Abstract
Bacterial growth as a biofilm on solid surfaces is strongly associated with the development of human infections. Biofilms on native heart valves (infective endocarditis) is a life-threatening disease as a consequence of bacterial resistance to antimicrobials in such a state. Enterococci have emerged as a cause of endocarditis and nosocomial infections despite being normal commensals of the gastrointestinal and female genital tracts. We examined the role of two-component signal transduction systems in biofilm formation by the Enterococcus faecalis V583 clinical isolate and identified the fsr regulatory locus as the sole two-component system affecting this unique mode of bacterial growth. Insertion mutations in the fsr operon affected biofilm formation on two distinct abiotic surfaces. Inactivation of the fsr-controlled gene gelE encoding the zinc-metalloprotease gelatinase was found to prevent biofilm formation, suggesting that this enzyme may present a unique target for therapeutic intervention in enterococcal endocarditis.
Bacterial virulence is one of many adaptive responses generally believed to be controlled through signal transduction mechanisms (39). Signal transduction in bacteria is mainly the prerogative of the so-called two-component systems consisting of a sensory histidine kinase that senses the signal and relays the adaptive response through the transfer of a phosphoryl group to a response regulator, generally a transcriptional regulator, that modulates gene expression in response to the signal received. Two-component signal transduction pathways are responsible for controlling gene expression in a wide variety of cellular processes including sporulation, virulence, biofilm formation, and antibiotic production and resistance (for reviews, see references 15 and 28). A total of 17 two-component systems and one orphan response regulator have been identified on the genome of the Enterococcus faecalis strain V583 (14). In a study aimed at the systematic inactivation of all two-component systems present in E. faecalis and the analysis of their role in virulence, we identified the fsr system as the only one affecting biofilm formation when inactivated (our unpublished data).
The fsr regulatory locus is comprised of three genes, designated fsrA, fsrB, and fsrC (Fig. 1) (33). Recently, this system has been identified as a quorum-sensing locus which responds to the extracellular accumulation of a peptide lactone encoded at the C terminus of the FsrB protein (27). Accumulation of the peptide in the extracellular space is likely sensed by the FsrC histidine kinase, leading to the activation of the response regulator and transcription factor FsrA. The fsr system and the products of the genes it regulates have been shown to be important for virulence in several infection models, including mouse peritonitis, Caenorhabditis elegans infection, and rabbit endophthalmitis (26, 32, 37). The FsrABC proteins are necessary for the production of two secreted proteases, gelatinase (GelE) and serine protease (SprE) (33). Here, we show that the E. faecalis fsr quorum-sensing system controls biofilm development through the production of gelatinase.
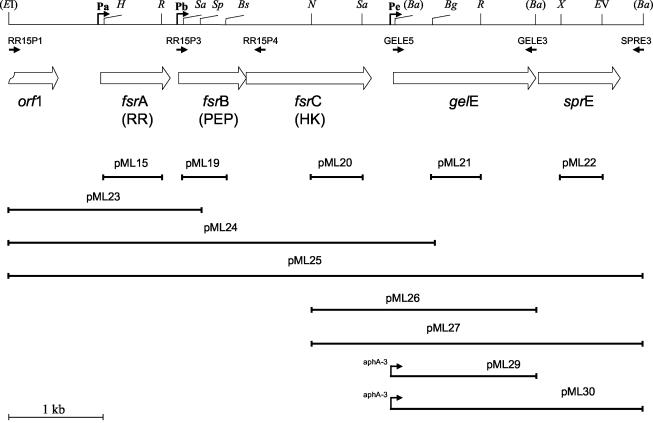
FIG. 1.
Genetic organization of the E. faecalis V583 fsr locus which includes fsrA (response regulator), fsrB (signaling peptide), fsrC (histidine kinase), gelE (gelatinase), and sprE (serine protease) (32). The open arrows specify the coding region of each gene. The gene designated orf1 encodes a putative N-acetylmuramidase; only the 3′ end of the gene is shown. The extents of the fragments cloned in the plasmids are indicated by the solid lines. For plasmids pML29 and pML30, the aphA-3 promoter was cloned upstream of the fragment shown, and it is depicted as an elevated arrow. The small solid arrows represent the position of the oligonucleotide primers used in the study. The positions of the promoters identified in this region are indicated by arrows and the symbols Pa, Pb, and Pe. Only restriction sites relevant to this study are shown. Restriction sites in parenthesis were introduced by the oligonucleotides used in PCR amplification reactions. Restriction enzyme symbols: EI, EcoRI; H, HincII; R, RsaI; Sa, Sau3A; Sp, SphI; Bs, BsmAI; N, NdeI; Ba, BamHI; Bg, BglII; X, XmnI; EV, EcoRV.
(The data reported here were partially presented [L. E. Hancock and M. Perego, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol., abstr. B078, 2003] and fully presented at the Functional Genomics of Gram-Positive Microorganisms, 12th International Conference on Bacilli, Baveno, Italy, 22 to 27 June 2003 [L. E. Hancock and M. Perego, abstr. T55]).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. faecalis strains and plasmids used in this study are listed in Tables 1 and 2. Strains were cultured in Todd-Hewitt broth (THB), brain heart infusion broth, or M17 medium (Difco Laboratories). Escherichia coli DH5α was used for plasmid constructions and propagation. Strains were cultivated in Luria-Bertani broth. Antibiotics used for selection in E. coli and E. faecalis were spectinomycin (150 and 1,000 μg/ml, respectively) and tetracycline (15 μg/ml). Screening for gelatinase production was carried out on THB agar plates containing 3% gelatin or 1.5% skim milk. Zymography to detect serine protease activity was carried out as described previously by Qin et al. (32). Electroporation was carried out as described previously (8).
TABLE 1.
E. faecalis strains used in this study
| Strain | Relevant genotype | Complementation in pAT28 | Relevant phenotype | Origin |
|---|---|---|---|---|
| V583 | Parental | GelE+ SprE+ | Clinical isolate | |
| JML101 | fsrA | GelE− SprE− Tetr | pML15→V583 | |
| JML102 | fsrB | GelE− SprE− Tetr | pML19→V583 | |
| JML103 | fsrC | GelE− SprE− Tetr | pML20→V583 | |
| JML104 | gelE | GelE− SprE− Tetr | pML21→V583 | |
| JML105 | sprE | SprE− Tetr | pML22→V583 | |
| JML106 | fsrA | GelE− SprE− Tetr Spcr | pAT28→JML101 | |
| JML107 | fsrA | fsrA | GelE+ SprE+ Tetr Spcr | pML23→JML101 |
| JML108 | gelE | GelE− SprE− Tetr Spcr | pAT28→JML104 | |
| JML109 | gelE | gelE | GelE+ SprE− Tetr Spcr | pML26→JML104 |
| JML110 | gelE | gelE sprE | GelE+ SprE+ Tetr Spcr | pML27→JML104 |
| FA2-2 | fsrC | GelE− SprE− | Lab strain | |
| JML111 | fsrC | GelE− SprE− Spcr | pAT28→FA2-2 | |
| JML112 | fsrC | fsrA | GelE− SprE− Spcr | pML23→FA2-2 |
| JML113 | fsrC | fsrABC | GelE+ SprE+ Spcr | pML24→FA2-2 |
| JML114 | fsrC | fsrABC gelE sprE | GelE++ SprE+ Spcr | pML25→FA2-2 |
| JML115 | fsrC | aphA-3 promoter | GelE− SprE− Spcr | pML28→FA2-2 |
| JML116 | fsrC | aphA-3 promoter-gelE | GelE++ SprE− Spcr | pML29→FA2-2 |
| JML117 | fsrC | aphA-3 promoter-gelE sprE | GelE++ SprE+ Spcr | pML30→FA2-2 |
| JML120 | fsrA | aphA-3 promoter | GelE− SprE− Tetr Spcr | pML28→JML101 |
| JML121 | fsrA | aphA-3 promoter-gelE | GelE+ SprE− Tetr Spcr | pML29→JML101 |
| JML122 | fsrA | aphA-3 promoter-gelE sprE | GelE+ SprE+ Tetr Spcr | pML30→JML101 |
TABLE 2.
