Abstract
Lysogenization of Mycoplasma arthritidis with the MAV1 bacteriophage increases the virulence of the mycoplasma in rats. The MAV1 vir gene is one of only two constitutively transcribed phage genes in the lysogen. We show here that Vir is a lipoprotein and is located on the outer surface of the cell membrane. To investigate whether Vir is a virulence factor, the vir gene was cloned into the transposon vector Tn4001T and inserted in the genome of the nonlysogen strain 158. The virulence of the resulting transformants was no different from that of the parent strain. Interestingly, all vir-containing transformants were resistant to infection by MAV1. Vir had no effect on MAV1 adsorption. We conclude that Vir is not a virulence factor but functions to exclude superinfecting phage, possibly by blocking the injection of phage DNA into the bacterial cytoplasm.
Mycoplasmas are pathogens of humans and a variety of animals and plants (9). Among the members of the animal kingdom, primary sites of mycoplasmal infections are the membranes of the respiratory and urogenital tracts and joints (5). Mycoplasmas have some of the smallest genomes (as small as 0.58 Mb) in the prokaryotic kingdom (10). Consequently, their coding capacity is severely limited. The mycoplasmas lack a functional tricarboxylic acid cycle, a cytochrome-mediated electron transport chain system, and pathways for de novo biosynthesis of purines (9). They cannot synthesize cholesterol but require it for growth (6). Hence, mycoplasmas are dependent on their host for numerous nutrients and have a parasitic lifestyle.
Mycoplasma arthritidis is a natural rat pathogen, but disease can be induced experimentally in both rats and mice by intravenous injection of bacteria (21). Rats typically present with acute polyarthritis lasting 6 to 8 weeks, whereas disease in mice is a chronic arthritis with periods of remission and exacerbation that can persist for the life of the animal (4). Few virulence factors have been identified in mycoplasmas in general and in M. arthritidis in particular. The immunomodulating factor MAM (5), a soluble T-cell mitogen, and adhesins such as MAA1 and MAA2 (23) have been suggested as possible M. arthritidis virulence factors, but correlation between these proteins and virulence has not been established conclusively. All strains of M. arthritidis are thought to produce MAM, but many MAM-producing strains are of low virulence (4). Like MAM, the cytadhesins are found in strains of both high and low virulence (20). Therefore, other factors influencing disease must exist.
Unlike MAM and the MAA adhesins, the presence of MAV1 DNA in the bacterial chromosome strongly correlates with a virulent phenotype. In a previous study, the virulence of 20 M. arthritidis strains was examined (21). Ten of the strains were MAV1 lysogens and were highly virulent, whereas the other 10 strains were of low virulence and lacked the bacteriophage. For lysogenization, MAV1 DNA can insert into any of numerous sites in the mycoplasmal chromosome (17). A low-virulence strain, 158, acquired virulence upon lysogenization with MAV1, regardless of the particular site in which the prophage integrated (21). Therefore, the increase in pathogenicity upon MAV1 lysogenization is likely due to a phage-encoded virulence factor and not the activation or inactivation of bacterial genes flanking the inserted DNA.
MAV1 has a 16-kb genome composed of double-stranded DNA with 15 predicted genes. Only two MAV1 genes are constitutively transcribed in the lysogen (20). One is the imm gene that is thought to code for a transcriptional repressor. The other is the vir gene. The predicted Vir protein contains a lipoprotein signal sequence. MAV1 infectivity is resistant to chloroform and nonionic detergents (17), indicating that the virions are not lipid enveloped. Therefore, Vir should not be associated with the MAV1 virion and is most likely associated with the mycoplasma cell membrane. As a membrane protein, it is possible to envision a role in which Vir influences mycoplasma-host interactions and virulence.
We report here that Vir is indeed a lipoprotein on the surface of MAV1 lysogens. When inserted into the genome of a nonlysogenic strain, the vir gene did not increase the virulence of the mycoplasma but did render cells resistant to MAV1 infection. Vir had no effect on the adsorption of MAV1 to host cells, suggesting that it excludes superinfecting phage by another mechanism, such as interfering with phage DNA injection into the host.
MATERIALS AND METHODS
Mycoplasmal culture medium and strains.
Mycoplasmas were propagated in EA (agar) or EB (broth) medium as described previously (17, 21) unless otherwise indicated. M. arthritidis strain 158L3-1 is a highly virulent MAV1 lysogen of strain 158 (21). Strain 158-1 is a subclone of 158 (19). Strain 158-1L2 is a MAV1 lysogen of 158-1 that was obtained by stabbing a turbid plaque, assaying the cell mixture for CFU, and picking a well-separated colony to obtain a pure culture for further study. To confirm that 158-1L2 was a MAV1 lysogen, genomic DNA was isolated with the Easy-DNA kit (Invitrogen, Carlsbad, Calif.) and digested with Sau3A. The 16-kb MAV1 DNA genome is devoid of Sau3A sites (17). The M. arthritidis genome has numerous Sau3A sites, with the largest Sau3A fragment of strain 158 being only 4.4 kb (B. A. Methe and K. Dybvig, unpublished data). As expected for a MAV1 lysogen, 158-1L2 had a 16-kb Sau3A fragment that was lacking in strain 158-1. The presence of MAV1 DNA in the lysogen was confirmed by PCR amplification using the primers vir forward (5′-GCTAGGATCCGTAATGAGGAATTGGTTGC-3′) and vir reverse (5′-GTTGGATCCTGCGAAATCTTTTCAAGG-3′).
