Abstract
D-alanine (D-Ala) is an essential amino acid that has a key role in bacterial cell wall synthesis. Alanine racemase (Alr) is a unique enzyme that interconverts L-alanine and D-alanine in most bacteria, making this enzyme a potential target for antimicrobial drug development. Streptococcus mutans is a major causative factor of dental caries. The factors involved in the survival, virulence and interspecies interactions of S. mutans could be exploited as potential targets for caries control. The current study aimed to investigate the physiological role of Alr in S. mutans. We constructed alr mutant strain of S. mutans and evaluated its phenotypic traits and interspecies competitiveness compared with the wild-type strain. We found that alr deletion was lethal to S. mutans. A minimal supplement of D-Ala (150 μg·mL−1) was required for the optimal growth of the alr mutant. The depletion of D-alanine in the growth medium resulted in cell wall perforation and cell lysis in the alr mutant strain. We also determined the compromised competitiveness of the alr mutant strain relative to the wild-type S. mutans against other oral streptococci (S. sanguinis or S. gordonii), demonstrated using either conditioned medium assays or dual-species fluorescent in situ hybridization analysis. Given the importance and necessity of alr to the growth and competitiveness of S. mutans, Alr may represent a promising target to modulate the cariogenicity of oral biofilms and to benefit the management of dental caries.
Keywords: alanine racemase, biofilm, D-alanine, dental caries, Streptococcus mutans
Introduction
D-alanine (D-Ala) is an essential substance for the synthesis of peptidoglycan (PG), an important constituent of the cell wall of both Gram-negative and Gram-positive bacteria.1, 2 The synthesis of the cell wall is initiated with uridine diphosphate (UPD)-N-acetylmuramic acid, L-Ala, D-glutamic acid (D-Glu) and meso-diaminopimelic acid to generate UDP-N-acetylmuramyl-L-Ala-D-Glu-meso-diaminopimelate.1, 3 A D-Ala dipeptide is coupled to this intermediate through UDP-N-acetylmuramyl-tripeptide-D-Ala-D-Ala ligase to generate UDP-N-acetylmuramyl-tripeptide-D-Ala-D-Ala, which is subsequently incorporated into the growing PG peptide chain. The D-Ala dipeptide is synthesized by two enzymes: alanine racemase (Alr), which converts L-enantiomers of alanine to the D-counterparts, and D-Ala-D-Ala ligase, which generates the D-Ala dipeptide.3
Alr is a unique enzyme responsible for the racemization of L-Ala to D-Ala.4 Previous studies have revealed that Alr is essential for the survival of numerous Gram-positive bacteria, including Bacillus, Lactobacillus, Burkholderia pseudomallei, Burkholderia mallei and Mycobacterium tuberculosis. Knockout of the alr gene in these bacteria resulted in a strict exogenous D-Ala-dependent growth phenotype.5, 6, 7, 8 Similar growth arrest and extensive cell lysis were also observed in the alr mutant of Gram-negative Escherichia coli, which underwent rapid and extensive cell lysis when deprived of D-Ala.5, 9 Further in vitro and in vivo studies of M. tuberculosis have shown that Alr is a primary target of D-cycloserine, and the inhibition of Alr alone could reduce the viability and perseverance of this bacterium.8
Streptococcus mutans is the major caries-associated bacterium in humans. During cariogenic conditions (e.g., frequent sugar intake), S. mutans metabolizes carbohydrates, leading to acid accumulation and subsequent fall in pH in the dental biofilm.10 The acidic micro-environment selectively enriches acidogenic/aciduric species (e.g., mutans streptococci and lactobacilli) and suppresses less aciduric commensal residents (e.g., Streptococcus sanguinis and Streptococcus gordonii). This microbial disequilibrium leads to continuous pH decline to reach a critical pH, below which tooth hard-tissue demineralization begins and dental caries gradually occur.11, 12 Although Alr has been well documented as closely associated with the viability and survival of various bacteria,5, 6, 7, 8, 9 its physiological importance in S. mutans has not been explored, particularly in a biofilm context. In the present study, we constructed alr mutant strain and investigated the physiological role of alr in the cell growth, cell wall integrity and interspecies competitiveness of S. mutans. We found that alr is an essential factor to maintain the growth and cell wall integrity of S. mutans. Deletion of alr in S. mutans significantly compromised its competitiveness with other co-residents (e.g., S. sanguinis) in the oral biofilm. We concluded that Alr might represent a promising drug target to modulate the cariogenicity of oral biofilm and to benefit the management of dental caries.