Plasmids used in this study
| Plasmid | Description |
|---|---|
| p3TET | Integrational vector, Tetr derivative of p3ERM |
| pML15 | p3TET containing a 622-bp internal fragment of fsrA |
| pML19 | p3TET containing a 460-bp internal fragment of fsrB |
| pML20 | p3TET containing a 528-bp internal fragment of fsrC |
| pML21 | p3TET containing a 522-bp internal fragment of gelE |
| pML22 | p3TET containing a 444-bp internal fragment of sprE |
| pAT28 | Shuttle vector for E. coli and E. faecalis, specr |
| pML23 | pAT28 containing fsrA on a 2.0-kb EcoRI/SphI fragment |
| pML24 | pAT28 containing fsrABC on a 4.5-kb EcoRI/BglII fragment |
| pML25 | pAT28 containing fsrABC, gelE, and sprE on a 6.8-kb EcoRI/BamHI fragment |
| pML26 | pAT28 containing gelE on a 2.4-kb NheI/BamHI fragment |
| pML27 | pAT28 containing gelE and sprE on a 3.5-kb NheI/BamHI fragment |
| pML28 | pAT28 plus aphA-3 promoter on a 369-kb EcoRI/BamHI fragment |
| pML29 | pML28 containing a 1.6-kb BamHI fragment with gelE fused to the aphA-3 promoter |
| pML30 | pML28 containing a 2.8-kb BamHI fragment with gelE and sprE fused to the aphA-3 promoter |
Construction of insertion mutations.
The insertional inactivation vector p3TET was constructed from p3ERM (4) by replacing an MfeI/NaeI fragment carrying the erythromycin resistance determinant with the tetM gene from plasmid pFW16 (30). PCR products were obtained from the amplification of V583 genomic DNA with the primers indicated in Fig. 1 and Table 3. To obtain an internal fragment of fsrA, primers RR15P1 and RR15P4 were used, and a 622-bp internal HincII/RsaI fragment was ligated to p3TET digested with SmaI, resulting in plasmid pML15. For fsrB, primers RR15P3 and RR15P4 were used for amplification. The product was digested with BsmAI followed by Klenow treatment and then restricted with Sau3AI to liberate a 460-bp internal fsrB fragment which was ligated to p3TET cut with BamHI and SmaI, resulting in pML19. For fsrC, the PCR product obtained with primers RR15P3 and GELE3 was digested with NheI and Sau3AI, and a 528-bp internal fragment was ligated to p3TET cut with BamHI and XbaI, resulting in pML20. For gelE, primers GELE5 and GELE3 were used for amplification. The product, cut with BglII and RsaI to generate a 522-bp internal gelE fragment, was ligated to p3TET cut with BamHI and SmaI, resulting in pML21. For sprE, primers GELE5 and SPRE3 were used for amplification. A 444-bp XmnI/EcoRV internal sprE fragment was purified and ligated to p3TET cut with SmaI, resulting in pML22. All constructs were verified by sequence analysis. The resulting constructs were electroporated in E. faecalis V583 followed by selection for tetracycline on THB plates. The insertions were confirmed by diagnostic PCR. The growth curves and stability of single-crossover insertions were determined for cultures grown in liquid medium as previously described (42).
TABLE 3.
Oligonucleotides used in this study
| Primer | Sequence |
|---|---|
| RR15P1 | 5′-GAGAGAATTCCGTTCGGAAGCCAA |
| RR15P4 | 5′-GAGAAAGCTTCCTGTAAAAATAACGACTG |
| RR15P3 | 5′-GAGAATGCATTAGGGAGGGATAATGACT |
| GELE3 | 5′-TTTACGGATCCAACCATTTTTCTCACAATGCC |
| GELE5 | 5′-GGGAGGATCCAGCAATACTTTTGTTGG |
| SPRE3 | 5′-TTGGGGATCCTTTTTCATTCATTGACC |
Complementation studies.
To complement the fsrA mutation of strain JML101, a 2.0-kb EcoRI/SphI fragment was recovered from the PCR amplicon obtained by using primers RR15P1 and RR15P4. The fragment was cloned into the shuttle vector pAT28 (44) cut with EcoRI and SphI, resulting in plasmid pML23. Transformation of E. faecalis JML101 with pML23 resulted in strain JML107. For complementing the gelE mutation of strain JML104, two separate plasmids were constructed by ligating the 2.4-kb NheI/BamHI restriction fragment, recovered from the PCR product generated by using primers RR15P3 and GELE3, and the 3.5-kb NheI/BamHI fragment, recovered from the PCR product generated by using primers RR15P3 and SPRE3, to pAT28 cut with XbaI and BamHI, obtaining plasmids pML26 and pML27, respectively. Transformation of JML104 with pML26 and pML27 resulted in strains JML109 and JML110, respectively.
To complement E. faecalis strain FA2-2 for biofilm production, plasmid pML24 was constructed by ligating a 4.5-kb EcoRI/BglII fragment, obtained from the amplicon generated by using primers RR15P1 and SPRE3, to pAT28 digested with EcoRI and BamHI. Plasmid pML25 was obtained by ligating a 6.8-kb EcoRI/BamHI fragment, generated by using primers RR15P1 and SPRE3, to pAT28. Plasmid pML29 was constructed by ligating the 1.6-kb BamHI fragment from the PCR product, obtained by using primers GELE5 and GELE3, to pML28 cut with BamHI. Plasmid pML28 is a derivative of pAT28 carrying a 369-bp EcoRI-BamHI fragment containing the aphA-3 promoter from pTCV-lac (31). Plasmid pML30 was constructed by ligating the 2.8-kb BamHI fragment from the PCR product obtained by using primers GELE5 and SPRE3 to pML28. FA2-2 was transformed with pAT28, pML23, pML24, pML25, pML28, pML29, and pML30, obtaining strains JML111, JML112, JML113, JML114, JML115, JML116, and JML117, respectively. These strains were screened for protease complementation by streaking colonies onto THB skim milk agar plates.
Sequence analysis of PCR products.
The fsr region was amplified from chromosomal DNA of E. faecalis strains FA2-2 and V583 by using PCR Supermix High Fidelity enzyme (Invitrogen, Carlsbad, Calif.) with primers RR15P3 and GELE3 (Table 3). Sequencing reactions were carried out directly on the PCR products, and the results were compared to the V583 genomic DNA sequence available on the Institute for Genomic Research website (http://www.tigr.org).
Biofilm assay on polystyrene microtiter plates.
