Abstract
Motile prokaryotes use a sensory circuit for control of the motility apparatus in which ligand-responsive chemoreceptors regulate phosphoryl flux through a modified two-component signal transduction system. The chemoreceptors exhibit a modular architecture, comprising an N-terminal sensory module, a C-terminal output module, and a HAMP domain that connects the N- and C-terminal modules and transmits sensory information between them via an unknown mechanism. The sensory circuits mediated by two chemoreceptors of Bacillus subtilis have been studied in detail. McpB is known to regulate chemotaxis towards the attractant asparagine in a CheD-independent manner, whereas McpC requires CheD to regulate chemotaxis towards the attractant proline. Although CheD is a phylogenetically widespread chemotaxis protein, there exists only a limited understanding of its function. We have constructed chimeras between McpB and McpC to probe the role of CheD in facilitating sensory transduction by McpC. We found that McpC can be converted to a CheD-independent receptor by the replacement of one-half of its HAMP domain with the corresponding sequence from McpB, suggesting that McpC HAMP domain function is complex and may require intermolecular interactions with the CheD protein. When considered in combination with the previous observation that CheD catalyzes covalent modification of the C-terminal modules of B. subtilis receptors, these results suggest that CheD may interact with chemoreceptors at multiple, functionally distinct sites.
Motile prokaryotes regulate chemotactic responses to environmental stimuli by using a complex sensory circuit based on a two-component signal transduction system (for reviews, see references 4 and 17). The motility apparatus is controlled by modulating the flux of phosphoryl groups through the histidine kinase CheA and its cognate response regulator CheY. Environmentally responsive sensory input to CheA is initiated by receptors (methyl-accepting chemotaxis proteins or transducers) that undergo a conformational change upon ligand binding (5). For a given chemotactic organism, the repertoire and specificities of its receptors determine the chemoeffectors to which that organism responds.
Comparative sequence analysis of more than 120 transducers revealed that they exhibit a modular architecture (20). The receptors possess distinct structural and functional modules, including (i) a variable N-terminal module, responsible for sensing environmental stimuli, and (ii) a highly conserved C-terminal module, which receives sensory signals from the N-terminal module and transmits those signals to the downstream chemotactic machinery. The C-terminal module also mediates adaptation to the imposed stimulus via changes in the extent of its posttranslational modification. Because of the modular nature of the chemoreceptors, functional chimeric receptors can be readily constructed by exchanging N-terminal modules from different receptors (6, 11, 12, 15, 18) to yield chimeras that usually, but not always, respond to identical environmental signals as the parental receptor from which the N-terminal module was derived.
Intramolecular transmission of sensory information from the receptor N-terminal ligand binding module to the conserved C-terminal module involves a linker element now known as the HAMP domain (found in histidine kinases, adenylate cyclases, methyl-accepting chemotaxis proteins, and phosphatases) (3, 19, 20). The HAMP structural element is predicted to form two short amphipathic helices (denoted AS-1 and AS-2 for amphipathic sequences 1 and 2, respectively) (19) separated by a short connector of undefined structure (depicted in Fig. 1). The precise boundaries of the amphipathic helices are unknown. Although HAMP domains from different receptors often exhibit considerable variation at the level of primary sequence, the sequences comprising the amphipathic helices typically display a conserved, repeating pattern of seven residues, known as a heptad repeat, that is characteristic of helices known to form coiled coils (16, 19). HAMP domains from diverse receptors appear to use a similar mechanism to mediate the transmission of ligand-induced conformational signals between the N-terminal and C-terminal modules. This suggestion is based on the results of several studies in which the N- and C-terminal modules of heterologous receptors were exchanged to create chimeric receptors that were fully functional (6, 11, 12, 15, 18). It is worth noting that all ligand-responsive chimeras constructed thus far contain a HAMP domain derived entirely from a single parental receptor. Chimeras in which AS-1 was derived from the HAMP domain of one protein and AS-2 was derived from the HAMP domain of a heterologous protein did not respond to the presence of ligand (1), suggesting that sequence-specific contacts within a given HAMP domain are important for proper function. However, the precise molecular mechanism by which the HAMP domain passes sensory information between the receptor modules remains unknown.
