Abstract
The htx and ptx operons of Pseudomonas stutzeri WM88 allow for the use of the inorganic reduced phosphorus (P) compounds hypophosphite (P valence, +1) and phosphite (P valence, +3) as sole P sources. To support the proposed in vivo role for the htx and ptx operons, namely the use of phosphite and hypophosphite as alternative P sources, we used reporter gene fusions to examine their expression levels with respect to various P conditions. Expression of the htx and ptx operons was induced up to 17- and 22-fold, respectively, in cultures grown under phosphate starvation conditions relative to expression in medium with excess phosphate (Pi). However, the presence of the reduced P substrate hypophosphite, phosphite, or methylphosphonate, in addition to excess Pi, did not result in an increase in the expression of either operon. To provide further support for a role of the htx and ptx operons in Pi acquisition, we identified P. stutzeri phoBR homologs and constructed deletion mutants. Induction of the htx and ptx reporter gene fusions in response to growth on limiting Pi was abolished in ΔphoB, ΔphoR, and ΔphoBR mutants, demonstrating that htx and ptx expression is phoBR dependent. The putative LysR-type regulator encoded by ptxE has no apparent role in the expression of the htx and ptx operons, as no effect was observed on the level of induction of either operon in a ΔptxE mutant.
Despite the fact that phosphorus has long been considered the only essential element that does not partake in biologically catalyzed oxidation-reduction reactions, it has become increasingly evident that utilization of the inorganic reduced phosphorus compounds hypophosphite (P valence, +1) and phosphite (P valence, +3) as alternative phosphorus sources is common among microorganisms. Microbial oxidation of hypophosphite and phosphite has been documented in the literature for several decades (1, 6, 10, 12, 17, 19). Although these studies clearly established the microbial oxidation of these compounds, the processes by which this occurs remained largely unexplored in any detail on the genetic or biochemical level, until recently.
A genetic analysis of hypophosphite oxidation in Pseudomonas stutzeri WM88 led to the identification of two distinct regions of the chromosome, htxABCDEFGHIJKLM and ptxABCDE, that are required for the oxidation of hypophosphite and phosphite, respectively (20). Subsequent purification and biochemical characterization of the putative P-oxidizing enzymes HtxA and PtxD demonstrated that the two enzymes form a biochemical pathway for the oxidation of hypophosphite to Pi (5, 34). Genetic and biochemical data support the hypothesis that the htx and ptx genes serve the purpose of providing the organism with alternative sources of phosphorus.
In many bacteria, genes involved in the assimilation of Pi from various phosphorus compounds in the environment are phosphate starvation inducible (Psi). Collectively, such genes comprise a phosphate (Pho) regulon that is controlled by the two-component signal transduction system PhoBR (13-16, 30). The Pho regulon of Escherichia coli, for example, includes genes that encode transport systems for the uptake of Pi and a variety of alternate phosphorus sources, such as organophosphates and phosphonates, as well as genes that encode enzymes required for the utilization of alternative phosphorus sources (pstSCAB, ugpBAEC, phoA, and phnC-phnP) (2, 27, 33, 36). Under conditions of Pi starvation, phosphorylated PhoB binds to a highly conserved sequence called a Pho box located within the promoters of the genes that it activates (30). Although considerable data exist regarding the regulation of the E. coli phn genes required for the use of phosphonates (P valence, +3), no information is yet available on the regulation of genes required for the utilization of other reduced phosphorus compounds such as hypophosphite and phosphite.
Although the genes within the htx and ptx operons are clearly responsible for hypophosphite and phosphite oxidation in P. stutzeri, the physiological relevance of such a process with respect to phosphorus acquisition is less clear. To our knowledge, neither hypophosphite nor phosphite has ever been measured in the natural environment. However, note that a recent study demonstrated that previously used methods were inadequate for this task (22). To further clarify the in vivo role of the htx and ptx operons in P. stutzeri with respect to the oxidation of these compounds, we examined the regulation of expression of both operons. Here we report an expression analysis of htx and ptx in response to Pi starvation in P. stutzeri and demonstrate the dependence of this expression on phoBR, supporting a role for these genes in phosphorus acquisition through the oxidation of hypophosphite and phosphite.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains used for this study are shown in Table 1. E. coli DH5αλpir or BW20767 was used as a host for molecular cloning experiments. BW20767 is a tra+ strain that was also used as a donor for conjugations between E. coli and P. stutzeri strains. Plasmids pAH120 and pLA2 (8) were obtained from Barry Wanner (Purdue University, Lafayette, Ind.).