Biofilm formation on polystyrene was quantified with the crystal violet staining method essentially as previously described (43). Each assay was performed in triplicate and repeated twice. For 48-h biofilms, cultures were grown in M17 medium for 24 h in the microtiter plate, at which time medium was removed and the adherent film was rinsed three times with phosphate-buffered saline (PBS) before the addition of fresh M17 medium. After an additional 24 h of growth, the biofilms were processed as described above.
Confocal laser scanning microscopy.
Confocal microscopy was performed on E. faecalis biofilms grown on glass coverslips. Sterile glass coverslips were placed on the bottom of 6-well tissue culture plates and submerged with 5 ml of M17 broth, seeded with a 1:100 dilution from an overnight culture (approximately 5 × 106 to 10 × 106 CFU), and grown for 24 h at 37°C. For 48-h biofilms, cultures were grown for 24 h, at which time medium was removed and the adherent film was rinsed three times with PBS before the addition of fresh M17 medium. The optical density (OD) of the cultures was measured at the 24-h time point, and no differences were observed between the two strains. Both the 24- and 48-h biofilms were washed three times with PBS and then stained with 5 ml of acridine orange (100 μg/ml) for 15 min at room temperature. Unbound stain was removed by gentle washing three times with PBS. The coverslips were mounted onto a microscope slide by using a Prolong Antifade kit (Molecular Probes) and sealed with clear nail polish. Slides were visualized by using an inverted confocal microscope (Zeiss Instruments).
Gelatinase purification.
Two liters of THB was inoculated with 20 ml of an overnight culture of E. faecalis strain JML116. Bacteria were grown as a standing culture at 37°C for 20 h. Cells were removed by centrifugation for 30 min at 15,000 × g at 4°C. Nucleic acid present in the supernatant was removed by precipitation with 0.9% protamine sulfate by the dropwise addition of 200 ml of 10% protamine sulfate in 50 mM Tris-HCl and 1 mM CaCl2, pH 7.8. After 2 h of standing at 4°C, precipitants were removed by centrifugation (30 min at 27,500 × g at 4°C). Gelatinase was precipitated from the remaining supernatant with ammonium sulfate at 60% saturation by slowly adding 858 g of ammonium sulfate powder to the 2.2 liters of supernatant. After overnight precipitation at 4°C, the mixture was centrifuged for 30 min at 27,500 × g at 4°C. Pellets were resuspended in 200 ml of 50 mM Tris-HCl and 1 mM CaCl2, pH 7.8. The 200-ml sample was applied to a phenyl-Sepharose CL-4B column (2.5 by 17 cm) and washed with 6 column volumes of 50 mM Tris-HCl and 1 mM CaCl2, pH 7.8. The enzyme was eluted from the column by washing with 3 column volumes of 50% (vol/vol) ethylene glycol in 50 mM Tris-HCl and 1 mM CaCl2, pH 7.8. Five-milliliter fractions were collected, and those fractions displaying protease activity on a 1.5% skim milk agar plate were pooled and dialyzed extensively against 50 mM Tris-HCl and 1 mM CaCl2, pH 7.8, with an Mr cutoff of 12,000 to 14,000. The protein was concentrated to 4 ml with Centricon-10 membrane filters, and its purity was analyzed by sodium dodecyl sulfate (SDS)-acrylamide gel electrophoresis (>90% pure) and mass spectrometry (only the peak for gelatinase was detected). Amino acid sequence analysis of the first six amino-terminal residues revealed the correct sequence, VGSEVT, of the mature protein.
Site-directed mutagenesis.
Plasmid pML29 was subjected to site-directed mutagenesis by using a QuickChange II site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) and mutagenic primers E137A (5′ TAG ATG TAG TTG GTC ATG CAA TGA CAC ATG GTG TGA C 3′) and E137A-antisense (5′ GTC ACA CCA TGT GTC ATT GCA TGA CCA ACT ACA TCT A 3′) according to the manufacturer's recommendations. The resulting plasmid, pML31, which contains the nucleotide substitution GAA to GCA which results in a glutamic acid-to-alanine substitution at amino acid residue 137 in the mature gelatinase sequence, was confirmed by DNA sequence analysis.
Construction of expression plasmids.
Plasmid vector pET21 (Novagen) was used for constructing plasmids pET-MATH6 and pET-E137AH6. Plasmid pET-MATH6 was constructed by cloning the PCR-amplified mature gelE coding sequence obtained by using primers MGELNT-NdeI (5′ ATT TAA CGC ATA TGG TCG GTA GTG AAG TAA CG 3′) and MGELCT-XhoI (5′ GAG ACT CGA GTT CAT TGA CCA GAA CAG ATT C 3′), modified to carry an NdeI site at the 5′ end and an XhoI site at the 3′ end (underlined), into the NdeI-XhoI sites of pET21. The amplicon contains an artificially introduced Met codon followed by the codon for valine, as the mature protease sequence is known to begin at Val-192 (40). The XhoI site replaced the stop codon of gelE generating a fusion to leucine and glutamic acid codons followed by six histidine codons in the vector. Plasmid pET-E137AH6 was constructed by replacing the wild-type gelE sequence by digestion with NdeI-XhoI and replacement with the gelE gene amplified from plasmid pML31. Both constructs were verified by DNA sequence analysis.
Protein expression and purification.
Plasmids pET-MATH6 and pET-E137AH6 were transformed into the E. coli expression strain BL21(DE3) (Novagen). One-liter cultures were grown at 37°C with shaking at 220 rpm in LB medium supplemented with 100 μg of ampicillin per ml to an OD at 600 nm (OD600) of 0.6 to 0.8. The cultures were induced with isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM final concentration) for 3 h and then harvested by centrifugation at 6,000 × g. Cell pellets were resuspended in 35 ml of cold lysis buffer (5 mM imidazole, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 50 mM Tris-HCl, pH 8.0). The suspension was French pressed, and cellular debris was removed by centrifugation at 35,000 × g for 30 min. The soluble lysate was mixed with nickel-nitrilotriacetic acid agarose resin (QIAGEN) and incubated overnight on a rocker before column packing. The column was washed with 10 volumes of binding buffer (5 mM imidazole, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 50 mM Tris-HCl, pH 8.0). His-tagged proteins were eluted by using a stepwise imidazole elution gradient in binding buffer (50 mM Tris-HCl, 100 mM NaCl, 300 mM imidazole). Fractions were analyzed by 10% Tris-glycine SDS-polyacrylamide gel electrophoresis, and those fractions containing mature GelE were pooled and dialyzed against 50 mM Tris-Cl, pH 8.0. Proteins were concentrated by using Amicon Centriprep 10 centrifugal concentrators. Proteins were ∼90% pure as assessed by SDS-polyacrylamide gel electrophoresis and Coomassie staining. Proteins were stored at 4°C.
Complementation of FA2-2 with purified proteins.
E. faecalis strain FA2-2 (biofilm negative) was grown overnight in M17 broth at 37°C and diluted 1:100 into fresh M17 broth. Two hundred microliters of the diluted culture was used to inoculate a U-bottom polystyrene microtiter plate. Ten microliters of purified gelatinase or His-tagged mature wild-type or E137A mutant protein (range, 0.001 to 1.0 μg) was added to triplicate wells, and the plates were incubated for 24 h at 37°C prior to processing as described above. Based on the work of Makinen et al. (24), the concentration of gelatinase in culture supernatants is approximately 0.5 μg/ml.