Materials and methods
Bacterial strains, growth media and reagents
S. mutans UA159 was obtained from the Dental Research Institute, University of Toronto13 and was routinely anaerobically (90% N2, 5% CO2, 5% H2) or aerobically (95% air, 5% CO2) incubated at 37 °C in brain heart infusion (BHI) broth (Difco, Sparks, MD, USA). For the transformation experiments, the cells were maintained in Todd-Hewitt medium (Difco, Sparks, MD, USA) supplemented with 3 ġL−1 yeast extract (THYE; Difco, Sparks, MD, USA). The competence-stimulating peptide used for S. mutans transformation was custom-synthesized by Sangon Biotech (Shanghai, China). For the selection of antibiotic-resistant colonies, BHI plates were supplemented with erythromycin (erm, 12.5 μġmL−1). D-Ala (150 μg·mL−1) was added to the BHI broth to promote the growth of the S. mutans alr mutant strain. The optical density (OD) of the cell culture was measured at 600 nm (OD600).
Taq DNA polymerase, restriction enzymes and T4 DNA ligase were all purchased from New England Biolabs (Ipswich, MA, USA). Taq DNA polymerase was used for overlapping polymerase chain reaction (PCR).
Construction of the alr mutant strain
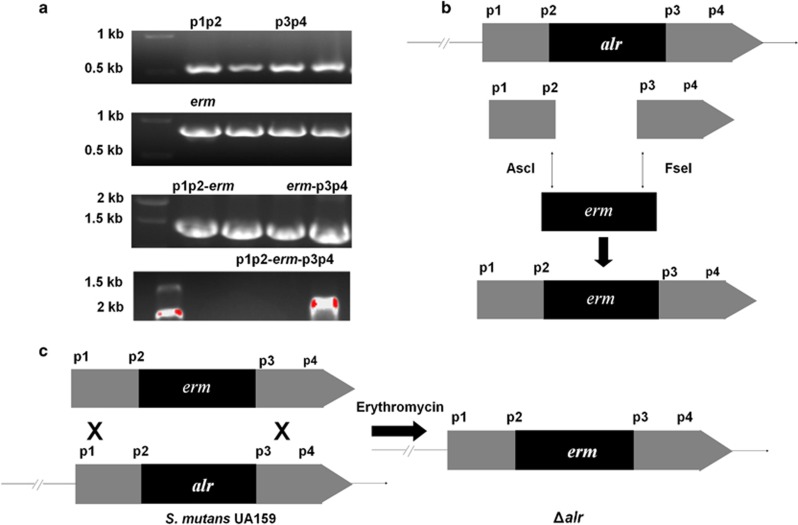
The primers used in this study are shown in Table 1. Two 500 bp fragments (up- and down-stream of alr) were generated through PCR using the primer pair alr-up-f/ldh and alr-down-f/ldh. The erm fragment (876 bp) was amplified with primer pair ldh-f/erm-r by PCR.14 The three pairs of primers were specifically designed for subsequent ligation based on the sequences adjacent to mutagenesis site. The restriction enzymes AscI (New England Biolabs, Ipswich, MA, USA) and FseI were used to digest the up- and down-stream fragments, respectively, and the aforementioned enzymes were also used to digest the purified erm segment. The three digested fragments were subsequently mixed, and T4 DNA ligase was added to generate the proposed segment (Figure 1).15, 16, 17 The resulting 1.876 kb fragment was transformed into S. mutans, and transformants were selected on BHI plates containing 12.5 μg·mL−1 of erythromycin. The alr deletion mutant was confirmed using sequencing. All primers used are listed in Table 1.
Table 1. Oligonucleotide primers used for the construction of the S. mutans alr mutant.
| Primers | Nucleotide sequence (5′-3′) |
|---|---|
| alr-up-f | ATCCCATGAACATCAGTTTATGTC |
| alr-up-ldh | GGCGCGCCCTAGCAATCATAAGTTATTCTCCTCC |
| alr-down-r | GGCCGGCCGTTAAGTGGCTGAACTTTTTTGGC |
| alr-down-erm | CTGATCAAGCTCCACCTTATCTTG |
| Idh-f | GGCGCGCCCCGGGCCCAAAATTTGTTTGAT |
| erm-r | GGCCGGCCAGTCGGCAGCGACTCATAGAAT |
The nucleotide sequence indicated in bold and underlined is the restriction enzyme site of AscI, and the sequence indicated in bold is the restriction enzyme site of FseI.
Figure 1.
The S. mutans alr mutant was constructed using homologous recombination. (a) Two 500 bp fragments were generated (p1p2: up-stream, and p3p4: down-stream of alr) using the primer pair alr-up-f/ldh and alr-down-f/ldh. An erm fragment (876 bp) was amplified using the primer pair ldh-f/erm-r. (b) The three digested fragments ligated using T4 DNA ligase to generate the proposed segment. (c) This fragment was transformed into S. mutans, and the transformants were selected on BHI broth plates containing 12.5 μg·mL−1 of erythromycin. Δalr, S. mutans alr mutant; BHI, brain heart infusion.
Growth of the alr mutant
S. mutans UA159 and the alr mutant strain were cultivated overnight in BHI broth. Stationary phase cultures were diluted 1:20 in BHI broth and incubated at 37 °C until the OD600 reached 0.2. A 20 μL aliquot of the cell culture and 180 μL of BHI broth were added to each well of a 96-well plate. The OD of the bacteria culture was measured at intervals over a period of 1 h. The cells were diluted to 1 × 106 CFU·mL−1, plated onto BHI broth agar plates, and incubated at 37 °C for 24 h.