TABLE 1.
Bacterial strains used for this study
| Species and strain | Relevant characteristics | Construction or reference |
|---|---|---|
| Escherichia coli | ||
| DH5α/λpir | λpir φ80dlacZΔM15 Δ(lacZYA-argG)U169 recA1 hsdr17 deoR thi-1 supE44 gyrA96 relA1 | 21 |
| S17-1 | RP4-2-Tc::Mu-1 Kan::Tn7 integrant recA1 proA creB510 hsdR17 endA1 supE44 thi | 25 |
| BW20767 | RP4-2-Tc::Mu-1 Kan::Tn7 integrant leu-63::IS10 recA1 zbf-5 creB510 hsdR17 endA1 thi uidA (ΔMluI)::pir+ | 18 |
| Pseudomonas stutzeri | ||
| WM567 | Spontaneous Strr mutant of P. stutzeri WM536, Hpt+ Pt+ | 20 |
| WM2033 | ptxE::lacZ Strr Hpt+ Pt+ | Sucr WM567 segregant of pAW30a |
| WM2757 | ΔptxE Strr Hpt+ Pt+ | Sucr WM567 segregant of pAW28a |
| WM2106 | ΔptxE::lacZ Strr Hpt+ Pt+ | Sucr WM567 segregant of pAW29a |
| WM2940 | pAW41 integrants Strr Hpt+ Pt+ | Kanr WM567 integrant of pAW41 |
| WM3021 | pAW41 integrants ΔptxE Strr Hpt+ Pt+ | Kanr WM2757 integrant of pAW41 |
| WM4275 | ΔphoB Strr Hpt− Pt− | Sucr WM567 segregant of pAW84a |
| WM4296 | ΔphoR Strr Hpt− Pt− | Sucr WM567 segregant of pAW86a |
| WM4294 | ΔphoBR Strr Hpt− Pt− | Sucr WM567 segregant of pAW85a |
| WM4268 | ptxE::lacZ ΔphoB Strr Hpt− Pt− | Sucr WM2033 segregant of pAW84a |
| WM4261 | ptxE::lacZ ΔphoR Strr Hpt− Pt− | Sucr WM2033 segregant of pAW86a |
| WM4300 | ptxE::lacZ ΔphoBR Strr Hpt− Pt− | Sucr WM2033 segregant of pAW85a |
| WM4340 | pAW41 integrants ΔphoB Strr Hpt− Pt− | Kanr WM4275 integrant of pAW41 |
| WM4341 | pAW41 integrants ΔphoR Strr Hpt− Pt− | Kanr WM4296 integrant of pAW41 |
| WM4342 | pAW41 integrants ΔphoBR Strr Hpt− Pt− | Kanr WM4294 integrant of pAW41 |
| WM4269 | ΔptxE::lacZ ΔphoB Strr Hpt− Pt− | Sucr WM2106 segregant of pAW84a |
| WM4292 | ΔptxE::lacZ ΔphoR Strr Hpt− Pt− | Sucr WM2106 segregant of pAW86a |
| WM4288 | ΔptxE::lacZ ΔphoBR Strr Hpt− Pt− | Sucr WM2106 segregant of pAW85a |
Integration and segregation of pAW19-derived plasmids harboring the sacB gene were done as described in reference 20.
Media and growth of cultures.
The media used throughout were previously reported (32). Tryptone-yeast extract-agar containing an appropriate antibiotic was used for the selection of transformants and exconjugants of strain constructions unless otherwise indicated. A 0.2% glucose-MOPS [3-(N-morpholino)propanesulfonic acid] minimal medium was used for the growth of P. stutzeri strains on various phosphorus sources and for the screening and selection of proline auxotrophs. Antibiotics were used at the following concentrations for plasmid propagation and strain construction in E. coli: kanamycin, 50 μg/ml; streptomycin, 100 μg/ml. For the integration and maintenance of pAW41 in the P. stutzeri WM2940 chromosome, kanamycin was used at 10 μg/ml.