FIG. 1.
Schematic of the HAMP regions of wild-type and chimeric receptors used in this study. The single-letter amino acid sequences of the relevant receptor portions (the C-terminal ends of transmembrane helix 2 [TM-2] and the HAMP domains) are shown. At the ends of the appropriate lines, residue numbers from full-length McpB or full-length McpC are indicated. Sequences derived from McpB are in plain type, whereas sequences derived from McpC are presented on a gray background. The positions of residues located in helices AS-1 and AS-2 of the HAMP domains (with arbitrarily designated endpoints) are indicated below the receptor sequences, according to a previously described numbering scheme (2).
Sensory responses mediated by two chemoreceptors of Bacillus subtilis have been characterized in detail. McpB is the sole receptor for the amino acid attractant asparagine (8), whereas McpC is the sole receptor for the amino acid attractant proline (12, 14) and for carbohydrate attractants (7, 12). Although most elements of the sensory pathways connecting ligand binding by these receptors to flagellar control are identical (for example, both receptors signal through the kinase-regulator pair CheA/CheY), there exists at least one difference in the respective sensory pathways. In particular, McpC exhibits an absolute requirement for the cheD gene product in order to transmit ligand-induced signals to the CheA kinase, whereas McpB is capable of doing so in the absence of CheD (10), albeit less efficiently than in the presence of CheD. This receptor-specific requirement for CheD function suggests that the mechanism of intramolecular signal transduction by McpC differs from that of McpB in some way.
Recent studies focusing on CheD have begun to elucidate its role in chemotactic sensory transduction. CheD homologs have been discovered within chemotaxis-like operons of many bacterial and archaeal species (10), suggesting that CheD plays an important role in chemoreceptor-mediated sensory transduction for many organisms. Because the only homologs of CheD in the publicly available databases are proteins for which no function has been described, no function for CheD can be inferred. However, CheD was recently shown to be a chemoreceptor-glutamine deamidase, catalyzing modification at specific sites within the C-terminal modules of several B. subtilis receptors (13). In light of this enzymatic activity, we hypothesized that the McpC-specific requirement for CheD was a result of CheD-mediated deamidation of glutaminyl residues in the C-terminal module of McpC.
To test this hypothesis and to probe the role of CheD in modulating receptor function, we constructed a set of chimeric receptors, by exchanging corresponding receptor modules from McpB and McpC (Fig. 1), and assessed the ability of these chimeras to support chemotaxis in the presence and absence of CheD. The plasmids pAIN402 and pAIN450 carry cloned versions of the genes for mcpC and mcpB, respectively, each with a BglII restriction site inserted into a conserved receptor sequence in such a manner that the amino acid sequence of the translated product is not altered from the wild type (12). Using pAIN402 and pAIN450 as sources of the genes for mcpC and mcpB, we created mcpC317B and mcpB324C by subcloning the BglII/NotI fragment carrying the C-terminal module from a given receptor into the BglII/NotI sites of the reciprocal plasmid, thereby replacing the original wild-type C-terminal module. In all cases where a chimera is designated with a number (e.g., McpB324C), the number represents the final residue of the initial receptor indicated (McpB in this example) that is present in the chimera, with the remainder of the chimera contributed by sequences from the second receptor indicated (McpC in this example). We note that the fusion point between receptors in the chimeras described above occurs at a conserved sequence in the connector region of the HAMP domain, between AS-1 and AS-2 (shown in Fig. 1). The mcpCBC chimera was created in two steps: first, a preliminary chimera (mcpC296B) was constructed using a method identical to that previously described (12), with pAIN402 and pAIN450 as starting materials. In McpC296B, the entire predicted cytoplasmic sequence of McpC (including the HAMP domain and C-terminal module) was replaced by the corresponding sequence from McpB. In the second step of mcpCBC construction, the BglII/NotI fragment of mcpC296B carrying the McpB C-terminal module was replaced with a BglII/NotI fragment carrying the C-terminal module of McpC. All chimeras were subcloned and integrated in single copy, according to a previously described method (21), into the chromosomes of either a B. subtilis strain carrying mutations in all 10 known chemoreceptors (OI3535 [9], referred to as Δ10) or an isogenic strain harboring a replacement of the cheD gene with a chloramphenicol resistance cassette (OI3628 [13], referred to as Δ10 cheD). Expression of all chimeras was analyzed by using antisera raised against the cytoplasmic portion of McpB or McpC, as previously described (13), and we found that all chimeras were expressed at levels similar to those for the wild-type receptors (data not shown).