RESULTS
Analysis of biofilm formation in fsr, gelE, and sprE mutants.
In a systematic gene inactivation study of the 18 response regulators identified in the E. faecalis strain V583 (reference 14 and our unpublished data), we found that an fsrA insertion mutant was significantly impaired in its ability to form a biofilm on polystyrene microtiter plates. Since fsrA is part of a regulatory operon (Fig. 1) known to control the production of two secreted proteases, gelatinase and serine protease, we investigated whether the gene products of the fsrB, fsrC, gelE, and sprE genes also contributed to biofilm formation. Insertion mutants of each gene were obtained by single-crossover integration of an internal fragment cloned into the insertion vector p3TET (see Materials and Methods), and they were compared to the parental strain V583. All strains had similar doubling times, reached similar cell densities at stationary phase, and stably maintained the tetracycline resistance associated with the gene inactivation event when tested in THB medium at 37°C (data not shown) (see Materials and Methods). An examination of the biofilm-forming capacity of these mutants on polystyrene revealed that all were attenuated in their ability to form a biofilm, except for the strain defective in serine protease JML105 (sprE) (Fig. 2A). Detectable gelatinase and serine protease activity was absent from mutants in fsrB, fsrC, and gelE genes (data not shown). The fact that JML104 (gelE) lacked both gelatinase and serine protease activity confirmed that the insertion in gelE exerted a polar effect on sprE expression (33). Furthermore, strain JML105 (sprE) still exhibited proteolytic activity on 1.5% skim milk agar plates, suggesting that gelatinase accounts for the predominant proteolytic activity on this substrate (data not shown).
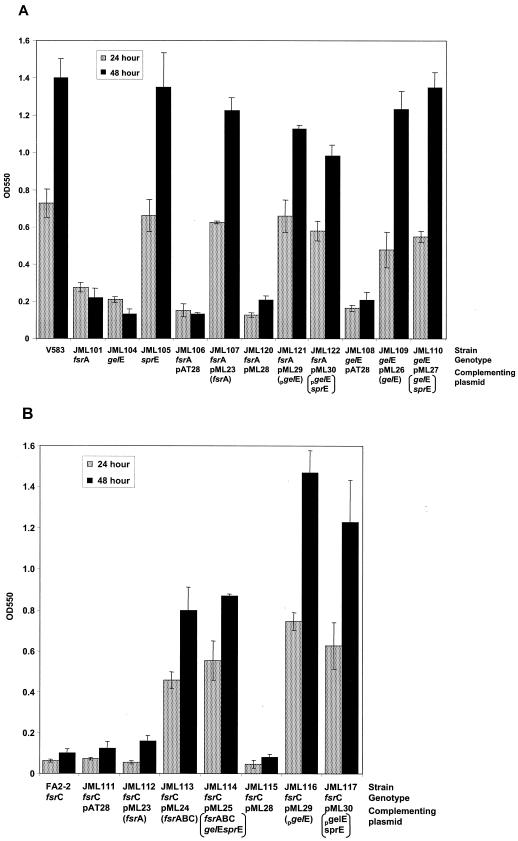
FIG. 2.
Biofilm formation by parental and mutant E. faecalis strains. The OD550s of solubilized crystal violet from microtiter plate assays at 24 h (stippled) and 48 h (solid) are shown for the strains listed. (A) All strains are derivatives of the E. faecalis V583 clinical isolate. Indicated is the name of the strain, its relevant genotype, the name of the plasmid it carries, and the complementing gene present on the plasmid (in parentheses). Strains JML102 and JML103 were omitted, as the level of biofilm formation was comparable to the level observed with strain JML101. (B) All strains are derivatives of the E. faecalis FA2-2 laboratory strain and thus carry the fsrC mutation. Complementing plasmids are indicated as described above.
To demonstrate that the phenotypes associated with the insertion mutations were due to insertion in the intended target sequence and not to mutations elsewhere on the chromosome, complementation studies were performed for strains JML101 (fsrA) and JML104 (gelE). JML101 was complemented with the replicative vector pAT28 (44) containing the fsrA gene (pML23) (Fig. 1) giving rise to strain JML107. As strain JML104 possessed an insertion in gelE which also prevented sprE expression, we constructed two separate derivatives of pAT28, plasmid pML26 containing gelE and plasmid pML27 containing both gelE and sprE (Fig. 1), to determine the role of gelatinase with and without serine protease production in the complementation of strain JML104. Strain JML104 complemented with pML26 and pML27 was designated JML109 and JML110, respectively. An examination of the proteolytic activity of these complemented strains on gelatin or skim milk agar plates indicated that the proteolytic activity could be complemented in trans (data not shown). The complemented strains also reacquired the ability to form biofilms on a polystyrene surface (Fig. 2A). The presence of the sprE gene in addition to gelE did not quantitatively affect the formation of biofilms. These results confirmed that the phenotypes observed were due to a mutation of the intended target and that complementation by the gelE gene restores biofilm formation even in the double mutant gelE sprE.
Localization of strain FA2-2 biofilm defect to the fsrC gene.
In the course of examining our E. faecalis strain collection for biofilm formation on polystyrene, we found that strain FA2-2 was unable to produce a biofilm. FA2-2 is also nonproteolytic on gelatin and casein substrates (40), suggesting a defect in the fsr or gelE gene or in their expression. To determine whether fsr and gelE were present in strain FA2-2, we performed PCR with DNA templates from strains FA2-2 and V583 with primers RR15P1 and RR15P4 as well as primers RR15P3 and GELE3 (Fig. 1). Amplified fragments were of the predicted size from both strains, suggesting that the fsr and gelE genes are present in FA2-2 (data not shown). To determine the location of the defect in these genetic loci in FA2-2, we performed complementation studies with a series of pAT28 derivatives containing fsrA, fsrABC, or fsrABC plus gelE and sprE (pML23, pML24, and pML25, respectively) (Fig. 1). These constructs were chosen to include the three different RNA transcripts found in this region (33). An analysis of complementation of proteolytic activity demonstrated that fsrA alone was insufficient to restore protease activity (strain JML112), whereas FA2-2 derivatives transformed with fsrABC or fsrABC gelE sprE carrying plasmids (strains JML113 and JML114, respectively) were restored for protease activity on gelatin and skim milk substrates (data not shown). These strains and the control strain JML111 (FA2-2 complemented with vector only) were examined for their ability to form a biofilm on polystyrene. The results depicted in Fig. 2B revealed a significant difference (P = 0.002) in adherence to polystyrene between strains JML111 and JML112 compared to JML113 and JML114, with the latter strains exhibiting four- to eightfold better adherence to polystyrene as assessed by crystal violet staining.
The fact that fsrA alone did not complement the defect in FA2-2 but the presence of fsrABC restored both proteolytic and biofilm-positive phenotypes to FA2-2 indicated that the fsr defect in FA2-2 localized to the fsrBC genes. We amplified and sequenced this region from strain FA2-2 with primers RR15P3 and GELE3 (Fig. 1). Sequence analysis and comparison to the V583 sequence (http://www.tigr.org) revealed four base changes in the FA2-2 fsrB sequence, two of which resulted in silent mutations and the other two of which resulted in a leucine-to-phenylalanine substitution at amino acid position 39 and a methionine-to-isoleucine substitution at position 111. However, the primary sequence of the peptide lactone (QNSPNIFGQWMG) (27) remained unchanged. In the fsrC gene, we found three nucleotide changes that resulted in missense mutations. Since complementation studies with a plasmid expressing the fsrB gene alone from V583 did not rescue the gelatinase production in FA2-2, it is likely that the mutations in the fsrC gene inactivate the FsrC protein in this strain (data not shown). Characterization of these mutations is ongoing.