Transmission electron microscopy
Transmission electron microscopy (TEM) was performed as previously described.18 Approximately 10 mL of cell culture was harvested by low-speed centrifugation (3 000g, 10 min), washed twice in 200 mmol̇L−1 sodium cacodylate buffer, pre-fixed with 2.5 ġL−1 glutaraldehyde and fixed with 10 ġL−1 OsO4. Samples were embedded in Epon resin, and thin sections (60 nm) were prepared using a microtome. The sections were stained with 40 ġL−1 uranyl acetate and subsequently with 4 ġL−1 lead citrate and were examined using a Tecnai G2 F20 S-TWIN electron microscope (FEI, Hillsboro, OR, USA).
Conditioned media assay
To investigate whether other commensal colonizers such as S. sanguinis and S. gordonii can support the growth of the alr-deficient mutant under exogenous D-Ala-deprived conditions, we performed a conditioned media assay as previously described.19 Briefly, the culture supernatants of S. sanguinis and S. gordonii at the mid-exponential phase were collected and filter sterilized as a conditioned medium for the growth of the alr mutant. After aerobic incubation (5% CO2) for 24 h, the OD600 values of the bacterial cultures were determined to evaluate the effect of conditioned medium on the growth of the alr mutant. We also diluted the conditioned medium 1:2 with fresh BHI broth medium to evaluate the dose-dependent effects of conditioned medium. In addition, an exogenous D-amino acid oxidase (0.5 U·mL−1, Sigma, St Louis, MO, USA) was added to the conditioned medium to investigate whether the growth compensatory effects of conditioned medium on the alr mutant could be attenuated.
Fluorescent in situ hybridization
Fluorescent in situ hybridization (FISH) was performed according to Zheng et al.19 Biofilm specimens established on saliva-coated glass coverslips were rinsed with distilled water and dried for 10 min at 46 °C. To enhance probe penetration, the specimens were treated with 1 mL of lysis buffer (100 mmol̇L−1 tris(hydroxymethyl)aminomethane (Tris)-HCl, 50 mmol̇L−1 ethylene diamine tetraacetic acid, and 30 mg·mL−1 lysozyme (Sigma), pH 8.0) for 20 min at 37 °C. The specimens were subsequently rinsed with distilled water, serially dehydrated in ethanol (50%, 80% and 96% 3 min each), dried for 10 min at 46 °C, exposed to 20 μL of hybridization buffer (0.9 mol̇L−1 NaCl, 20 mmol̇L−1 Tris-HCl, 0.01% sodium dodecyl sulfate (SDS) and 20% formamide) containing the designated oligonucleotide probes (2 nmol̇L−1), and incubated at 46 °C for 90 min. After hybridization, the specimens were washed in buffer (20 mmol̇L−1 Tris-HCl, 0.01% SDS, 5 mmol̇L−1 EDTA and 215 mmol̇L−1 NaCl) for 15 min at 48 °C in a water bath and were subsequently rinsed in ice-cold nuclease-free water.
The specific probe for S. sanguinis was designed using ARB software (Linux release arb_5.3, Bremen, Germany). The S. mutans-specific probe was synthesized according to the sequence provided in a previous study.20 The probe sequences are listed in Table 2. Biofilms were examined using an Olympus BX3-CBH fluorescence microscope (Olympus, Tokyo, Japan) with an SpGr-B Filter (Semrock, Rochester, NY, USA) for Alexa Fluor 488 and an SpRed-B Filter (Semrock, Rochester, NY, USA) for Alexa Fluor 594. Black-and-white micrographs from at least five randomly selected positions of each sample were obtained using an Andor iXon3 camera (Andor Technology, Beijing, China) and were processed using Cell Sens Dimension (Olympus, Tokyo, Japan) and Photoshop CS 4.0 (Adobe, San Jose, CA, USA) without any qualitative changes to the raw images. The ratio of S. mutans/S. sanguinis was calculated based on the coverage area of each bacterium analysed using Image Pro Plus 6.0 (Media Cybernetics, Silver Spring, MD, USA).
Table 2. Oligonucleotide probes used for fluorescent in situ hybridization.
| Probes | Nucleotide sequence (5′-3′) | Target gene |
|---|---|---|
| S. mutans | (Alexa Fluor 488)-ACTCCAGACTTTCCTGAC | 16S rRNA |
| S. sanguinis | (Alex Fluor 594)-GCATACTATGGTTAAGCCACAGCC | 16S rRNA |
Statistical analysis
All experiments were performed in triplicate. The statistical analysis was performed using SPSS16.0 software (SPSS, Chicago, IL, USA). Differences were considered as significant when the two-tailed P-value was <0.05.