Intramolecular signal transduction by the chimeric chemoreceptors was monitored by observing the ability of these chimeras to support chemotaxis in soft-agar swarm assays, essentially as previously described (12), except that the swarm plates were inoculated with 1 μl of a suspension of cells adjusted to equivalent optical density at 600 nm in phosphate-buffered saline. In the soft-agar swarm experiment, cells are inoculated at a single point in soft-agar plates (0.25% agar) consisting of a minimal medium supplemented with the attractant to be tested (asparagine, for receptors containing the N-terminal module of McpB, or proline, for receptors containing the N-terminal module of McpC). As the cells metabolize the attractant in the surrounding medium, they create an attractant gradient in their environment. If the cells can perform chemotaxis towards the attractant, they follow this gradient away from the point of inoculation, creating an expanding, clearly defined ring of cell density visible in the agar. In contrast, if the cells cannot perform chemotaxis towards the attractant due to the loss of cognate receptor function, random motility results in diffuse spreading away from the point of inoculation (manifest as an apparently solid disk of cell density visible in the agar) but does not result in formation of a discrete ring. Thus, the presence of a discrete ring can be attributed to a chemotactic response (see Fig. 3A for a representative example of this difference; compare Δ10 with Δ10 expressing mcpC).
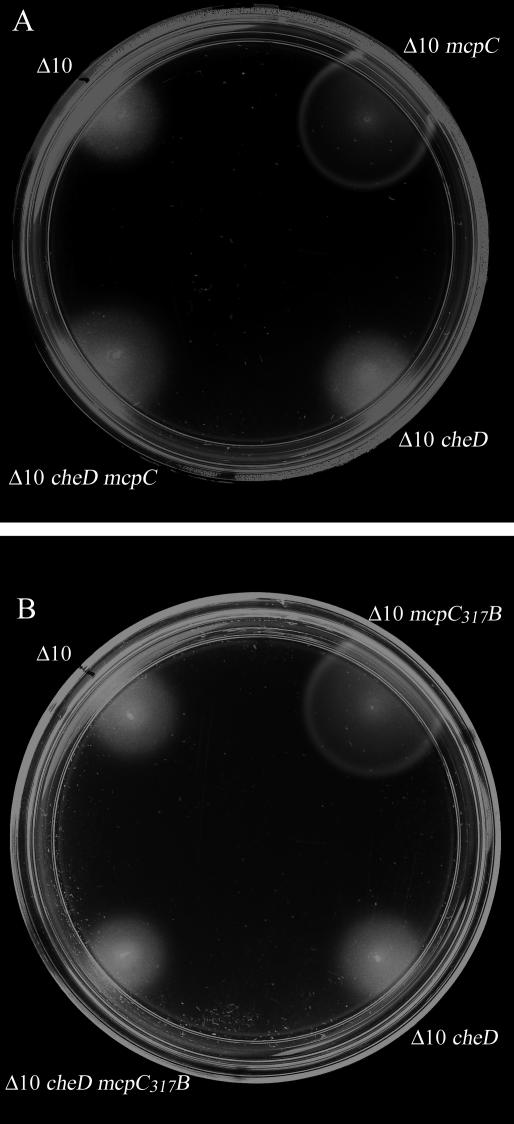
FIG. 3.