Biofilm analysis by confocal laser scanning microscopy.
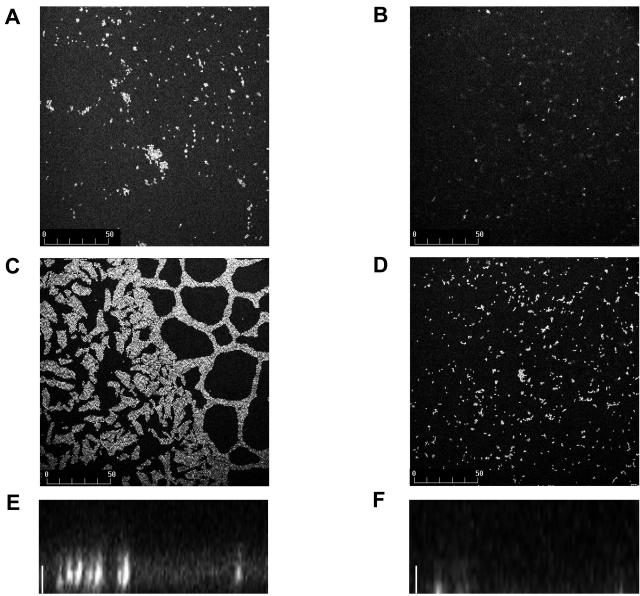
Experiments with glass coverslips as a substrate on which to form biofilms confirmed the defect of strain JML101 (fsrA) in biofilm formation compared to the parental strain V583. Using confocal laser scanning microscopy, we observed first a difference in the formation of initial foci of attachment between the parental strain V583 and the fsrA mutant JML101, with the former showing bigger and more numerous surface-attached microcolonies than the latter (Fig. 3A and B). At 48 h we then observed a well-developed biofilm for V583 with a heterogeneous distribution, consisting of microcolonies of bacterial cells encased in an extracellular matrix separated by interstitial voids (water channels) (Fig. 3C). This was in contrast to the JML101 (fsrA) strain, which appeared as clusters of small numbers of cells and lacked the three-dimensional architecture possessed by V583 (Fig. 3D, E, and F). Thus, the fsrA mutation seems to affect both the primary attachment to a solid surface and the growth of the biofilm.
FIG. 3.
Confocal laser scanning microscopy of E. faecalis biofilms. The 24-h (A and B) and 48-h (C and D) biofilms were stained with acridine orange and visualized by confocal laser scanning microscopy as described in Materials and Methods. Standard projections of the biofilm through the x-y plane for strain V583 (A and C) and strain JML101 (fsrA) (B and D) are shown. Bars (A to D) indicate size in microns. Projections of the biofilms through the x-z plane at 48 h for strains V583 (E) and JML101 (fsrA) (F) are also shown. Bars (E and F), 10 μm.
Expression of gelatinase is sufficient to restore biofilm formation.
As gelatinase expression is dependent on an intact fsr regulatory system (33) and because FA2-2 possessed a defective fsr system, we addressed the question of whether gelatinase expression alone or in combination with serine protease could influence biofilm formation independent of fsr. To this aim, we placed the gelE or gelE together with the sprE coding sequences under the control of the constitutive aphA-3 promoter in the replicative vector pAT28 (44). The aphA-3 gene encodes a 3′,5"-aminoglycoside phosphotransferase responsible for kanamycin resistance, which is functional in both gram-positive and -negative bacteria (44). The plasmids obtained, pML29 and pML30, restored protease activity to strain FA2-2 as well as to strain JML101 (fsrA) compared to the nonproteolytic strains JML120 and JML115 carrying the pAT28 plasmid with the aphA-3 promoter alone (pML28) (data not shown). Thus, gelatinase and serine protease could be expressed independently of a functional fsr locus. Examination of the biofilm-forming capacity of these strains demonstrated that gelatinase alone, or in combination with serine protease, could also restore a biofilm-positive phenotype to FA2-2 (strains JML116 and JML117) (Fig. 2B) and JML101 (strains JML121 and JML122) (Fig. 2A), even in the absence of a functional fsr system.
Gelatinase enzymatic activity is required for biofilm formation.
As expression of the gelE gene was required for biofilm formation, we wondered whether the gelatinase was required for its enzymatic activity or as a protein per se. If the former were the case, then one can envision the existence of a substrate(s) whose processing is necessary for the formation of a biofilm. If the latter possibility were the case, then either the full-length gelatinase somehow contributes to the process of biofilms formation or it is itself a substrate for processing by an unknown enzyme with the resulting peptides having a role in attachment and/or development of the biofilm.
To address this question, we attempted to purify a mature wild-type form of the gelatinase from an overexpressing E. coli strain and a control gelatinase protein presumably inactive due to a glutamate-to-alanine substitution at residue 137 in the active site of the enzyme. Although both proteins were soluble and thus purified as described in Materials and Methods, the wild-type gelatinase did not show any enzymatic activity in a casein plate assay. The mutated form was not expected to have any activity. The reason for this unexpected result is not known at this time. In order to test an active gelatinase enzyme, we then purified the protein from the supernatant of the gelatinase producer strain JML116 (see Materials and Methods). The protein thus purified was active as determined by the casein plate assay, and its N-terminal end sequence was as expected for the mature form of the enzyme (i.e., VGSEVT) as determined by amino acid sequence analysis.
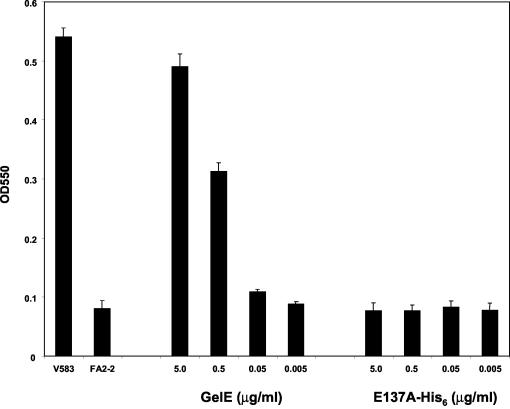
The active gelatinase enzyme purified from the E. faecalis culture supernatant and the two inactive forms isolated from E. coli were then tested for their ability to promote biofilm formation when added to the culture medium of a strain unable to make biofilm (FA2-2). As shown in Fig. 4, 5 μg of purified active gelatinase/ml (final concentration) could induce biofilm formation by strain FA2-2 to a level comparable to the one observed for strain V583. The wild-type inactive (data not shown) or the mutated gelatinase proteins from E. coli did not promote biofilm formation when used at the same concentrations.
FIG. 4.
Extracellular complementation of E. faecalis FA2-2 biofilm formation by purified gelatinase. OD550s of solubilized crystal violet from the microtiter plate assay at 24 h are shown. Biofilm formation by E. faecalis strains V583 and FA2-2 at 24 h was measured as a control. Strain FA2-2 was complemented extracellularly with purified gelatinase (GelE) from E. faecalis culture supernatants or the catalytic site mutant E137A expressed in E. coli as a His tag fusion. The values indicated on the x axis correspond to the amount of purified protein added to 200 μl of culture. All assays were performed in triplicate and repeated at least twice. Representative data from two independent experiments are shown with mean values and standard deviations (error bars).