Results
Growth of the alr- mutant is strictly dependent on exogenous D-Ala
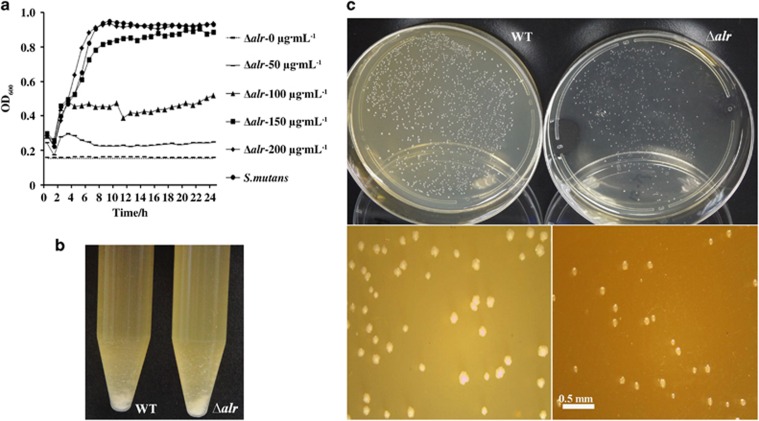
As shown in Figure 2a, alr deletion without the addition of exogenous D-Ala was lethal to S. mutans. The optimal growth of the alr mutant strain was observed after adding no less than 150 μg·mL−1 of D-Ala to the BHI medium (Figure 2a).
Figure 2.
Phenotypic traits of the alr mutant compared with S. mutans. (a) Growth curve of S. mutans and the alr mutant in the presence of an increasing dose of D-Ala for 24 h (0–200 μg·mL−1). (b) Growth of planktonic cultures of S. mutans (left) and the alr mutant strain (right) at the exponential phase in tubes. No significant difference was observed. (c) Representative stereoscopic micrographs of colonies of S. mutans and the alr mutant strain grown on agar plates. The results were averaged from three independent experiments, shown as the mean±standard deviation. Ala, alanine; Δalr, S. mutans alr mutant; WT, wild-type S. mutans.
In addition, no significant difference with respect to the planktonic culture (Figure 2b), colony shape, colour and transparency (Figure 2c) of the alr mutant strain was observed relative to that of the wild-type UA159 strain, although the colony size of the alr mutant strain was relatively smaller (Figure 2c).
D-Ala starvation causes cell morphology alterations of the alr mutant
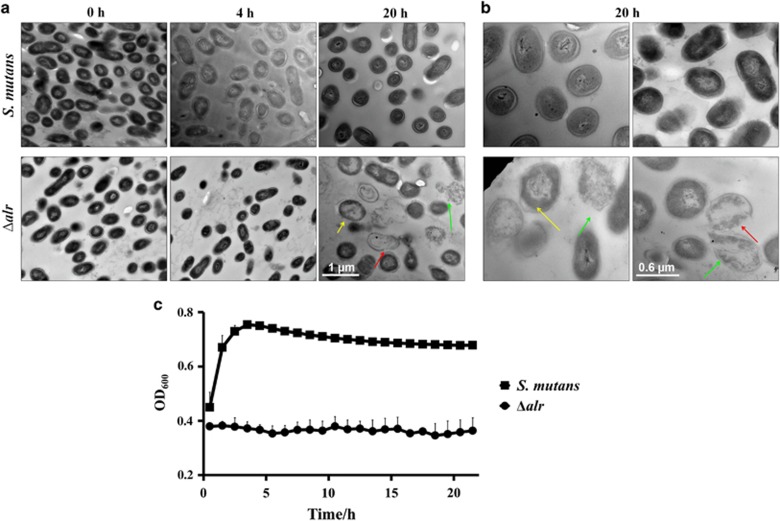
To determine how alr deletion affects the growth of S. mutans, we further examined the kinetic cell morphological alterations of the alr mutant during D-Ala depletion using TEM. As shown in Figure 3a, when incubated in BHI without exogenous D-Ala, initial D-Ala starvation (4 h) did not result in significant morphological changes in the alr mutant relative to those of the wild-type control. However, the alr mutant strain exhibited pronounced morphological alterations after prolonged D-Ala starvation (20 h). Specifically, conspicuous cell lysis was observed in the alr mutant strain relative to the wild-type control (Figure 3b). These morphological alterations indicate the pronounced cell lysis of the alr mutant, potentially reflecting compromised cell wall stability and integrity after the deletion of alr in a D-Ala-deprived environment.
Figure 3.
Morphological alterations and growth of the alr mutant under D-Ala starvation. (a, b) Representative transmission electron micrographs of S. mutans and alr mutant strain (Δalr) under D-Ala starvation for 0, 4 and 20 h. The red arrows indicate plasmolysis, and the green arrows indicate damaged cells. (c) The growth curve of S. mutans and Δalr under D-Ala starvation. The results were averaged from three independent experiments, presented as the mean±standard deviation. Ala, alanine; Δalr, S. mutans alr mutant.
Consistent with increased cell lysis of the alr mutant, kinetic monitoring of the bacterial growth in the BHI medium without exogenous D-Ala also showed the significant growth arrest of the alr mutant, reflected by the unchanged OD600 values compared with the wild-type strain (Figure 3c).