Insertion of the MAV1 vir gene into the chromosome of strains 158 and 158-1.
Plasmid pIVT contains the transposon Tn4001T, a previously described derivative of Tn4001 that contains the tetracycline resistance determinant tetM (8). No plasmid vectors are known to replicate in M. arthritidis, but genes such as vir can be incorporated into the M. arthritidis genome and expressed by using Tn4001T as a vector. To construct Tn4001T-vir, the MAV1 vir gene and its putative promoter Pv were amplified from the genomic DNA of strain 158L3-1 by using the vir forward and reverse primers described above and the proofreading polymerase Pwo (Roche Applied Science, Indianapolis, Ind.). Both primers contained BamHI restriction sites incorporated into their 5′ ends to facilitate cloning of the vir PCR product (1.3 kb) into the BamHI site of pIVT. The resulting plasmid, pIVT-vir, was maintained in Escherichia coli strain JM109 (selected at 100 μg of ampicillin/ml). The nucleotide sequence of the vir portion of pIVT-vir was determined to ensure that no errors had been introduced during PCR amplification and cloning.
Strains 158 and 158-1 were transformed with pIVT-vir DNA by using the polyethylene glycol-mediated transformation procedure described previously (19), with 3 μg of tetracycline per ml for antibiotic selection. Plasmid DNA was modified in vitro by incubation with the AluI DNA methyltransferase (New England BioLabs, Beverly, Mass.) prior to transformation to protect the plasmid from the M. arthritidis restriction enzyme MarI (19), which is an isoschizomer of AluI. Mycoplasmal transformants were confirmed by PCR to contain the vir gene. Strain 158 was also transformed with pIVT, generating transformants that had Tn4001T but not vir.
Identifying open reading frames (ORFs) disrupted by Tn4001T or Tn4001T-vir in M. arthritidis transformants.
A drawback to using a transposon as a cloning vector is that a gene in the recipient genome may be disrupted, creating a mutation that may have unexpected consequences. Therefore, the precise nucleotide position of the transposon in the mycoplasmal chromosome was determined for each transformant chosen for subsequent study. Mapping of the transposon's position in the genome was accomplished by amplifying one end of the transposon along with the adjacent host DNA by inverse PCR. Details of the mapping strategy and the primers used for inverse PCR and subsequent sequencing were as described elsewhere (16). Briefly, genomic DNA was digested with either NlaIII or TaiI, the restriction fragments circularized by incubation with T4 DNA ligase, the ligation product PCR amplified to obtain amplicons containing the transposon-mycoplasma junction, and the nucleotide sequence of the PCR product determined. Sequence analysis of the PCR product identified the junction between the transposon and the mycoplasma chromosome. A comparison of the junction sequence to the complete genome sequence of M. arthritidis strain 158L3-1 (B. A. Methe and K. Dybvig, unpublished) identified the nucleotide position of the transposon in the genome. The nucleotide position of the transposon in each transformant used in this study is provided in Table 1.
TABLE 1.
Genomic location of transposons in transformants of strains 158 and 158-1
| Transformant | Parent strain | Transposon | Genomic nucleotide positiona | Disrupted gene product, if any |
|---|---|---|---|---|
| TvirM1 | 158-1 | Tn4001T-vir | 803310 | FMN oxidoreductase |
| TvirM2 | 158-1 | Tn4001T-vir | 756252 | Sugar ABC transporter, ATP-binding protein RbsA-2 |
| TvirM3 | 158-1 | Tn4001T-vir | 726085 | S1 RNA binding domain protein Tex |
| Tvir3 | 158 | Tn4001T-vir | 66304 | Hypothetical protein (238 amino acids) |
| Tvir4 | 158 | Tn4001T-vir | 627511 | Intergenic |
| Tvir1 | 158 | Tn4001T-vir | 330744 | Conserved hypothetical protein |
| Tvir7 | 158 | Tn4001T-vir | 736742 | Conserved hypothetical protein |
| Tvir8 | 158 | Tn4001T-vir | 326554 | Serpentine membrane protein |
| Tn1 | 158 | Tn4001T | 804828 | Surface-located membrane protein |
| Tn4 | 158 | Tn4001T | 568018 | ATP-dependent protease La (Lon) |
| Tn5 | 158 | Tn4001T | 625418 | Hypothetical protein (388 amino acids) |
| Tn6 | 158 | Tn4001T | 610640 | Membrane protein |
The nucleotide positions correspond to the completed M. arthritidis genome sequence as determined by collaboration between The Institute for Genomic Research (Rockville, Md.) and B. A. Methe and K. Dybvig (unpublished).