These results strongly suggest that the enzymatic activity of gelatinase is required for its role in biofilm development.
DISCUSSION
Gelatinase is a member of the thermolysin-like M4 family of proteases (TLPs) that includes enzymes from pathogens such as Legionella, Listeria, Clostridium, Staphylococcus, Pseudomonas, and Vibrio (2). Many of these bacterial metalloproteases have been found to be associated with virulence, while eukaryotic members of the family (so-called matrix metalloproteases) (16) were shown to be involved in a number of processes in humans, including the processing of precursors that play modulation roles in the formation of tumors (20, 25). Thus, metalloproteases of the M4 family have attracted increasing attention as model proteins for the development of specific inhibitors that can be applied to disease treatment (41). Our finding of the role of gelatinase in biofilm development in E. faecalis raises the possibility that protease inhibitors may also be relevant, synergistically with antibiotics, in the treatment of biofilm-based pathogeneses such as infective endocarditis.
Gelatinase has been implicated in E. faecalis virulence by both epidemiological data and animal model studies (7, 11, 26, 29, 37, 38). Biochemical analysis of purified GelE showed that it cleaved a number of substrates in vitro, including insulin-β chain, Azocoll, insoluble collagen fragments, and endothelin-1, at primarily hydrophobic amino acids (24, 25, 48). It was also found to degrade the pheromone and inhibitor peptides involved in conjugative plasmid transfer in E. faecalis (24). Recently, it was shown that GelE functions to clear the bacterial cell surface of misfolded proteins (48). A role for GelE in the activation of an autolysin and in the degradation of polymerized fibrin was also suggested by the same study (48). Those authors proposed that GelE acts to increase the dissemination of E. faecalis in high-density environments by fitting the two-step model of Streptococcus pyogenes infection described by Rasmussen and Bjorck (34). In the first phase, at low bacterial cell density, a low level of bacterial proteolytic activity leads to attachment mediated by surface proteins. In the second phase, at high bacterial cell density, high levels of proteolytic activity promote bacterial dissemination by cleaving bacterial attachment proteins and host tissue proteins. Our finding that GelE is required for bacterial attachment to a surface and the growth in biofilms argues against that proposal. Certainly, the absence of GelE in E. faecalis is highly pleiotropic and affects many phenotypic aspects of the pathogenicity of this organism. The present results show that GelE is required for the formation of biofilm, and it promotes the aggregation of the cells in microcolonies to form the primary attachment site and subsequent development into a three-dimensional structure. We envisioned two possibilities for this function of GelE. Either the gelatinase proteolytic activity is required to efficiently initiate the attachment of E. faecalis to surfaces or the physical presence, not the enzymatic activity, of GelE is the crucial requirement. It seems intuitive that cleavage of a bacterial surface protein(s) may be required to promote the attachment of the cells to a given surface to form the biofilms (the abiotic surfaces used in our experiment should not presumably provide any substrate involved in the process). Since GelE is known to play a role in cell separation (48), the surface of a gelE-deficient cell may be sufficiently altered by this lack of enzymatic activity to prevent the cell from attaching to hydrophobic surfaces such as plastic or glass. Perhaps the uncleaved surface protein(s) is more hydrophilic and lacks affinity for such surfaces. While it is widely inferred that GelE is a virulence factor for E. faecalis, proof of this notion in vivo is not definitive (7, 38). The modification of surface hydrophobicity by its presence, suggested by our results, is another piece of circumstantial evidence that GelE or its inhibition could be important in the initiation of infection in susceptible individuals. This would be keeping with the hypothesis of Rasmussen and Bjorck (34), who suggested that host proteinase inhibitors inhibit proteolysis by bacterial enzymes early in infection. In the E. faecalis situation, the recruitment of host proteinase inhibitors to GelE would lead to altered surface characteristics and altered presentation to the immune system and other host defenses. In support of this hypothesis, Makinen et al. in fact showed inhibition of gelatinase activity by a host inflammatory exudate (24).
Our inability to generate an active gelatinase from an overexpressing E. coli system (despite numerous attempts and conditions or even in the presence of the propeptide sequence) did not allow us to test whether active site mutants of GelE would be active or inactive in promoting biofilm formation. Certainly, the purified enzyme from E. faecalis was sufficient to complement gelE and the fsr defect in biofilm formation, while an inactive gelE protein purified from E. coli was not. These observations strongly support the hypothesis that the processing of a substrate(s) by GelE is required to form a biofilm. The fact that an active GelE protein could not be purified from E. coli suggests that perhaps this organism lacks some crucial component for correct folding or modification of the protease. A precedent for an additional requirement for the folding and maturation of an extracellular protease was found in Streptococcus pyogenes; for its activation, the SpeB extracellular cysteine proteinase requires the streptococcal M1 protein, a fibrous surface protein and virulence determinant (6), in addition to the peptidyl-prolyl isomerase activity of the cytoplasmic trigger factor chaperone (22, 23).
Clearly, proteolysis and its regulation are important aspects of bacterial pathogenesis, and the role of GelE in enterococcal biofilm formation confirms this notion. Unlike GelE, the serine protease encoded by the E. faecalis sprE gene is not involved in biofilm formation and does not have an additive effect with GelE, at least in our assay conditions. However, SprE was reported to have a role in enterococcal infection when a mouse peritonitis model (32) or the nematode C. elegans killing model (37) was used. In the latter model, an additive effect of GelE and SprE was also noted.
Our results represent the first documented link between a two-component system (Fsr), a molecular entity (GelE), and biofilm formation in E. faecalis. A previous report suggested the product of the esp gene, a cell surface protein, was critical for biofilm formation by E. faecalis based on epidemiological studies (43). The same study, however, also showed that esp-deficient mutants of E. faecalis were fully capable of forming biofilms, thus leaving open the question of whether Esp has a role in this process or not (19). The E. faecalis V583 strain used in our study was shown to be esp deficient, as it lacks the region within the pathogenicity island that carries this gene in other strains (36). Yet strain V583 is fully capable of forming biofilms on abiotic surfaces, thus confirming that Esp is not a requirement for this type of bacterial growth, at least in the conditions used here, although we cannot rule out the possibility of a synergistic effect with GelE. Esp-independent biofilm formation was also reported by Kristich et al. with strain OG1RF (19). The same study incidentally reported the enhancement of biofilm formation by GelE.