Deletion of alr in S. mutans compromises its interspecies competitiveness
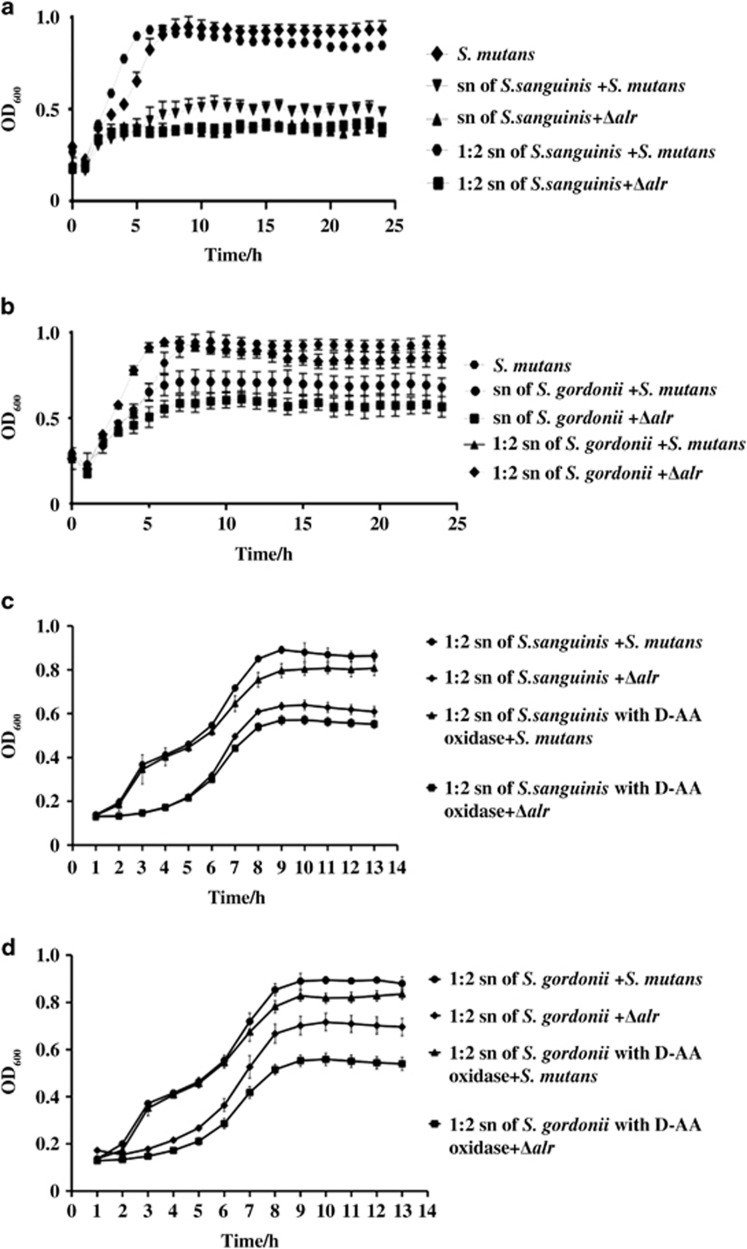
We further investigated the interspecies competitiveness of the alr mutant against other common oral streptococci, such as S. sanguinis and S. gordonii. We observed that the alr mutant strain could survive in the conditioned media of S. sanguinis or S. gordonii without exogenous D-Ala (Figure 4a and 4b), although its growth in the conditioned media was significantly inhibited relative to that of the wild-type parental strain. The addition of D-amino acid oxidase into the conditioned media further attenuated the growth of alr mutant, but had no significant effect on the growth of the wild-type S. mutans strain (Figure 4c and 4d). These data suggest that although the alr mutant might feed on the metabolites derived from other oral streptococci (D-Ala) to partially compensate for the scarce endogenous D-Ala, its perseverance in the co-existence of other oral streptococci remained compromised.
Figure 4.
Effect of conditioned medium derived from other commensal bacteria on the growth of the alr mutant. (a, b) Effect of S. sanguinis-derived and S. gordonii-derived conditioned medium on the growth of the Δalr with or without exogenous D-Ala, respectively. (c, d) Effect of S. sanguinis-derived and S. gordonii-derived conditioned medium supplemented with D-AA oxidase on the growth of Δalr. The results were averaged from three independent experiments and presented as the mean±standard deviation. “sn”: supernatant from S. sanguinis or S. gordonii culture; “1:2 sn”: 1:2 diluted supernatant with fresh BHI broth media. AA, amino acid; Ala, alanine; Δalr, S. mutans alr mutant; BHI, brain heart infusion.