Bacteriophage preparation.
Stocks of MAV1 were prepared as described previously (17) by incubating 105 PFU of MAV1 with 500 μl of log-phase 158 cells for 45 min, adding the mixture to top agar (41°C), and pouring the contents onto a prewarmed EA plate. After incubation at 37°C for 2 days to allow nearly confluent plaques to develop, the top agar overlay was collected and combined with 2 ml of EB containing 100 μl of chloroform, which served to lyse the mycoplasmas. Debris was removed by centrifugation (8,000 × g for 10 min), and the supernatant was filtered through a 0.2-μm-pore-size Acrodisc syringe filter (Gelman Sciences, Ann Arbor, Mich.) and stored at 4°C.
Megaplaque assay.
The susceptibility of mycoplasma strains to infection with MAV1 was determined by a megaplaque assay (18). Host cells (20 μl of a culture in logarithmic growth phase) were added to 1.5 ml of top agar at 41°C, mixed, and poured onto a prewarmed EA plate. Immediately thereafter, 10 μl of MAV1 phage stock (3 × 107 PFU/ml) was spotted onto the center of the plate. The plates were incubated at 37°C for 24 to 48 h. The presence of a zone of clearing at the site of the bacteriophage spotting indicates MAV1 infection. The plates were stained with Dienes (Becton, Dickinson, and Co., Franklin Lakes, N.J.) to enhance photographic imaging.
Phage adsorption assay.
Host cells (6 ml of culture) of either 158-1 or 158-1L2 were grown in a modified EB medium containing 20% GG-free horse serum (Gibco, Grand Island, N.Y.) in place of whole horse serum. Although adsorption occurred in the presence of whole horse serum, a greater percentage of MAV1 bound to cells in medium with GG-free serum, suggesting that immunoglobulin G antibodies interfere with adsorption. The cells were harvested by centrifugation at 9,000 × g for 5 min, and the cell pellet was suspended in 6 ml of fresh GG-free EB medium. Analysis of a 10-μl sample revealed the culture contained 1.2 × 109 to 2.6 × 109 CFU/ml, depending on the experiment. MAV1 (1.5 × 107 PFU) was added to the culture, creating a multiplicity of infection of approximately 0.0015. Samples (1 ml) were removed after incubation at 37°C for 0 and 45 min and were centrifuged (5 min at 12,000 × g) to harvest cells and adsorbed phage. The supernatant was assayed for PFU on lawns of strain 158-1 as described previously (18). The fraction of unabsorbed phage was calculated as the ratio of PFU obtained after 45 min of incubation with host cells to the initial PFU at time zero. Results are averaged from three independent experiments.
Generation of polyclonal antisera to Vir.
A synthetic peptide (CGTDRKDASDWIHDSYKDK) corresponding to a region of Vir predicted to be antigenic and surface exposed was synthesized and conjugated to keyhole limpet hemocyanin. The peptide was used to immunize two rabbits (A2031 and A2032) with four injections per rabbit to generate polyclonal Vir-specific antibodies. Peptide synthesis and antisera production were performed by AnaSpec (San Jose, Calif.) and included prebleed serum from each animal. A 1:1,500 dilution of the final bleed from rabbit A2032 was used for Western analysis. At this concentration, the prebleed serum did not detect Vir but did detect other M. arthritidis proteins that were absent in conjugate controls in which the primary antibody was omitted.
Metabolic labeling and TX-114 fractionation of M. arthritidis.
M. arthritidis strains 158L3-1 and 158 were labeled with [3H]palmitic acid as previously described (7). EB (20 ml) was inoculated with 20 μl of stock culture and was incubated at 37°C for 48 h. Mycoplasmas were recovered by centrifugation, suspended in 2 ml of EB containing 1 mCi of [3H]palmitic acid (Perkin-Elmer Life Sciences, Inc., Boston, Mass.), and incubated an additional 18 h at 37°C. Cells were washed once in phosphate-buffered saline and fractionated into aqueous and hydrophobic phases by Triton X-114 (TX-114; Sigma) extraction (7). A sample of each fraction was subjected to sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE), transferred to a nitrocellulose membrane, and reacted with the anti-Vir polyclonal rabbit serum. The blot was also exposed to Biomax MS film (Kodak, Rochester, N.Y.) for 2 weeks to visualize labeled proteins.
Trypsin treatment of intact cells.
A trypsin treatment of intact cells was adapted from that of Duffy et al. (7). A 20-ml culture of 158L3-1 in late logarithmic growth phase was centrifuged at 12,000 × g for 30 min and was washed three times in 10 ml of Tris-saline buffer (50 mM Tris-HCl, 145 mM NaCl, pH 7.4). Cells were suspended in 600 μl of trypsin digest buffer (1 mM CaCl2, 50 mM Tris-HCl, pH 7.6), divided into six tubes, and incubated at 37°C for 45 min with trypsin (Sigma) in a dilution series. The final enzyme concentrations were 0, 7.5, 15, 30, 60, and 120 μg/ml. After 45 min, samples were harvested by centrifugation and suspended in 15 μl of 2× SDS sample buffer. Samples were subjected to SDS-PAGE and Western analysis as described above.