Screening for phosphate starvation induction of alkaline phosphatase in E. coli was done on 0.2% glucose-MOPS minimal medium containing 0.1 mM Pi and 60 μg of 5-bromo-4-chloro-3-indolyl-phosphate (XP) (Research Products International Corp., Mt. Prospect, Ill.)/ml. Screening for phosphate starvation induction of fusions to the lacZ gene, which encodes β-galactosidase, was done on 0.2% glucose-MOPS minimal medium containing 0.1 mM Pi and 32 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Research Products International Corp.)/ml. For reporter gene fusion analysis, P. stutzeri strains harboring a lacZ reporter gene fusion were grown in glucose-MOPS minimal medium containing either 0.12% glucose and 2 mM Pi (excess Pi) or 1.0% glucose and a 0.1 mM concentration of one of the following phosphorus sources (limiting Pi): Pi, hypophosphite, phosphite, or methylphosphonate. All phosphorus sources were purchased from Sigma (St. Louis, Mo.), and solutions were made immediately prior to use and then filter sterilized. Cultures were harvested at stationary phase (optical density at 600 nm [OD600], ca. 1.0) and cell extracts were made as described below.
DNA methods.
Standard methods for the isolation and manipulation of chromosomal and plasmid DNAs were used throughout (3). DNA hybridization reactions were done by using the DIG system (Roche, Mannheim, Germany) according to the manufacturer's instructions. DNA sequencing was performed by using an ABI Prism BigDye Terminator cycle sequencing reaction kit (Applied Biosystems, Foster City, Calif.) per the manufacturer's instructions and were analyzed at the W. M. Keck Center for Comparative and Functional Genomics, University of Illinois, Urbana.
Identification and cloning of P. stutzeri phoBR.
The phoBR operons and flanking sequences of six pseudomonad species were aligned with ClustalW (28). Highly conserved regions of DNA sequence were used as the basis for degenerate primer design to amplify the phoBR operon from the P. stutzeri chromosome. The P. stutzeri phoBR operon and flanking sequence were amplified by a PCR using Accuzyme DNA polymerase (Bioline USA Inc., Randolph, Mass.) and the following degenerate primers: 5′-AATTYCGTTATCTAATGCG-3′, which anneals to the P. stutzeri phoBR region 53 bp upstream of the putative PhoB translational start site, and 5′-CRAGYYGAAGGGTCCATG-3′, which anneals to the P. stutzeri phoBR region 115 bp downstream of the translational stop codon of PhoR, resulting in the amplification of a 2,224-bp fragment. The resulting PCR fragment was cloned into the pCR4-TOPO vector by use of a TOPO TA cloning kit (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions, creating plasmid pAW83. The inserted PCR fragment was sequenced initially by using M13 reverse and forward standard primers (Invitrogen) followed by sequencing with sequence-specific internal primers.
Plasmid constructions.
Plasmid pAW41 harbors an htxA::lacZ translational fusion and the oriT sequence for plasmid transfer by conjugation in a Kanr CRIM plasmid (8). In the first step of construction, oriT was amplified by the use of Pfu Turbo DNA polymerase (Invitrogen) as described previously (11). The resulting PCR fragment was digested with ClaI and inserted into the same sites of pAH120 (8) to create pAW38. In the second step, the 1.0-kbp region upstream of htxA, including the htxA translational start codon and ribosomal binding site, was amplified by a PCR using the following primers: 5′-GGCGCGCCCATATGGATGCTCCAAGGTCTTCCAA-3′ and 5′-GGCGCGCCCTGCAGTCTAGAGTGGCTATGTCCTGGGCGTT-3′, which insert NdeI and PstI sites, respectively (restriction sites are underlined). The resulting PCR product was digested with NdeI and PstI and inserted into the same sites of pLA2 (8) to construct a translational htxA::lacZ fusion. Finally, a BamHI-PstI fragment carrying the htxA::lacZ translational fusion was inserted into the same sites of pAW38 to create pAW41.