Soft-agar swarm assays were performed with swarm plates composed of minimal medium supplemented with proline as a chemoattractant. All strains were assayed a minimum of seven independent times. Results from a representative experiment are shown. Strains are designated as follows. (A) Δ10, OI3545; Δ10 mcpC, OI3545 expressing mcpC; Δ10 cheD, OI3628; Δ10 cheD mcpC, OI3628 expressing mcpC. (B) Δ10, OI3545; Δ10 mcpC317B, OI3545 expressing mcpC317B; Δ10 cheD, OI3628; Δ10 cheD mcpC317B, OI3628 expressing mcpC317B. The image was processed using Adobe Photoshop 5.5 to maximize contrast.
Using the soft-agar swarm assay, we found that most of the chimeras reported in this study were functional in the presence of CheD (Fig. 2-4). To our knowledge, this is the first report documenting the construction of ligand-responsive chimeric receptors in which AS-1 and AS-2 of the HAMP domain were derived from heterologous parental receptors. The nonfunctional chimera that we created is discussed in more detail below.
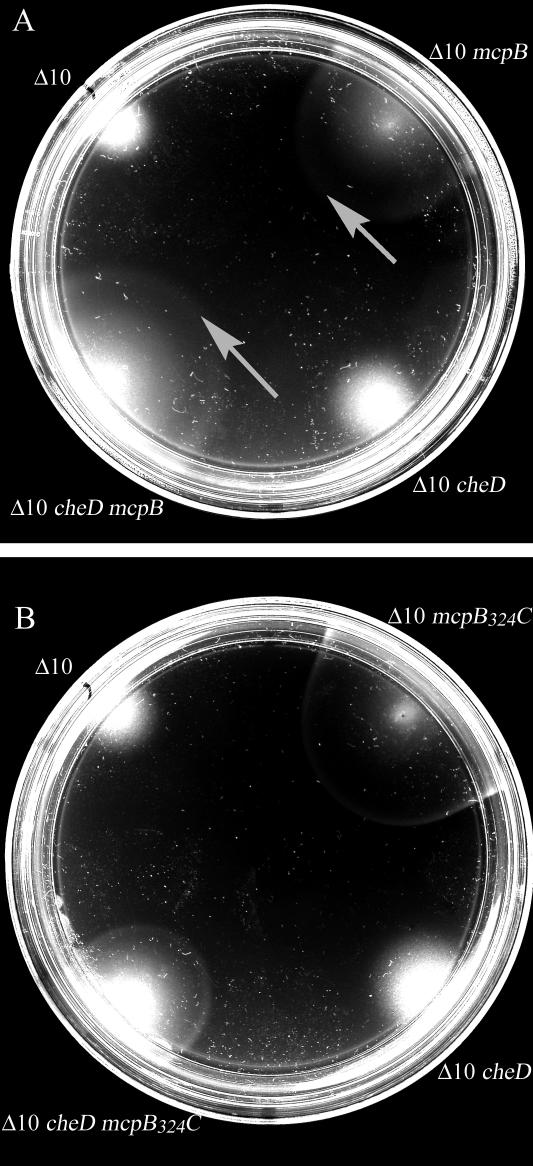
FIG. 2.
Soft-agar swarm assays were performed with swarm plates composed of minimal medium supplemented with asparagine as a chemoattractant. All strains were assayed a minimum of seven independent times. Results from a representative experiment are shown. Strains are designated as follows. (A) Δ10, OI3545 (parental strain carrying mutations in all 10 known chemoreceptors, but otherwise che+); Δ10 mcpB, OI3545 expressing mcpB; Δ10 cheD, OI3628 (nonpolar cat insertion in cheD, but otherwise isogenic to Δ10); Δ10 cheD mcpB, OI3628 expressing mcpB. (B) Δ10, OI3545; Δ10 mcpB324C, OI3545 expressing mcpB324C; Δ10 cheD, OI3628; Δ10 cheD mcpB324C, OI3628 expressing mcpB324C. The image was processed using Adobe Photoshop 5.5 to maximize contrast. The arrows demarcate the chemotactic rings.