Induction and assessment of arthritis in rats.
Male Lewis (LEW) rats (Charles River Laboratories Inc., Wilmington, Mass.) weighing an average of 180 g were divided into groups of eight for infection experiments and a control group of five for mock infection. Each rat was injected intravenously in the caudal vein with 200 μl of 109 CFU of the appropriate bacterial strain or 200 μl of EB for control animals. The experiment was repeated, yielding similar results.
The method to determine the numerical arthritis scores was similar to those described previously (3, 21). Peripheral joints were measured with a caliper and were assigned a score between 0 (no swelling) and 4 (>40 and >70% increase in diameter of the ankle and wrist joints, respectively). Interphalangeal joints and tail vertebrae were assigned a score between 0 and 1.5 based on a visual assessment of swelling. The total arthritic score for each rat was determined, and the average numeric arthritis score per rat was calculated for each group. To assess mobility, animals were assigned the following scores: 0 if they walked normally; 1 if they walked awkwardly; 2 if they refused to walk on one limb; 3 if they failed to walk and crawled; 4 if they were unable to crawl (3).
Statistical analysis.
Mobility, numerical arthritic scores, and weight data were analyzed with the software SigmaStat version 2.03 (SPSS Inc.). Data from groups of mycoplasma-infected rats were compared by two-way analysis of variance, with the strain of mycoplasma and time being the two variables. Differences were significant only for values of P < 0.05.
Transposon stability assay.
Upon completion of the 2-week rat experiment, lesions from infected areas were lanced and the exudates were collected. All material collected from an animal was pooled and placed in 700 μl of EB. Material from three rats infected with strain Tvir3, two rats infected with Tvir4, and five rats infected with Tn5 were analyzed. The material was disrupted by vigorous mixing with a vortex and was passed through a 0.45-μm-pore-size filter. Serial dilutions were assayed for CFU on EA plates with and without tetracycline selection at 3 μg per ml. The percentage of cells containing the transposon was determined by dividing the number of tetracycline-resistant colonies by the total number of colonies obtained with no antibiotic. Twenty colonies grown in the presence of antibiotic from each strain were further analyzed by inverse PCR as described above to determine whether the transposon resided in its original insertion site.
RESULTS
Vir is a lipoprotein.
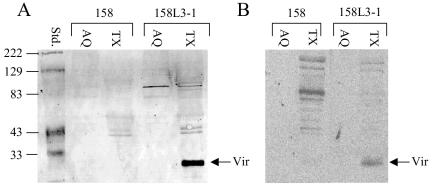
To determine whether Vir is indeed a lipoprotein, strains 158 and 158L3-1 were labeled with [3H]palmitate followed by fractionation of the cell lysate with TX-114. Generally, proteins associated with the membrane partition to the detergent phase, while cytoplasmic proteins segregated to the aqueous phase (2). The vir gene encodes a predicted protein of 25.4 kDa. Western blots of the detergent and aqueous fractions of 158 and 158L3-1 were reacted with polyclonal Vir antisera. A 25-kDa protein was detected in the detergent phase of the 158L3-1 extract, demonstrating that Vir is a hydrophobic molecule and is likely associated with the mycoplasma cell membrane (Fig. 1A). The protein was absent in extracts from 158 (Fig. 1A) and from transformants of 158 containing Tn4001T without the vir gene (data not shown), demonstrating that the protein is phage encoded. The immunoblot was exposed to radiographic film to identify radiolabeled lipoproteins. The protein recognized by the Vir antibody was labeled with [3H]palmitate, establishing that it is a lipoprotein (Fig. 1B). No other lipoproteins comigrated with Vir, as indicated by the absence of a radiolabeled protein of similar size in the detergent fraction of 158 lysates.
FIG. 1.
(A) Western blot of TX-114-extracted proteins reacted with polyclonal anti-Vir rabbit antibody. A protein present in 158L3-1 (and vir transformants; data not shown) corresponds to the predicted size of Vir, which is 25.4 kDa. Its presence in the detergent phase after fractionation shows that Vir is associated with the membrane. (B) Autoradiogram of [3H]palmitate-labeled, TX-114-extracted 158L3-1 proteins. The [3H]palmitate labeling of Vir indicates that it is a lipoprotein. Standard values are measured in kilodaltons, TX represents the triton phase, and AQ represents the aqueous phase.
Vir is exposed on the outer surface of the membrane.