Our study also indicates that through the control of gelE expression, the fsr two-component system is the only one among the 17 systems identified from E. faecalis V583 as being involved in biofilm formation (reference 14 and our unpublished data; Hancock and Perego, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol. 2003). Deletion of the other nonessential systems indeed did not affect biofilm growth, at least not at a detectable level in our assay. Although synergistic effects of two or more systems cannot be ruled out, it appears that GelE is a major requirement for biofilm development, as its sole expression can complement the lack of the FsrA response regulator or the lack of the FsrC histidine kinase (Fig. 2). Additionally, if one assumes that the basal OD of the fsrA mutant or of the fsrC mutant strain FA2-2 in the microtiter plate assay is equal to an inability to form a biofilm, as supported by the confocal microscopy images of Fig. 3B and D, then it is possible to conclude that the gelatinase enzymatic activity is necessary and sufficient (in the presence of a hypothetical substrate[s] and in the absence of the fsr system) for biofilm formation in E. faecalis V583. Furthermore, overexpression of gelE from the kanamycin promoter in a multicopy plasmid resulted in higher biofilm accumulation than in the control strain carrying a single copy of the gelE gene. (Fig. 2B). Accordingly, biofilm density was directly proportional to the concentration of GelE in the medium, as shown in Fig. 4. These observations, together with the knowledge that gelE expression is dependent upon an active FsrABC system (32), indicate that any physiological or environmental signal affecting the activation of the Fsr two-component system or the activity of its quorum-sensing, regulating peptide FsrB, if any, will have an impact on the efficiency of biofilm formation.
To this regard, it should also be pointed out that the role of the Fsr system in enterococcal biofilm formation does not seem to have any resemblance to the role of its counterpart systems AgrABCD of Staphylococcus aureus and ComCDE of streptococci in the respective ability of these organisms to develop biofilms. These three systems belong to the so-called agr-like subfamily of two-component signal transduction systems identified in gram-positive bacteria and are known as quorum-sensing systems for the presence of a third component, a secreted signaling peptide, in addition to a membrane-bound histidine kinase and an intracellular response regulator (18). This type of quorum-sensing system has been found to regulate a variety of physiological activities in gram-positive organisms, including competence development in streptococci and Bacillus subtilis, antibiotic biosynthesis in Lactococcus lactis, and induction of virulence factors in S. aureus (1, 5, 9, 10, 17). Recently, it was reported that the inactivation of the genes encoding the ComD histidine kinase or the ComE response regulator of Streptococcus mutans resulted in the formation of a biofilm with a reduced mass, while the inactivation of the comC gene encoding the signaling peptide gave rise to a biofilm with a defective architecture probably due to a defect in cell separation. An additive effect of the comDE and comC mutations was also reported previously (21). These observations suggested the existence of a second receptor for the ComC extracellular peptide in addition to the ComD competence histidine kinase. In contrast, the FsrB signaling peptide of E. faecalis does not seem to have additional targets other than the FsrC histidine kinase, at least for the biofilm phenotype, since an fsrBC mutant is quantitatively similar to fsrC or fsrA single mutants in biofilm formation. Additionally, the FsrB-containing supernatant of strain JML104 (gelE) did not improve the biofilm defect of the FA2-2 strain, thus ruling out the existence of a secondary pathway, at least in these strains (our unpublished observations).
For the process of biofilm formation in staphylococci, there is mounting evidence that the lack of a functional agr system increases the formation of biofilm despite the observation that proteins required for biofilm formation, such as alpha-toxin, are positively regulated by the Agr system (3, 47). Additionally, there is increasing evidence that whereas secreted virulence factors may be important for establishing infections, the loss of agr function may enhance the long-term survival of staphylococci in the host, thus contributing to persistent infections often associated with biofilm formation (35). The observations by Vuong et al. (46) that the disruption of the agr locus in Staphylococcus epidermidis resulted in increased biofilm formation and that the S. epidermidis O-47 clinical isolate is an agr mutant further support the notion that the role of the staphylococcal quorum-sensing system in biofilm formation may not be as obvious as generally anticipated. Furthermore, the observation that the expression of agr seen in certain animal models of infections does not significantly affect the expression of virulence factors, as would be expected from in vitro data, suggests that the role of quorum sensing in staphylococcal infections is not as clear as originally presented (12, 13, 49).
Clearly, the involvement, if any, of peptide-controlled systems in biofilm formation in gram-positive microorganisms is species specific and apparently requires the activation or inactivation of a myriad of known and unknown genes, making the general understanding of the pathway a very complex task. This also means that there will probably be the need for selective and targeted pharmaceutical approaches for the control of bacterial infections in cases of biofilm development.
Acknowledgments
This research was supported in part by Public Health Service grant GM55594 and GM19416 from the National Institute of General Medical Sciences, National Institutes of Health. The Stein Beneficial Trust supported in part oligonucleotide synthesis and DNA sequencing.
This is publication 15858-MEM from The Scripps Research Institute.
REFERENCES
- 1.Abdelnour, A., S. Arvidson, T. Bremell, C. Ryden, and A. Tarkowski. 1993. The accessory gene regulator (agr) controls Staphyloccoccus aureus virulence in a murine arthritis model. Infect. Immun. 61:3879-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett, A. J., N. D. Rawlings, and J. F. Woessner. 1998. Handbook of proteolytic enzymes, p. 350-369. Academic Press, Inc., New York, N.Y.
- 3.Caiazza, N. C., and G. A. O'Toole. 2003. Alpha-toxin is required for biofilm formation by Staphylococcus aureus. J. Bacteriol. 185:3214-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callegan, M. C., B. D. Jett, L. E. Hancock, and M. S. Gilmore. 1999. Role of hemolysin BL in the pathogenesis of extraintestinal Bacillus cereus infection assessed in an endophthalmitis model. Infect. Immun. 67:3357-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung, A. L., K. J. Eberhardt, E. Chung, M. R. Yeaman, P. M. Sullam, M. Ramos, and A. S. Bayer. 1994. Diminished virulence of a sar−/agr− mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Investig. 94:1815-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collin, M., and A. Olsen. 2000. Generation of a mature streptococcal cysteine proteinase is dependent on cell wall-anchored M1 protein. Mol. Microbiol. 36:1306-1318. [DOI] [PubMed] [Google Scholar]
- 7.Coque, T. M., J. E. Patterson, J. M. Steckelberg, and B. E. Murray. 1995. Incidence of hemolysin, gelatinase, and aggregation substance among enterococci isolated from patients with endocarditis and other infections and from feces of hospitalized and community-based persons. J. Infect. Dis. 171:1223-1229. [DOI] [PubMed] [Google Scholar]
- 8.Cruz-Rodz, A. L., and M. S. Gilmore. 1990. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol. Gen. Genet. 224:152-154. [DOI] [PubMed] [Google Scholar]
- 9.Cvitkovitch, D. G. 2001. Genetic competence and transformation in oral streptococci. Crit. Rev. Oral Biol. Med. 12:217-243. [DOI] [PubMed] [Google Scholar]
- 10.Dubnau, D., J. Hahn, M. Roggiani, F. Piazza, and Y. Weinrauch. 1994. Two-component regulators and genetic competence in Bacillus subtilis. Res. Microbiol. 145:403-411. [DOI] [PubMed] [Google Scholar]
- 11.Garsin, D. A., C. A. Sifri, E. Mylonakis, X. Qin, K. V. Singh, B. E. Murray, S. B. Calderwood, and F. M. Ausubel. 2001. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 98:10892-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goerke, C., S. Campana, M. G. Bayer, G. Doring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goerke, C., U. Fluckiger, A. Steinhuber, W. Zimmerli, and C. Wolz. 2001. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of α-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 40:1439-1447. [DOI] [PubMed] [Google Scholar]
- 14.Hancock, L., and M. Perego. 2002. Two-component signal transduction in Enterococcus faecalis. J. Bacteriol. 184:5819-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoch, J. A., and T. J. Silhavy. 1995. Two-component signal transduction. ASM Press, Washington, D.C.