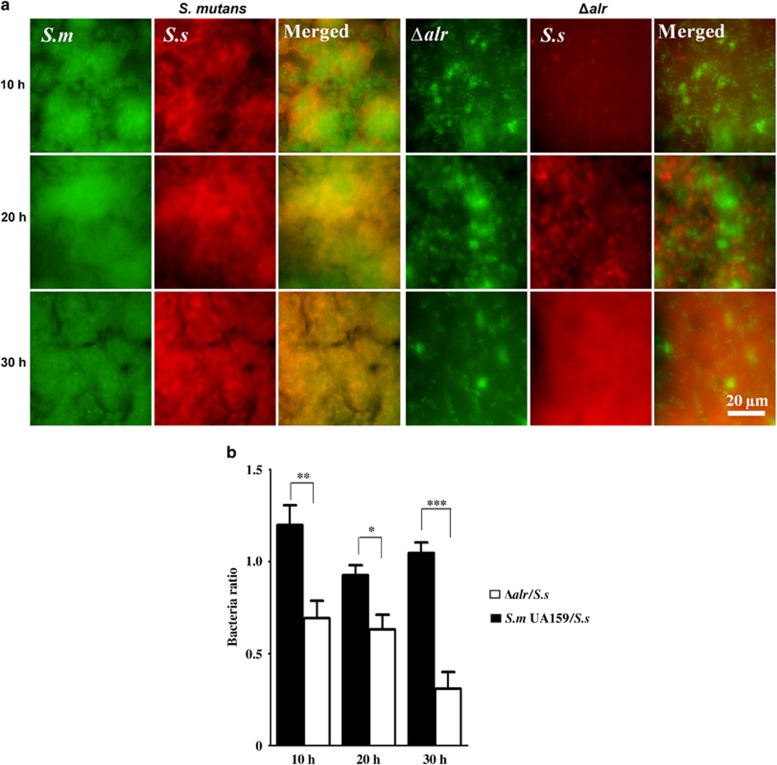
To further demonstrate the role of alr in the interspecies competition between S. mutans and S. sanguinis within biofilm, we used species-specific FISH to quantify the bacterial composition in a dual-species biofilm model (Figure 5a). Consistent with the data obtained from conditioned media assays, the colonization of the alr mutant was significantly reduced, as reflected by a significantly lower S. mutans/S. sanguinis ratio within the dual-species biofilm after 30 h compared with that of the wild-type S. mutans controls at corresponding time points (Figure 5b). These data further support that alr is critical for the interspecies competitiveness of S. mutans within biofilm.
Figure 5.
Interspecies competition between alr mutant and S. sanguinis in dual-species biofilms. (a) S. mutans (green), alr mutant strain (Δalr, green) and S. sanguinis (red) were labelled using species-specific fluorescent in situ hybridization probes. (b) The ratio of S. mutans/S. sanguinis quantified using the coverage area of each species. The results were averaged from at least five randomly selected positions of each sample and shown as the mean±standard deviation. *P<0.05, **P<0.01, ***P<0.0001. Δalr, S. mutans alr mutant; S.m, S. mutans; S.s, S. sanguinis.
Discussion
Alr is responsible for the production of D-Ala, which is used for PG biosynthesis in all bacteria, including harmful pathogens.1, 2, 21 Alr is encoded by alr, which is ubiquitous in prokaryotes but absent in eukaryotes, making this enzyme a great target for antimicrobial drug development. Previous knockout studies have shown that bacteria deficient in the alr gene require an external source of D-Ala for survival,6, 7, 8, 9, 22 further supporting the idea that alr could be a feasible target for antimicrobial drugs. In the present study, deleting the alr gene in S. mutans, a major caries-associated bacterium in the oral cavity of humans, revealed that the viability and growth of the alr mutant was strictly dependent on exogenous D-Ala derived from either the growth media or other commensal co-colonizers.
The observed kinetic growth of the alr mutant under D-Ala starvation up to 24 h is similar to that of the previously reported alr mutant of Lactobacillus plantarum.23 The relative static cell amount during D-Ala starvation indicates balanced cell lysis and proliferation. We speculated that dead cells under D-Ala starvation might not completely rupture, reflecting either osmo-protection in the nutrient-rich growth medium or the protection of relatively thick and rigid cell wall of S. mutans. In addition, the alr-deficient cells might feed on the recycled D-Ala from the cell wall of the lysed cells, thus maintaining a balanced cell mass under D-Ala starvation.
The pronounced cell lysis of the alr mutant was observed under D-Ala-deprived conditions. Cell lysis was also observed in the alr mutant of L. plantarum and E. coli,6, 9 reflecting the disturbed balance between PG degradation and biosynthesis during septation after the inactivation of Alr.23, 24 The PG peptide stem often contains D-Ala residues at the fourth and fifth positions.25 The disruption of the synthesis of these two residues could lead to the cell death of Gram-positive Bacillus and Lactobacillus.6, 26 In Gram-positive bacteria, such as Bacillus subtilis and Staphylococcus aureus, the inhibition of the D-alanylation on teichoic acid (TA) reflects the inactivation of dlt operon-stimulated autolysis.27, 28 The removal of D-alanyl residues from TA could result in the formation of cell surface holes, which likely confer a higher affinity to cationic PG hydrolases and thereby facilitating cell lysis.27 The observed cell wall damage and cell lysis of the alr mutant in the current study might also reflect the defective PG peptide stem and the D-alanylation of TA. The glutamate racemase (murl) has also been reported as an essential enzyme in the synthesis of the bacterial cell wall.29, 30 The murl mutant of E. coli lysed in the absence of D-glutamic acid,31 and the murl mutant of Mycobaterium smegmatis also showed aberrant cell shapes.32 Although the precise mechanisms remain unknown, the deletion of alr not only led to growth arrest but also to cell death, indicating that alr might be a potential bactericidal target for the inhibition of S. mutans.