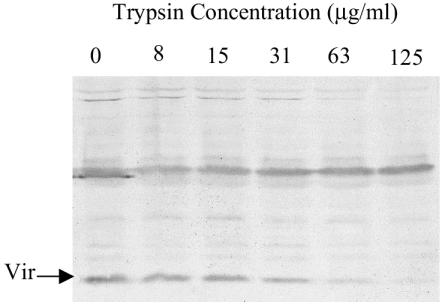
Intact 158L3-1 cells were treated with various concentrations of trypsin for 45 min, and proteins were subsequently examined by Western analysis. The amount of detectable Vir protein decreased as the concentration of trypsin increased, suggesting that Vir is exposed on the outer surface of the membrane (Fig. 2). A sample of each cell mixture was assayed for CFU after trypsin digestion. No loss of CFU was evident, even in samples with the highest concentration of trypsin. Therefore, the degradation of Vir was not the result of a generalized proteolysis following cell death.
FIG. 2.
Trypsin treatment of intact 158L3-1 cells. 158L3-1 cells were digested with increasing concentrations of trypsin as labeled and were subjected to Western analysis with Vir antisera.
Vir is produced by transformants containing Tn4001T-vir.
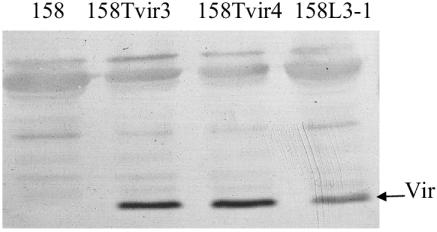
Transformants of strain 158 that contain Tn4001T-vir (158Tvir3 and -4) were examined to determine whether Vir was produced. An immunoblot of a gel loaded with equal amounts of CFU from 158L3-1, 158Tvir3, and 158Tvir4 was reacted with Vir antisera. All of the vir transformants expressed approximately the same amount of or slightly more Vir protein than the lysogen 158L3-1 (Fig. 3).
FIG. 3.
Immunoblot loaded with equal amounts of protein from 158L3-1, 158Tvir3, and 158Tvir4 reacted with Vir antisera.
Vir is not a virulence factor.
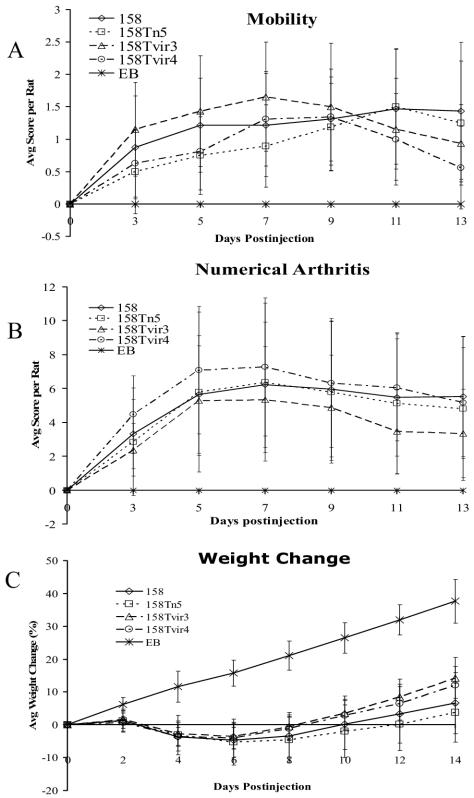
Previous studies showed that lysogenization of strain 158 by MAV1 resulted in a profound increase in virulence (21). To investigate whether vir was responsible for this increase, the arthritogenicity of transformants of 158 that contained Tn4001T-vir was examined. There were no significant differences in the mobility (Fig. 4A) and degree of arthritogenicity (Fig. 4B) between each of the infected animal groups. Throughout the 2-week course of infection, rats infected with the vir-containing transformants exhibited amounts of weight loss similar to those of animals infected with either 158 or transformants of 158 that contain Tn4001 without vir (Fig. 4C). If anything, statistical analysis of the data indicates that vir attenuated the weight loss. According to a two-way analysis of variance of the weight data, 158Tn5 is more virulent than both 158Tvir3 and 158Tvir4 (P < 0.001 and P = 0.003, respectively). Also, 158 is more virulent than 158Tvir3 (P = 0.005). Based on the criteria of mobility, arthritic scores, and weight loss, Vir did not enhance the virulence of the mycoplasma.
FIG. 4.
Development of arthritis in rats injected with M. arthritidis strains 158, 158Tn5, 158Tvir3, and 158Tvir4 and with EB as a control. Mobility (A), average arthritic score (B), and weight change (C) per rat are shown over a 2-week period. Each point represents the mean measurements taken from eight rats (five for EB) with standard deviations.