- 16.Johnson, L. L., Q. Z. Ye, L. L. Johnson, D. J. Hupe, D. F. Ortwine, J. B. Dunbar, Jr., J. B. Rubin, A. Pavlovsky, C. Humblet, and T. L. Blundell. 1996. Designing inhibitors of the metalloproteinase superfamily: comparative analysis of representative structures. Drugs Des. Discov. 13:3-14. [PubMed] [Google Scholar]
- 17.Kleerebezem, M., and L. E. Quadri. 2001. Peptide pheromone-dependent regulation of antimicrobial peptide production in Gram-positive bacteria: a case of multicellular behavior. Peptides 22:1579-1596. [DOI] [PubMed] [Google Scholar]
- 18.Kleerebezem, M., L. E. N. Quadri, O. P. Kuipers, and W. de Vos. 1997. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 24:895-904. [DOI] [PubMed] [Google Scholar]
- 19.Kristich, C. J., Y. H. Li, D. G. Cvitkovitch, and G. M. Dunny. 2004. Esp-independent biofilm formation by Enterococcus faecalis. J. Bacteriol. 186:154-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lennarz, W. J., and W. J. Strittmatter. 1991. Cellular functions of metallo-endoproteinases. Biochim. Biophys. Acta 1071:149-158. [DOI] [PubMed] [Google Scholar]
- 21.Li, Y.-H., N. Tang, M. B. Aspiras, P. C. Y. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyon, W. R., and M. G. Caparon. 2003. Trigger factor-mediated prolyl isomerization influences maturation of the Streptococcus pyogenes cysteine protease. J. Bacteriol. 185:3661-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makinen, P.-L., D. B. Clewell, F. An, and K. K. Makinen. 1989. Purification and substrate specificity of a strongly hydrophobic extracellular metalloendopeptidase (“gelatinase”) from Streptococcus faecalis (strain 0G1-10). J. Biol. Chem. 264:3325-3334. [PubMed] [Google Scholar]
- 25.Makinen, P., and K. K. Makinen. 1994. The Enterococcus faecalis extracellular metalloendopeptidase (EC 3.4.24.30; coccolysin) inactivates human endothelin at bonds involving hydrophobic amino acid residues. Biochem. Biophys. Res. Commun. 200:981-985. [DOI] [PubMed] [Google Scholar]
- 26.Mylonakis, E., M. Engelbert, X. Qin, C. D. Sifri, B. E. Murray, F. M. Ausubel, M. S. Gilmore, and S. B. Calderwood. 2002. The Enterococcus faecalis fsrB gene, a key component of the fsr quorum-sensing system, is associated with virulence in the rabbit endophthalmitis model. Infect. Immun. 70:4678-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakayama, J., Y. Cao, T. Horii, S. Sakuda, A. D. L. Akkermans, W. de Vos, and H. Nagasawa. 2001. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41:145-154. [DOI] [PubMed] [Google Scholar]
- 28.Olson, M. E., H. Ceri, D. W. Morck, A. G. Buret, and R. R. Read. 2002. Biofilm bacteria: formation and comparative susceptibility to antibiotics. Can. J. Vet. Res. 66:86-92. [PMC free article] [PubMed] [Google Scholar]
- 29.Pillai, S. K., G. Sakoulas, H. S. Gold, C. Wennersten, G. M. Eliopoulos, R. C. Moellering, Jr., and R. T. Inouye. 2002. Prevalence of the fsr locus in Enterococcus faecalis infections. J. Clin. Microbiol. 40:2651-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Podbielski, A., B. Spellerberg, M. Woischnik, B. Pohl, and R. Lütticken. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137-147. [DOI] [PubMed] [Google Scholar]
- 31.Poyart, C., and P. Trieu-Cuot. 1997. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to β-galactosidase in Gram-positive bacteria. FEMS Microbiol. Lett. 156:193-198. [DOI] [PubMed] [Google Scholar]
- 32.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68:2579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183:3372-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasmussen, M., and L. Bjorck. 2002. Proteolysis and its regulation at the surface of Streptococcus pyogenes. Mol. Microbiol. 43:537-544. [DOI] [PubMed] [Google Scholar]
- 35.Schwan, W. R., M. H. Langhorne, H. D. Ritchie, and C. K. Stover. 2003. Loss of hemolysin expression in Staphylococcus aureus agr mutants correlates with selective survival during mixed inflections in murine abscesses and wounds. FEMS Immunol. Med. Microbiol. 38:23-28. [DOI] [PubMed] [Google Scholar]
- 36.Shankar, N., A. S. Baghdayan, and M. S. Gilmore. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746-750. [DOI] [PubMed] [Google Scholar]
- 37.Sifri, C. D., E. Mylonakis, K. V. Singh, X. Qin, D. A. Garsin, B. E. Murray, F. M. Ausubel, and S. B. Calderwood. 2002. Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect. Immun. 70:5647-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh, K. V., X. Qin, G. M. Weinstock, and B. E. Murray. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 178:1416-1420. [DOI] [PubMed] [Google Scholar]
- 39.Stephenson, K., and J. A. Hoch. 2002. Virulence- and antibiotic resistance-associated two-component signal transduction systems of Gram-positive pathogenic bacteria as targets for antimicrobial therapy. Pharmacol. Ther. 93:293-305. [DOI] [PubMed] [Google Scholar]
- 40.Su, Y. A., M. C. Sulavik, P. He, K. K. Makinen, P.-L. Makinen, S. Fiedler, R. Wirth, and D. B. Clewell. 1991. Nucleotide sequence of the gelatinase (gelE) from Enterococcus faecalis subsp. liquefaciens. Infect. Immun. 59:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamaki, M., K. Tanzawa, S. Kurihara, T. Oikawa, S. Monma, K. Shimada, and Y. Sugimura. 1995. Synthesis and structure-activity relationships of gelatinase inhibitors derived from matlystatins. Chem. Pharm. Bull. (Tokyo) 43:1883-1893. [DOI] [PubMed] [Google Scholar]
- 42.Teng, F., L. Wang, K. V. Singh, B. E. Murray, and G. M. Weinstock. 2002. Involvement of PhoP-PhoS homologs in Enterococcus faecalis virulence. Infect. Immun. 70:1991-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toledo-Arana, A., J. Valle, C. Solano, M. J. Arrizubieta, C. Cucarella, M. Lamata, B. Amorena, J. Leiva, J. R. Penades, and I. Lasa. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67:4538-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1990. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in Gram-positive bacteria. Nucleic Acids Res. 18:4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trieu-Cuot, P., and P. Courvalin. 1983. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5"-aminoglycoside phosphotransferase type III. Gene 23:331-341. [DOI] [PubMed] [Google Scholar]
- 46.Vuong, C., C. Gerke, G. A. Somerville, E. R. Fischer, and M. Otto. 2003. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J. Infect. Dis. 188:706-718. [DOI] [PubMed] [Google Scholar]
- 47.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182:1688-1693. [DOI] [PubMed] [Google Scholar]
- 48.Waters, C. M., M. H. Antiporta, B. E. Murray, and G. M. Dunny. 2003. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J. Bacteriol. 185:3613-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yarwood, J. M., J. K. McCormick, M. L. Paustian, V. Kapur, and P. M. Schlievert. 2002. Repression of the Staphylococcus aureus accessory gene regulator in serum and in vivo. J. Bacteriol. 184:1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]