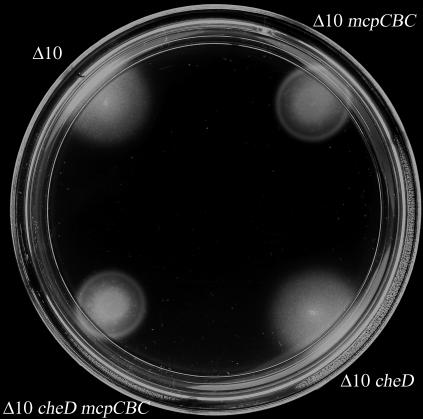
FIG. 4.
Soft-agar swarm assays were performed using swarm plates composed of minimal medium supplemented with proline as a chemoattractant. All strains were assayed a minimum of seven independent times. Results from a representative experiment are shown. Strains are designated as follows. Δ10, OI3545; Δ10 mcpCBC, OI3545 expressing mcpCBC; Δ10 cheD, OI3628; Δ10 cheD mcpCBC, OI3628 expressing mcpCBC. The image was processed using Adobe Photoshop 5.5 to maximize contrast.
The C-terminal module of McpC functions independently of CheD in signal transduction.
Because CheD is predicted to catalyze deamidation of one or more Gln residues in the C-terminal module of McpC (13), we hypothesized that this enzymatic processing was required for the McpC C-terminal module to transmit ligand occupancy signals to the downstream chemotactic machinery. To test this hypothesis, we examined whether the McpB324C chimera could support chemotactic ring formation in the absence of CheD function. Previous results (confirmed in Fig. 2A of this study) have shown that McpB is capable of supporting chemotactic ring formation in response to asparagine in the absence of CheD (10), demonstrating that both the McpB N- and C-terminal modules, as well as the HAMP domain, function independently of CheD. Surprisingly, we found that McpB324C was also able to support chemotactic ring formation in the absence of CheD (Fig. 2B), indicating that no sequence determinants in the C-terminal module of McpC (or in AS-2 of the McpC HAMP domain) are responsible for the CheD requirement exhibited by full-length McpC. In other words, the C-terminal module of McpC is capable of functioning in the absence of CheD. Therefore, although CheD-mediated deamidation likely occurs in the C-terminal module of McpC (13), this cannot be required for signal transduction by McpC and does not explain why McpC exhibits a CheD-dependent response to external stimuli. We speculate that CheD-mediated deamidation of glutaminyl side chains in the C-terminal module of McpC may serve to enhance adaptation by providing additional sites for reversible methylesterification by CheR and CheB.
CheD is required for intramolecular signal transduction by either the N-terminal module or AS1 of the HAMP domain of McpC.
Given that the chimera containing the McpC C-terminal module functioned independently of CheD (Fig. 2B), we examined whether the reciprocal chimera (McpC317B), containing only the N-terminal module and partial HAMP domain of McpC, could support chemotactic ring formation independently of CheD. As a control, we tested wild-type McpC in parallel. Figure 3A shows that, in the absence of CheD, the strain expressing wild-type McpC exhibits a dense disk of cell density without any chemotactic ring, indicating that wild-type McpC was unable to support chemotaxis towards proline in the absence of CheD function. This phenotype is consistent with previous results (10). We found that McpC317B, although fully capable of supporting chemotactic ring formation in the presence of CheD, was also not able to support chemotactic ring formation in the absence of CheD (Fig. 3B). Recall that both wild-type McpB and the McpB324C chimera are capable of supporting chemotactic ring formation in the absence of CheD (Fig. 2) (10), indicating that CheD function is not required for the activity of the C-terminal module of either McpB or McpC during transmission of ligand-induced signals. Thus, the inability of McpC317B to function in the absence of CheD indicates that a sequence determinant contained in the first 317 residues of McpC is responsible for the CheD requirement exhibited by the full-length McpC. The McpC sequences found in McpC317B include the N-terminal module as well as AS-1 of the HAMP domain. Therefore, taken together with the results shown in Fig. 2, these results argue that intramolecular signal transduction from the N-terminal module to the C-terminal module of wild-type McpC is dependent on CheD because a sequence determinant that falls within the first 317 residues of the receptor requires CheD activity for its function.