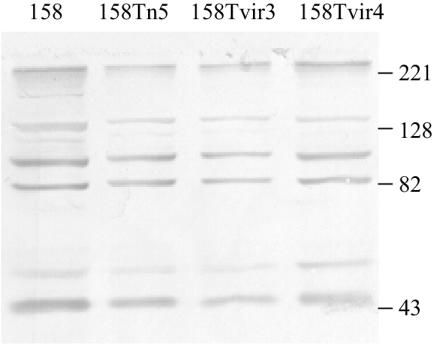
Mycoplasmas are notorious for their repertoire of size- and phase-variable proteins. It is possible that the transformants had a protein expression profile different from that of the parent strain, making them intrinsically less virulent regardless of whether Vir was produced. An immunoblot of proteins from strains 158, 158Tn5, 158Tvir3, and 158Tvir4 was reacted with serum collected from rats that had been infected with 158L3-1. No obvious antigenic differences were detected (Fig. 5). To determine whether transformants had an obvious growth defect, the colony diameters of 158, 158Tn5, 158Tvir3, and 158Tvir4 were compared after 1 week of growth. There were no significant differences in size.
FIG. 5.
Immunoblot of M. arthritidis strains used in animal experiments, reacted with anti-158L3-1 rat polyclonal sera.
One drawback of using transposons as a vector is the disruption of chromosomal genes by transposon insertion. The transposon in 158Tvir4 is inserted at an intergenic site between two divergent ORFs, 226 bp upstream of the beginning of ORF00717 (encoding a hypothetical protein of 518 amino acids) and 126 bp upstream of the start of ORF00718 (encoding ribosomal protein S4). It is unlikely that the transposon interferes with the expression of these adjacent genes. The transposons in 158Tvir3 and 158Tn5 disrupt hypothetical proteins (Table 1). Because 158Tvir3 caused a severity of arthritis similar to that caused by 158Tvir4, and because 158Tn5 had the same virulence as 158, the gene disruptions did not attenuate virulence.
The presence and stability of the transposon in vivo was also assessed. At the conclusion of the experiment, isolates were collected from each group of animals and were examined for their resistance to tetracycline. Essentially 100% of the isolates collected from animals infected with 158Tvir4, 158Tvir3, and 158Tn5 retained the transposon. Twenty tetracycline-resistant isolates from each group were subjected to inverse PCR to determine whether the transposon resided in its original insertion site. Ninety-five percent of the 158Tvir4 and 158Tn5 isolates and 100% of the 158Tvir3 isolates retained the transposon in its original location.
The MAV1 Vir protein confers resistance to MAV1 infection.
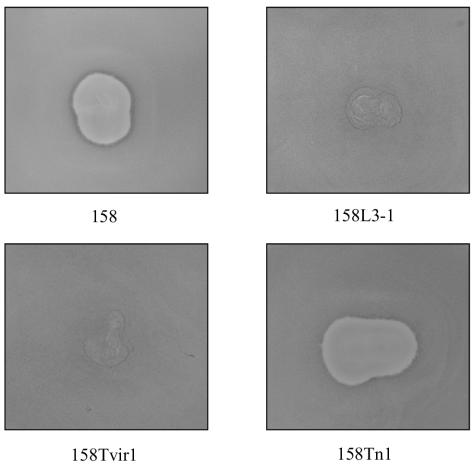
Five independent transformants of 158-1 and 158 containing vir (Table 1) were assayed for susceptibility to infection with MAV1 and were found to be resistant (Fig. 6). Because each transformant has a disruption in a different site within the chromosome, the resistance phenotype is due to the presence of the vir gene and not the disruption of the MAV1 receptor or other host proteins necessary for a successful viral infection. Controls consisting of four 158 transformants that contained the Tn4001T vector without vir remained susceptible to MAV1 infection.
FIG. 6.
Superinfection resistance to MAV1 infection. Strain 158 is susceptible to MAV1 infection, indicated by the large zone of clearing caused by phage-induced cell lysis. 158L3-1, a MAV1 lysogen of 158, is resistant to MAV1 infection, signified by confluent lawn growth. Five independent transformants containing the MAV1 vir gene (only 158Tvir1 is shown) were also resistant to MAV1. Transformants containing a transposon vector-only control (e.g., 158Tn1) were susceptible to MAV1 infection.
Cells expressing Vir allow phage adsorption.
To determine whether Vir confers phage resistance by interfering with adsorption of phage to host cells, adsorption assays were performed. MAV1 adsorbed to the lysogen 158-1L2 and the nonlysogen 158-1 with similar efficiency. After incubation of phage with host cells for 45 min, 88% ± 2% of the phage adsorbed to strain 158-1 and 76% ± 5% adsorbed to 158-1L2. Therefore, Vir does not block phage adsorption.
DISCUSSION
We began these experiments with the preconceived notion that Vir would enhance the virulence of M. arthritidis. The literature strongly indicates that the lysogenization of strain 158 with MAV1 increases the severity of experimentally induced arthritis in LEW rats, and the vir gene seemed the most likely candidate to be a virulence determinant because it was predicted to encode a lipoprotein. Indeed, we show in this study that Vir is a lipoprotein and is exposed on the outside surface of the mycoplasma. However, transformation of 158 with transposon Tn4001T-vir did not increase virulence. One possible explanation for the failure of the vir gene to act as a virulence determinant would be that it was not expressed. However, we show that the Vir protein was produced in the transformants and that the protein was functional in that resistance to phage infection was an acquired phenotype. It is possible that Vir has a minor role in pathogenesis that was not observed in our experiments, but the major function of Vir is clear. Vir serves to protect lysogens from superinfection by MAV1, making this the first example of a phage exclusion system in mycoplasma. In light of its function, the vir gene should perhaps be renamed to sie (superinfection exclusion gene).