Interestingly, we also observed that the alr mutant of S. mutans could survive in concert with other common oral streptococci, such as S. sanguinis and S. gordonii. The partially recovered growth of the alr-deficient mutant in conditioned medium from either S. sanguinis or S. gordonii could be further attenuated through D-amino acid oxidase. These results and data showing that the alr mutant requires exogenous D-Ala for survival, indicating that although the alr mutant might feed on either exogenous D-Ala or bacterial metabolites from other co-residents, the growth and competitiveness of the alr mutant strain are significantly compromised compared with the parental strain. This finding further supports the hypothesis that alr might be a promising target to control the prevalence of cariogenic S. mutans in a multispecies microbial consortium. Because Alr is universal in all bacteria, the utilization of this enzyme to develop antimicrobial drugs specific for S. mutans in multispecies microbial consortium is worthy of further investigation. Strategies as proposed by Professor Shi's group33, 34, 35 might be used to link Alr small-inhibitors with a targeting peptide (e.g., Competence stimulating peptide) and to achieve S. mutans-specific killing, even in a complex microbial consortium.
Taken together, the results of the present study provide the first evidence that alr is critical for the viability, growth and interspecies competitiveness of S. mutans. Given the ecological importance of S. mutans in initiating oral microbial disequilibrium and dental caries, Alr could be exploited as an antimicrobial target for S. mutans, thereby contributing to the control of dental caries.
Acknowledgments
This work was financially supported through grants from the National Natural Science Foundation of China (81400501 to Ming-Yun Li, 81371135 to Ji-Yao Li and 81430011 to Xue-Dong Zhou); the International Science and Technology Cooperation Programme of China (2014DFE30180 to Xue-Dong Zhou); the Talented Young Investigator Award of Sichuan University (2082604184224 to Xin Xu); and the Special Fund of State Key Laboratory of Oral Diseases, Sichuan University (SKLOD201525 to Ming-Yun Li).
References
- Lovering AL, Safadi SS, Strynadka NC. Structural perspective of peptidoglycan biosynthesis and assembly. Annu Rev Biochem 2012; 81: 451–478. [DOI] [PubMed] [Google Scholar]
- Ollivaux C, Soyez D, Toullec JY. Biogenesis of D-amino acid containing peptides/proteins: where, when and how? J Pept Sci 2014; 20 (8): 595–612. [DOI] [PubMed] [Google Scholar]
- Shaw JP, Petsko GA, Ringe D. Determination of the structure of alanine racemase from Bacillus stearothermophilus at 1.9–a resolution. Biochemistry 1997; 36 (6): 1329–1342. [DOI] [PubMed] [Google Scholar]
- Wasserman SA, Daub E, Grisafi P et al. Catabolic alanine racemase from Salmonella typhimurium: DNA sequence enzyme purification and characterization. Biochemistry 1984; 23 (22): 5182–5187. [DOI] [PubMed] [Google Scholar]
- Ferrari E, Henner DJ, Yang MY. Isolation of an alanine racemase gene from Bacillus subtilis and its use for plasmid maintenance in B. subtilis. Biotechnology 1985; 3 (11): 1003–1007. [Google Scholar]
- Hols P, Defrenne C, Ferain T et al. The alanine racemase gene is essential for growth of Lactobacillus plantarum. J Bacteriol 1997; 179 (11): 3804–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajdowicz SL, Jones-Carson J, Vazquez-Torres et al. Alanine racemase mutants of Burkholderia pseudomallei and Burkholderiamallei and use of alanine racemase as a non-antibiotic-based selectable marker. PLoS One 2011; 6 (6): e21523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthy D, Bharath S, Subbulakshmi V et al. Alanine racemase mutants of Mycobacterium tuberculosis require D-alanine for growth and are defective for survival in macrophages and mice. Microbiology 2012; 158 (Pt 2): 319–327. [DOI] [PubMed] [Google Scholar]
- Wijsman HJ. The characterization of an alanine racemase mutant of Escherichia coli. Genet Res 1972; 20 (3): 269–277. [DOI] [PubMed] [Google Scholar]
- Decker EM, Klein C, Schwindt D et al. Metabolic activity of Streptococcus mutans biofilms and gene expression during exposure to xylitol and sucrose. Int J Oral Sci 2014; 6 (4): 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology 2003; 149 (Pt 2): 279–294. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Nyvad B. The role of bacteria in the caries process ecological perspectives. J Dent Res 2011; 90 (3): 294–303. [DOI] [PubMed] [Google Scholar]