Replacement of AS1 in McpC with the corresponding sequence from McpB renders the chimeric receptor independent of CheD.
The subcellular locations of CheD and the various receptor sequences suggested to us a possible target for CheD activity. Because both CheD and AS-1 of McpC are expected to be cytoplasmic, whereas the majority of the N-terminal module of McpC is predicted to be located outside the cytoplasmic membrane, we hypothesized that the short cytoplasmic sequence of AS-1 was the target of the CheD activity that is required for intramolecular signal transduction by McpC. To test this hypothesis, we created a chimeric receptor in which the entire sequence encoding AS-1 of McpC (plus four adjacent residues predicted to be part of the connector) was replaced by the corresponding segment of McpB (McpCBC) (Fig. 1) and examined whether this chimera could support chemotactic ring formation towards proline in the absence of CheD activity (Fig. 4). In contrast to wild-type McpC, McpCBC was capable of supporting chemotactic ring formation in both the presence and absence of CheD, indicating that replacement of the wild-type McpC AS-1 sequence with the equivalent sequence from the CheD-independent receptor McpB was sufficient to alleviate the CheD requirement for intramolecular signal transduction by McpC. These results suggest that CheD activity is specifically required for AS-1 of wild-type McpC to function in transmission of a chemotactic signal from the N-terminal module to the C-terminal module of the receptor.
To test if AS-1 of McpC could confer a CheD-dependent signaling phenotype to a previously CheD-independent receptor, we constructed a variant of McpB containing AS-1 of McpC and analyzed its ability to support chemotactic ring formation in response to Asn. A procedure analogous to that used for the construction of mcpCBC was used to create this variant of McpB, in which the residues corresponding to AS-1 were replaced with those from McpC to create mcpBCB. Based on the success we observed with the McpCBC chimera, we expected McpBCB to be functional in the presence of CheD and, if our hypothesis was correct, to be unable to mediate signal transduction in the absence of CheD. However, we found that although McpBCB was expressed at levels similar to those for the other chimeras used in this study, it was completely unable to mediate chemotactic ring formation to Asn, even in the presence of CheD (data not shown). At the present time, we do not know why this particular chimera is unable to mediate signal transduction in the presence of CheD. Due to this technical limitation, we have not been able to test whether the introduction of AS-1 from McpC into a CheD-independent receptor will generate a CheD-dependent variant.
CheD-mediated deamidation of glutaminyl residues in AS1 of McpC is not required for intramolecular signal transduction.
Comparison of the amino acid sequences of AS-1 from McpC and McpB (Fig. 1) revealed that AS-1 of McpC contains a Gln-Gln pair (residues 304 and 305; these correspond to positions AS-1.3 and AS-1.4 of the HAMP domain, numbered according to a previously described scheme [2]) positioned such that they occupy the solvent-exposed b and c sites of the AS-1 heptad repeat motif. CheD has recently been shown to catalyze deamidation of such a Gln-Gln pair located at a specific site in the C-terminal module of a B. subtilis chemoreceptor (13), suggesting that the Gln-Gln pair in the HAMP domain of McpC may be a target for CheD enzymatic activity. However, covalent modification of HAMP domain residues has not been reported previously. In all instances in which posttranslational chemoreceptor modification has been investigated (either glutaminyl deamidation or reversible glutamyl methylesterification), these posttranslational receptor modifications have been found to occur exclusively in the C-terminal receptor modules, at positions corresponding to the b and c sites of the heptad repeat. Despite this, given that McpC exhibits the AS-1-specific requirement for CheD described above, we hypothesized that CheD-mediated deamidation of the Gln-Gln pair at the b and c sites of AS-1 in McpC might be required for McpC to transduce a chemotactic signal. To test this hypothesis, we used a previously described method of site-directed mutagenesis (21) to convert the corresponding Gln codons to Glu in the context of an otherwise wild-type McpC receptor, yielding the McpC305QE single mutant and the McpC304QE 305QE double mutant. Conversion of Gln to Glu in these mutant receptors mimics deamidation at these sites. Using the soft-agar swarm assay, we found that both mutants were capable of forming a chemotactic ring towards proline in the presence of CheD, but neither was able to do so in the absence of CheD (data not shown), indicating that the AS-1-specific requirement for CheD exhibited by wild-type McpC was retained by these mutants. CheD-mediated deamidation at Q304 or Q305 would yield Glu at these sites—therefore, our substitutions yield mutant receptors that mimic, in the absence of CheD activity in the cell, the receptor state in which the wild-type McpC would exist following modification by CheD. Because these mutants retain the CheD-dependent chemotactic phenotype, our results demonstrate that CheD-mediated deamidation of either McpC Q304 or Q305, whether or not it occurs, is not required for signal transduction by McpC and cannot explain why McpC exhibits a CheD-dependent phenotype.