The initial step of the infectious process for bacteriophages is the attachment of the phage virion to a specific receptor located on a bacterial surface, followed by the injection of the phage genome through the membrane and into the cytoplasm. Some bacteriophages possess superinfection exclusion systems that can abort these initial steps. Unlike immunity systems that use soluble repressors capable of preventing phage replication, phage exclusion systems do not participate in preserving the lysogenic state. One possible mechanism for phage exclusion is interference with phage adsorption, as has been described for the Cor proteins of bacteriophages N15 and φ80 (22). In this model, the phage protein integrates into the membrane where it can associate with the phage receptor, masking residues important for phage binding or inducing a conformational change in the receptor. Phage adsorption assays show that MAV1 adsorbs to MAV1 lysogens. Thus, Vir does not appear to function in this manner.
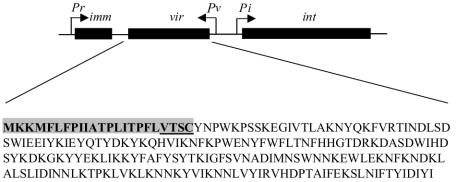
We favor a model in which Vir permits phage adsorption but prevents phage DNA penetration similar to the Sie2009 protein of lactococcal phage Tuc2009 (14). The vir gene is located between the putative MAV1 transcriptional repressor and the integrase gene, forming a lysogeny module with a gene organization similar to that of Tuc2009 and other lactococcal phages (Fig. 7). A gene within the lysogeny module at the position of vir is usually associated with resistance to superinfection in lactococcal phages (14), and it seems this generalization applies to MAV1.
FIG. 7.
Diagram of the MAV1 lysogeny module. The putative promoters are indicated by the letter P and an arrow showing direction of transcription. The immunity (imm), vir, and integrase (int) genes are represented by black bars. The deduced amino acid sequence of Vir is shown with the lipoprotein signal sequence highlighted and the lipobox underlined.
Analogous phage-encoded integral membrane proteins have been described for the SieA protein of the Salmonella enterica serovar Typhimurium phage P22 (11) and the Imm protein of the E. coli bacteriophage T4 (12). However, the additional outer membrane and a periplasmic space may alter the means by which phage exclusion is carried out in gram-negative bacteria. The glo gene of the Vibrio cholerae phage K139 (15) and the sim gene of the E. coli bacteriophage P1 (13) encode soluble proteins localized in the periplasmic space. Phage adsorb to the cell and inject their DNA into the periplasm. The phage-encoded proteins are thought to prevent the translocation of phage DNA into the host cytoplasm. The mechanism by which blockage of DNA injection occurs is poorly understood, and one can only speculate as to ways by which Vir may function. It is possible that Vir interacts with the MAV1 receptor to form a molecular plug that prevents the escape of phage DNA. Perhaps Vir interacts with receptor-bound phage and inactivates the DNA injection apparatus.
More exotic phage exclusion systems involving multiple phage-encoded proteins have also been described (1), but it is unlikely that a phage with a genome as small as 16 kb would have an elaborate exclusion system. Due to its extracellular location, it is also unlikely that Vir functions as a nuclease to degrade incoming DNA or interferes with any of the intracellular stages of infection, such as synthesis of phage proteins or transcription and replication of phage DNA.
Genomic sequencing revealed φMFV1, a putative Mycoplasma fermentans bacteriophage that is closely related to MAV1 (15a). MAV1 and MFV1 have the same general genetic organization, and most of the MFV1-predicted proteins share significant homology with MAV1 proteins. MFV1 lacks vir, but in its place in the phage genome is mem, which encodes a predicted transmembrane protein. It seems likely that Mem is part of a lysogeny module and has a role in phage exclusion similar to that of Vir, but this idea will be difficult to test until procedures to propagate MFV1 in a mycoplasmal host have been established.
Acknowledgments
This work was supported by Public Health Service grant AR44252 from the National Institutes of Health.
We thank Portia Caldwell for technical assistance.
Preliminary sequence data were obtained from The Institute for Genomic Research through their website at http://www.tigr.org.