For the construction of P. stutzeri strains with a chromosomal ptxE::lacZ fusion, pAW27 and pAW30 were constructed as derivatives of pAW19. Plasmid pAW19 is a Kanr derivative of the suicide plasmid pWM91 that can be transferred by conjugation and that carries the sacB gene for counterselection of sucrose-resistant plasmid segregants (18). Plasmid pAW30 harbors a ptxE::lacZ transcriptional fusion and was created by inserting the lacZ gene (including its own ribosomal binding site) between the 1.0-kbp sequence directly upstream of, and including, the ptxE translational stop codon and the 1.0-kbp sequence directly downstream of the ptxE translational stop codon. Both the upstream and downstream sequences were amplified by PCRs using Taq DNA polymerase (Invitrogen). Primers 5′-GGCGGCACTAGTACATAGGGTCGGCAGTGCGC-3′ and 5′-GGCGGCGCGGCCGCTTATCCAGCTAGATCCGCCT-3′, which introduce a SpeI and a NotI site, respectively, were used to amplify the upstream sequence. The 1.0-kbp downstream fragment was amplified with primers 5′-GGCGGCGCGGCCGCGGTGATGGATGGTTGCGATC-3′ and 5′-GGCGGCGAGCTCCAGCGTGGCGTAGAGCTGCG-3′, which incorporate a NotI and a SacI site, respectively. The resulting PCR products were digested with the appropriate restriction enzymes and were inserted into the SpeI and SacI sites of pAW19 in a three-fragment ligation to create pAW27. The lacZ gene was amplified from E. coli S17-1 genomic DNA with the primers 5′-GGCGGCGCGGCCGCAGGAAACAGCTATGACCATG-3′ and 5′-GGCGGCGCGGCCGCTTATTTTTGACACCAGACCA-3′, which introduce NotI sites immediately upstream of the ribosomal binding site of the lacZ gene and immediately downstream of the lacZ translational stop codon. The resulting PCR fragment was digested with NotI and inserted into the same sites of pAW27 to create pAW30.
For the construction of phoB, phoR, and phoBR deletion mutants of P. stutzeri, plasmids pAW84, pAW85, and pAW86 were constructed from pAW19. Plasmid pAW84 carries ca. 300 bp of 5′ phoB and its upstream flanking sequence ligated to ca. 300 bp of 3′ phoB and its downstream flanking sequence, resulting in an in-frame 46-amino-acid deletion of the P. stutzeri phoB gene. Both the upstream and downstream phoB sequences were amplified by PCRs using Platinum Pfx polymerase (Invitrogen). Primers 5′-GGATCCACTAGTTAATTTCGTTATCTAATGCC-3′ and 5′-GGATCCGCGGCCGCTGAGCATGATGATCGGCGTGTCG-3′, which incorporate SpeI and NotI sites, respectively, were used to amplify the upstream phoB sequence. The downstream phoB sequence was amplified with the following primers: 5′-GGATCCGCGGCCGCGGCGGCCTGCTGCTCGATCC-3′ and 5′-GGATCCGAGCTCTCAGCTTTTGCTGGAGAAACG-3′, which incorporate NotI and SstI sites, respectively. The resulting PCR fragments were digested with the appropriate restriction enzymes and were inserted into the SpeI and SstI sites of pAW19 in a three-fragment ligation to create pAW84. Plasmid pAW86 carries a 234-amino-acid in-frame deletion of PhoR and was constructed in a similar manner. Primers 5′-GGATCCACTAGTTTGAATCAGGACTGGCAAGG-3′ and 5′-GGATCCGCGGCCGCGGCGCGATCGATGATGCCTTGC-3′, which incorporate SpeI and NotI sites, respectively, were used to amplify the upstream phoR fragment, and primers 5′-GGATCCGCGGCCGCGTACACGCCCGATGGTGGC-3′ and 5′-GGATCCGAGCTCTCAGCGTTCGGACACCTGGC-3′, which incorporate NotI and SstI restriction sites, respectively, were used to amplify the phoR downstream fragment. Plasmid pAW85 carries a 1,512-bp internal deletion of the phoBR operon in which only the 5′-most 250 bp of phoB and the 3′-most 298 bp of phoR remain. This plasmid was constructed by inserting the SpeI-NotI upstream phoB fragment of pAW84 and the NotI-SstI downstream phoR fragment of pAW86 into the SpeI and SstI sites of pAW19 in a three-way ligation.
Genetic techniques.