Concluding remarks.
In order to probe the role of B. subtilis CheD in modulating chemoreceptor function, we used two B. subtilis receptors to construct chimeric molecules and analyzed the ability of these chimeras to support chemotaxis. Our results demonstrate that the CheD-dependent receptor, McpC, can be converted to a CheD-independent variant by replacement of a 21-residue segment of the McpC HAMP domain, encoding AS-1, with the corresponding sequence from the CheD-independent receptor, McpB. These results suggest that CheD may be involved, directly or indirectly, in facilitating the ability of AS-1 in McpC to participate in intramolecular signal transduction between the N- and C-terminal receptor modules. However, our results show that CheD does not influence the function of the HAMP domain by catalyzing deamidation of the AS-1-encoded residue Q304 or Q305, because the site-directed McpC mutants that we constructed to mimic a CheD-processed receptor retained the CheD-dependent phenotype exhibited by wild-type McpC. Therefore, CheD may play another critical role in enabling the HAMP domain of McpC to pass sensory information between receptor modules. It is presently unknown whether or not the catalytic activity of CheD is required for its role in facilitating intramolecular signal transduction by McpC. However, we note that aside from Q304 and Q305, there are no other residues in AS-1 that can serve as substrates for deamidation by CheD. Thus, it seems unlikely that the known catalytic activity of CheD plays a direct role in modulating the function of the HAMP domain of McpC. We speculate that CheD possesses an additional activity, independent of deamidation, which is required to facilitate the transmission of sensory information from the N-terminal module to the C-terminal module of McpC. Perhaps this activity involves direct intermolecular interaction of CheD with AS-1 of McpC to assist the HAMP domain in assuming a correct three-dimensional conformation or in facilitating a structural transition that must occur during transmission of the ligand-induced signal to the C-terminal module. To the best of our knowledge, there have previously been no reports of heterologous protein-protein interactions that occur to regulate HAMP domain function for any receptor. Taken together with the previously described activity of CheD (deamidation of residues located within chemoreceptor C-terminal modules), these results suggest that CheD may interact with McpC at multiple, functionally distinct sites. This intriguing possibility warrants further investigation.
Acknowledgments
We thank Tim Leonard for photography and technical assistance. We also thank Gary Dunny, in whose laboratory a portion of this work was performed.
This work was supported by a grant from the National Institutes of Health to G.W.O. (GM54365).