REFERENCES
- 1.Allison, G. E., and T. R. Klaenhammer. 1998. Phage resistance mechanisms in lactic acid bacteria. Int. Dairy J. 8:207-226. [Google Scholar]
- 2.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 3.Butler, S. H., J. Weil-Fugazza, F. Godefroy, and J. M. Besson. 1985. Reduction of arthritis and pain behaviour following chronic administration of amitriptyline or imipramine in rats with adjuvant-induced arthritis. Pain 23:159-175. [DOI] [PubMed] [Google Scholar]
- 4.Cole, B. C. 1991. The immunobiology of Mycoplasma arthritidis and its superantigen MAM. Curr. Top. Microbiol. Immunol. 174:107-119. [DOI] [PubMed] [Google Scholar]
- 5.Cole, B. C., and C. L. Atkin. 1991. The Mycoplasma arthritidis T-cell mitogen, MAM: a model superantigen. Immunol. Today 12:271-276. [DOI] [PubMed] [Google Scholar]
- 6.Dahl, J. 1993. The role of cholesterol in mycoplasma membranes. Subcell. Biochem. 20:167-188. [DOI] [PubMed] [Google Scholar]
- 7.Duffy, D. F., A. H. Noormohammadi, N. Basseggio, G. F. Browning, and P. F. Markham. 1998. Immunological and biochemical characterization of membrane proteins, p. 279-298. In R. Nicholas (ed.), Methods in molecular biology, vol. 104. Humana Press, Inc., Totowa, N.J. [DOI] [PubMed] [Google Scholar]
- 8.Dybvig, K., C. T. French, and L. L. Voelker. 2000. Construction and use of derivatives of transposon Tn4001 that function in Mycoplasma pulmonis and Mycoplasma arthritidis. J. Bacteriol. 182:4343-4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dybvig, K., and L. L. Voelker. 1996. Molecular biology of mycoplasmas. Annu. Rev. Microbiol. 50:25-57. [DOI] [PubMed] [Google Scholar]
- 10.Fraser, C. F., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, J. L. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J.-F. Tomb, B. A. Dougherty, K. F. Bott, P.-C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 11.Hofer, B., M. Ruge, and B. Dreiseikelmann. 1995. The superinfection exclusion gene (sieA) of bacteriophage P22: identification and overexpression of the gene and localization of the gene product. J. Bacteriol. 177:3080-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu, M. J., Y. D. Stierhof, and U. Henning. 1993. Location and unusual membrane topology of the immunity protein of the Escherichia coli phage T4. J. Virol. 67:4905-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maillou, J., and B. Dreiseikelmann. 1990. The sim gene of Escherichia coli phage P1: nucleotide sequence and purification of the processed protein. Virology 175:500-507. [DOI] [PubMed] [Google Scholar]
- 14.McGrath, S., G. F. Fitzgerald, and D. van Sinderen. 2002. Identification and characterization of phage-resistance genes in temperate lactococcal bacteriophages. Mol. Microbiol. 43:509-520. [DOI] [PubMed] [Google Scholar]
- 15.Nesper, J., J. Blass, M. Fountoulakis, and J. Reidl. 1999. Characterization of the major control region of Vibrio cholerae bacteriophage K139: immunity, exclusion, and integration. J. Bacteriol. 181:2902-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Roske, K., M. J. Calcutt, and K. S. Wise. 2004. The Mycoplasma fermentans prophage φMFV1: genome organization, mobility and variable expression of an encoded surface protein. Mol. Microbiol. 52:1703-1720. [DOI] [PubMed] [Google Scholar]
- 16.Teachman, A. M., C. T. French, H. Yu, W. L. Simmons, and K. Dybvig. 2002. Gene transfer in Mycoplasma pulmonis. J. Bacteriol. 184:947-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voelker, L. L., and K. Dybvig. 1998. Characterization of the lysogenic bacteriophage MAV1 from Mycoplasma arthritidis. J. Bacteriol. 180:5928-5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voelker, L. L., and K. Dybvig. 1998. Demonstration of extrachromosomal elements. Methods Mol. Biol. 104:239-246. [DOI] [PubMed] [Google Scholar]
- 19.Voelker, L. L., and K. Dybvig. 1996. Gene transfer in Mycoplasma arthritidis: transformation, conjugal transfer of Tn916, and evidence for a restriction system recognizing AGCT. J. Bacteriol. 178:6078-6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voelker, L. L., and K. Dybvig. 1999. Sequence analysis of the Mycoplasma arthritidis bacteriophage MAV1 genome identifies the putative virulence factor. Gene 233:101-107. [DOI] [PubMed] [Google Scholar]
- 21.Voelker, L. L., K. E. Weaver, L. J. Ehle, and L. R. Washburn. 1995. Association of lysogenic bacteriophage MAV1 with virulence of Mycoplasma arthritidis. Infect. Immun. 63:4016-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vostrov, A. A., O. A. Vostrukhina, A. N. Svarchevsky, and V. N. Rybchin. 1996. Proteins responsible for lysogenic conversion caused by coliphages N15 and phi80 are highly homologous. J. Bacteriol. 178:1484-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Washburn, L. R., and E. J. Weaver. 1997. Protection of rats against Mycoplasma arthritidis-induced arthritis by active and passive immunizations with two surface antigens. Clin. Diagn. Lab. Immunol. 4:321-327. [DOI] [PMC free article] [PubMed] [Google Scholar]