Plasmids pAW30 and pAW41 were introduced into P. stutzeri WM567 by conjugation as previously described (11). The desired deletion and reporter gene fusion strains resulting from double recombination events were acquired by sacB counterselection as described previously (20).
For the construction of an htxA::lacZ fusion in P. stutzeri, an exconjugant resulting from the integration of pAW41 via homologous recombination at the htx promoter region was isolated on glucose-MOPS minimal medium containing 0.1 mM Pi, 10 μg of kanamycin/ml, and X-Gal. This strain carries both the htxA::lacZ translational fusion and an intact htx operon. Correct construction of the chromosomal deletions and reporter gene fusions in P. stutzeri was verified by DNA hybridization analysis (data not shown).
RT-PCR.
Total RNAs were isolated from cultures of P. stutzeri WM88 grown to mid-logarithmic phase (OD600, ca. 0.6) in 0.2% glucose-MOPS minimal medium with 0.5 mM hypophosphite as the sole source of phosphorus. RNAs were isolated with an RNeasy mini kit containing an RNAprotect bacterial reagent (Qiagen Inc., Valencia, Calif.) per the manufacturer's instructions. For the removal of contaminating chromosomal DNA, the RNA preparation was digested with amplification-grade DNase I (Invitrogen). DNase I-treated RNA was then used as a template in a reverse transcription (RT) assay by using SuperScript II RNase H− reverse transcriptase (Invitrogen) according to the manufacturer's protocol. PCR amplification of the cDNA from the RT reaction was performed by using Platinum Pfx DNA polymerase (Invitrogen) per the manufacturer's instructions. Both a positive control, in which only chromosomal DNA was added to the PCR, and a negative control, in which only RNA without the RT step was used in the PCR, were run under identical PCR amplification conditions. The primers used to amplify each ptx junction sequence are listed in Table 2.
TABLE 2.
Oligonucleotide primers used for the amplification of ptx junction sequences
| Amplified junction | Primer set (5′-3′) | Predicted product size (bp) |
|---|---|---|
| ptxAB | ATGAGCCGGTAGCCAGTCT, | 561 |
| AAATACGCCAGGTCGATACG | ||
| ptxBC | GGGCAGGACTACGAACAACA, | 611 |
| TCGATAGCCCGAAAAGTCTG | ||
| ptxCD | CATGGTCGGCAAGTTCTTC, | 563 |
| CCGACTACACGCAGCTCA | ||
| ptxDE | GAGCTGCTTGCCCTCGTA, | 598 |
| CCATGCAGGGCTTCTAGC |
Enzymatic assays.
β-Galactosidase specific activities were determined by continuous assaying in 1-ml volumes and are reported in standard units (micromoles per minute per milligram). Extracts were made from P. stutzeri cultures grown as described above. Cells were harvested by centrifugation and the entire cell pellet was resuspended in 50 mM Tris-Cl, pH 8.0. The cells were lysed by sonication with two 30-s pulses at 4°C or by passage through a French press at 13,000 lb/in2. The resulting crude cell extract was centrifuged at 15,000 × g for 20 min and the supernatant was removed for activity assays. β-Galactosidase assays were carried out in 50 mM Tris-HCl buffer, pH 8.0, containing 10 mM KCl, 1 mM MgSO4, and 50 mM β-mercaptoethanol, with 2.7 mM o-nitrophenyl-β-d-galactoside (ONPG) (Sigma) as a substrate. The release of o-nitrophenol was monitored as an increase in the absorbance at 420 nm, and an extinction coefficient of 4,112 M−1 cm−1 was used to calculate o-nitrophenol production. Protein concentrations were determined by using the Coomassie Plus protein assay reagent (Pierce, Rockford, Ill.) as recommended.
Nucleotide sequence accession numbers.
The GenBank accession number for the P. stutzeri WM88 phoBR DNA sequence determined for this study is AY590886.
RESULTS
The genes within the ptx locus form a transcriptional unit.
