOI] [PubMed] [Google Scholar]
- 12.Errington, J., and C. Scazzocchio. 2003. Growth and development. Microbial development—regulation in space and time. Curr. Opin. Microbiol. 6:531-533. [DOI] [PubMed] [Google Scholar]
- 13.Feuchet, A., I. Lucet, M. D. Yudkin, and J. Errington. 2001. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol. Microbiol. 40:115-125. [DOI] [PubMed] [Google Scholar]
- 14.Gueiros-Filho, F. J., and R. Losick. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gullbrand, B., and K. Nordström. 2000. FtsZ ring formation without subsequent cell division after replication run out in Escherichia coli. Mol. Microbiol. 36:1349-1359. [DOI] [PubMed] [Google Scholar]
- 16.Harry, E. J., J. Rodwell, and R. G. Wake. 1999. Co-ordinating DNA replication with cell division in bacteria: a link between the early stages of a round of replication and mid-cell Z ring assembly. Mol. Microbiol. 33:33-40. [DOI] [PubMed] [Google Scholar]
- 17.Harry, E. J. 2001. Bacterial cell division: regulating Z-ring formation. Mol. Microbiol. 40:759-803. [DOI] [PubMed] [Google Scholar]
- 18.He, X. S., Y. T. Shu, S. Nathoo, S. L. Wong, and R. H. Doi. 1991. Construction and use of a Bacillus subtilis mutant deficient in multiple protease genes for the expression of eukaryotic genes. Ann. N. Y. Acad. Sci. 646:60-77. [DOI] [PubMed] [Google Scholar]
- 19.Horton, R. M., Z. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 20.Huang, W. C. 2000. Identification and characterization of the functional domains of Bacillus subtilis σA factor. Master's thesis. National Chung-Hsing University, Taiwan, Republic of China.
- 21.Jurgen, B., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 22.Kang, M. S., S. R. Kim, P. Kwack, B. K. Lim, S. W. Ahn, Y. M. Rho, I. S. Seong, S. C. Park, S. H. Eom, G. W. Cheong, and C. H. Chung. 2003. Molecular architecture of the ATP-dependent CodWX protease having an N-terminal serine active site. EMBO J. 22:2893-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karoui, M. E., and J. Errington. 2001. Isolation and characterization of topological specificity mutants of minD in Bacillus subtilis. Mol. Microbiol. 42:1211-1221. [DOI] [PubMed] [Google Scholar]
- 24.Keilty, S., and M. Rosenberg. 1987. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J. Biol. Chem. 262:6389-6395. [PubMed] [Google Scholar]
- 25.Leighton, T. J., and R. H. Doi. 1971. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J. Biol. Chem. 246:3189-3195. [PubMed] [Google Scholar]
- 26.Levin, P. A., I. G. Kurster, and A. D. Grossman. 1999. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 96:9642-9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin, P. A., R. L. Schwartz, and A. D. Grossman. 2001. Polymer stability plays an important role in the positional regulation of FtsZ. J. Bacteriol. 183:5449-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao, C. T., Y. D. Wen, W. H. Wang, and B. Y. Chang. 1999. Identification and characterization of a stress-responsive promoter in the macromolecular synthesis operon of Bacillus subtilis. Mol. Microbiol. 33:377-388. [DOI] [PubMed] [Google Scholar]
- 29.Lutkenhaus, J., and S. G. Addinall. 1997. Bacterial cell division and the Z-ring. Annu. Rev. Biochem. 66:93-116. [DOI] [PubMed] [Google Scholar]
- 30.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 31.Marston, A. L., H. B. Thomaides, D. H. Edwards, M. E. Sharpe, and J. Errington. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 12:3419-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Mulder, E., and C. L. Woldringh. 1989. Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J. Bacteriol. 171:4303-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi, F. X. 1990. Regulation of the sigA operon and its function during sporulation of Bacillus subtilis. Ph.D. thesis. University of California, Davis, Calif.
- 35.Regamy, A., E. J. Harry, and R. G. Wake. 2000. Mid-cell Z ring assembly in the absence of entry into the elongation phase of the round of replication in bacteria: coordinating chromosome replication with cell division. Mol. Microbiol. 38:423-434. [DOI] [PubMed] [Google Scholar]
- 36.Rothfield, L., S. Justice, and J. Garcia-Lara. 1999. Bacterial cell division. Annu. Rev. Genet. 33:423-448. [DOI] [PubMed] [Google Scholar]
- 37.Stricker, J., P. Maddox, E. D. Salmon, and H. P. Erickson. 2002. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. USA 99:3171-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, Q., X. C. Yu, and W. Margolin. 1998. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol. Microbiol. 29:491-503. [DOI] [PubMed] [Google Scholar]
- 39.Tabor, S. 1990. Expression using the T7 RNA polymerase/promoter system, p. 16.2.1-16.2.11. In F. A. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Greene Publishing & Wiley Interscience, New York, N.Y.
- 40.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]