REFERENCES
- 1.Appleman, J. A., L. L. Chen, and V. Stewart. 2003. Probing conservation of HAMP linker structure and signal transduction mechanism through analysis of hybrid sensor kinases. J. Bacteriol. 185:4872-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appleman, J. A., and V. Stewart. 2003. Mutational analysis of a conserved signal-transducing element: the HAMP linker of the Escherichia coli nitrate sensor NarX. J. Bacteriol. 185:89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 4.Armitage, J. P. 1999. Bacterial tactic responses. Adv. Microb. Physiol. 41:229-289. [DOI] [PubMed] [Google Scholar]
- 5.Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng, X., J. W. Baumgartner, and G. L. Hazelbauer. 1997. High- and low-abundance chemoreceptors in Escherichia coli: differential activities associated with closely related cytoplasmic domains. J. Bacteriol. 179:6714-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garrity, L. F., S. L. Schiel, R. Merrill, J. Reizer, M. H. Saier, Jr., and G. W. Ordal. 1998. Unique regulation of carbohydrate chemotaxis in Bacillus subtilis by the phosphoenolpyruvate-dependent phosphotransferase system and the methyl-accepting chemotaxis protein McpC. J. Bacteriol. 180:4475-4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanlon, D. W., and G. W. Ordal. 1994. Cloning and characterization of genes encoding methyl-accepting chemotaxis proteins in Bacillus subtilis. J. Biol. Chem. 269:14038-14046. [PubMed] [Google Scholar]
- 9.Hou, S., R. W. Larsen, D. Boudko, C. W. Riley, E. Karatan, M. Zimmer, G. W. Ordal, and M. Alam. 2000. Myoglobin-like aerotaxis transducers in Archaea and Bacteria. Nature 403:540-544. [DOI] [PubMed] [Google Scholar]
- 10.Kirby, J. R., C. J. Kristich, M. M. Saulmon, M. A. Zimmer, L. F. Garrity, I. B. Zhulin, and G. W. Ordal. 2001. CheC is related to the family of flagellar switch proteins and acts independently from CheD to control chemotaxis in Bacillus subtilis. Mol. Microbiol. 42:573-585. [DOI] [PubMed] [Google Scholar]
- 11.Krikos, A., M. P. Conley, A. Boyd, H. C. Berg, and M. I. Simon. 1985. Chimeric chemosensory transducers of Escherichia coli. Proc. Natl. Acad. Sci. USA 82:1326-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kristich, C. J., G. D. Glekas, and G. W. Ordal. 2003. The conserved cytoplasmic module of the transmembrane chemoreceptor McpC mediates carbohydrate chemotaxis in Bacillus subtilis. Mol. Microbiol. 47:1353-1366. [DOI] [PubMed] [Google Scholar]
- 13.Kristich, C. J., and G. W. Ordal. 2002. Bacillus subtilis CheD is a chemoreceptor modification enzyme required for chemotaxis. J. Biol. Chem. 277:25356-25362. [DOI] [PubMed] [Google Scholar]
- 14.Muller, J., S. Schiel, G. W. Ordal, and H. H. Saxild. 1997. Functional and genetic characterization of mcpC, which encodes a third methyl-accepting chemotaxis protein in Bacillus subtilis. Microbiology 143:3231-3240. [DOI] [PubMed] [Google Scholar]
- 15.Repik, A., A. Rebbapragada, M. S. Johnson, J. O. Haznedar, I. B. Zhulin, and B. L. Taylor. 2000. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36:806-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh, M., B. Berger, P. S. Kim, J. M. Berger, and A. G. Cochran. 1998. Computational learning reveals coiled coil-like motifs in histidine kinase linker domains. Proc. Natl. Acad. Sci. USA 95:2738-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stock, J. B., and M. G. Surette. 1996. Chemotaxis, p. 1103-1129. In F. C. Neidhardt, R. I. Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella. Cellular and molecular biology. ASM Press, Washington, D.C.
- 18.Weerasuriya, S., B. M. Schneider, and M. D. Manson. 1998. Chimeric chemoreceptors in Escherichia coli: signaling properties of Tar-Tap and Tap-Tar hybrids. J. Bacteriol. 180:914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams, S. B., and V. Stewart. 1999. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol. Microbiol. 33:1093-1102. [DOI] [PubMed] [Google Scholar]
- 20.Zhulin, I. B. 2001. The superfamily of chemotaxis transducers: from physiology to genomics and back. Adv. Microb. Physiol. 45:157-198. [DOI] [PubMed] [Google Scholar]
- 21.Zimmer, M. A., J. Tiu, M. A. Collins, and G. W. Ordal. 2000. Selective methylation changes on the Bacillus subtilis chemotaxis receptor McpB promote adaptation. J. Biol. Chem. 275:24264-24272. [DOI] [PubMed] [Google Scholar]