- Senadheera MD, Guggenheim B, Spatafora GA et al. A VicRK signal transduction system in Streptococcus mutans affects gtfBCDgbpB, and ftf expression, biofilm formation, and genetic competence development. J Bacteriol 2005; 187 (12): 4064–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Okinaga T, Qi F et al. Cloning-independent and counterselectable markerless mutagenesis system in Streptococcus mutans. Appl Environ Microbiol 2011; 77 (22): 8025–8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatum FM, Briggs RE. Construction of in-frame aroA deletion mutants of Mannheimia haemolytica, Pasteurella multocida, and Haemophilus somnus by using a new temperature-sensitive plasmid. Appl Environ Microbiol 2005; 71 (11): 7196–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Qi F, Merritt J. Cloning-independent plasmid construction for genetic studies in streptococci. J Microbiol Methods 2013; 94 (2): 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Nonaka H, Tsuge Y et al. New multiple-deletion method for the Corynebacterium glutamicum genome, using a mutant lox sequence. Appl Environ Microbiol 2005; 71 (12): 8472–8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zituni D, Schütt-Gerowitt H, Kopp M et al. The growth of Staphylococcus aureus and Escherichiacoli in low-direct current electric fields. Int J Oral Sci 2014; 6 (1): 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Zhang K, Zhou X et al. Involvement of gshAB in the interspecies competition within oral biofilm. J Dent Res 2013; 92 (9): 819–824. [DOI] [PubMed] [Google Scholar]
- Paster BJ, Bartoszyk IM, Dewhirst PE. Identification of oral streptococci using PCR-based, reverse-capture, checkerboard hybridization. Methods Cell Sci 1998; 20 (1): 223–231. [Google Scholar]
- Radkov AD, Moe LA. Bacterial synthesis of D-amino acids. Appl Microbiol Biotechnol 2014; 98 (12): 5363–5374. [DOI] [PubMed] [Google Scholar]
- Heaton MP, Johnston RB, Thompson TL. Controlled lysis of bacterial cells utilizing mutants with defective synthesis of D-alanine. Can J Microbiol 1988; 134 (3): 256–261. [DOI] [PubMed] [Google Scholar]
- Palumbo E, Favier CF, Deghorain M et al. Knockout of the alanine racemase gene in Lactobacillus plantarum results in septation defects and cell wall perforation. FEMS Microbiol Lett 2004; 233 (1): 131–138. [DOI] [PubMed] [Google Scholar]
- Heidrich C, Templin MF, Ursinus et al. Involvement of N-acetylmuramyl-L L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol 2001; 41 (1): 167–178. [DOI] [PubMed] [Google Scholar]
- Barreteau H, Kovac A, Boniface et al. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol Rev 2008; 32 (2): 168–207. [DOI] [PubMed] [Google Scholar]
- Steen A, Palumbo E, Deghorain M et al. Autolysis of Lactococcuslactis is increased upon D-alanine depletion of peptidoglycan and lipoteichoic acids. J Bacteriol 2005; 187 (1): 114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wecke J, Madela K, Fisher W. The absence of D-alanine from lipoteichoic acid and wall teichoic acid alters surface charge, enhances autolysis and increase susceptibility to methicillin in Bacillus subtilis. Microbiology 1997; 143(9): 2953–2960. [DOI] [PubMed] [Google Scholar]
- Nakao A, Imai S, Takano T. Transposon-mediated insertional mutagenesis of the D-alanyl-lipoteichoic acid (dlt) operon raises methicillin resistance in Staphylococcus aureus. Res Microbiol 2000; 151 (10): 823–829. [DOI] [PubMed] [Google Scholar]
- Double P, van Heijenoort J, Bohin JP et al. The murI gene of Escherichia coli is an essential gene that encodes a glutamate racemase activity. J Bacteriol 1993; 175 (10): 2970–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SL. Glutamate racemase as a target for drug discovery. Microb Biotechnol 2008; 1 (5): 345–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg EJ, Wijsman HJ, van Zaane D. Properties of a D-glutamic acid-requirng mutant of Escherichia coli. J Bacteriol 1973; 114 (2): 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Mortuza R, Milligan DL et al. Investigation of the essentiality of glutamate racemase in Mycobaterium smegmatis. J Bacteriol 2014; 196 (24): 4239–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert R, He J, Yarbrough DK et al. Targeted killing of Streptococcus mutans by a pheromone-guided "smart" antimicrobial peptide. Antimicrob Agents Chemother 2006; 50 (11): 3651–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Yarbrough DK, Kreth J et al. Systematic approach to optimizing specifically targeted antimicrobial peptides against Streptococcus mutans. Antimicrob Agents Chemother 2010; 54 (5): 2143–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, McLean JS, Yang Y et al. Precision-guided antimicrobial peptide as a targeted modulator of human microbial ecology. Proc Natl Acad Sci USA 2015; 112 (24): 7569–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]