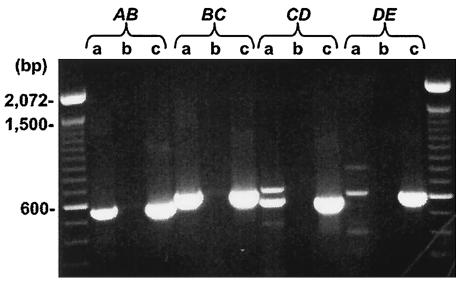
All of the open reading frames in the ptx locus either overlap one another or are separated by at most nine bases. This suggests that the ptxABCDE genes form an operon, but this had not been experimentally verified. We determined that the ptx genes are cotranscribed by performing RT-PCRs with the junction sequences between each of the genes (Fig. 1). Primers were designed to amplify ca. 300 bp upstream and downstream of the intergenic regions of each gene to yield amplification products of ca. 600 bp. Such products would be obtained only if the mRNAs spanned the junction of the two genes, indicating that they were cotranscribed. Although several additional bands were present in some of the reactions due to nonspecific amplification, a significant PCR product corresponding to the predicted size was amplified, supporting the conclusion that the ptx genes form an operon. With the same method, the genes in the htx locus were also determined to be cotranscribed (34a).
FIG. 1.
RT-PCRs with total RNAs prepared from P. stutzeri WM567 grown on hypophosphite as the sole P source to determine the operon structure of ptx. Lanes a, complete RT reactions; lanes b, negative controls for which no reverse transcriptase was added to the reaction; lanes c, PCR-positive controls in which chromosomal DNA was used as the template. The left- and rightmost lanes contain a 100-bp ladder. The junction sequences amplified are indicated above the reactions. For a list of primers used and the predicted PCR product size for each reaction, refer to Table 2.
Expression of htx and ptx operons is induced under phosphate starvation conditions.
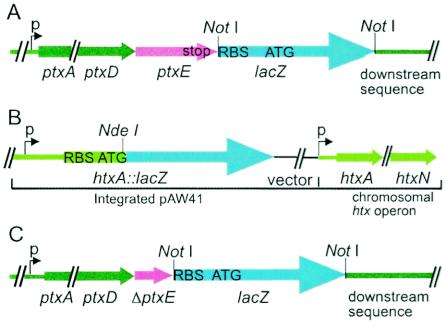
To examine the regulation of expression of the ptx operon in P. stutzeri, we constructed a chromosomal ptxE::lacZ transcriptional fusion (Fig. 2A) (strain WM2033) and used it to measure ptx gene expression. WM2033 was grown to stationary phase in triplicate cultures with different phosphorus sources and with either excess or limiting Pi, and β-galactosidase activities in the cell extracts were measured (Table 3). To verify that the cultures were starved for Pi, we also measured the activity of native phosphatase in each of the extracts, as the expression of phosphatase is only induced upon Pi starvation (S. E. Neuhaus, A. K. White, and W. W. Metcalf, unpublished data). Expression of the ptx operon was induced 14-fold during growth on limiting Pi and up to 22-fold during growth on phosphite relative to the expression levels on excess Pi. Thus, expression of the ptx operon is induced by Pi starvation. Similar expression levels were observed for a ptxA::lacZ translational fusion in E. coli in response to Pi starvation (data not shown).
FIG. 2.
Structures of chromosomal reporter gene fusions in P. stutzeri. (A) Structure of ptxE::lacZ transcriptional fusion. (B) Structure of htxA::lacZ translational fusion, showing the integrant structure formed by integration of pAW41 at the htx promoter region and the native promoter. (C) Structure of ΔptxE::lacZ transcriptional fusion. The diagram was not drawn to scale.
TABLE 3.
Expression of a ptxE::lacZ transcriptional fusion and an htxA::lacZ translational fusion in P. stutzeri in response to growth on different P sources
| P sourcec (concn [mM]) |
htxA::lacZ expressiona
|
ptxE::lacZ expressionb
|
||
|---|---|---|---|---|
| β-Galactosidase activityd | Fold inductione | β-Galactosidase activityd | Fold inductione | |
| Pi (2) | 0.17 ± 0.05 | 1 | 0.01 ± 0.00 | 1 |
| Pi (0.1) | 1.80 ± 0.77 | 10.6 | 0.15 ± 0.02 | 15 |
| Pt (0.1) | 2.25 ± 0.95 | 13.2 | 0.20 ± 0.03 | 20 |
| Hpt (0.1) | 2.98 ± 0.64 | 17.5 | 0.17 ± 0.02 | 17 |
| Pi (2) + Pt (0.1) | 0.18 ± 0.07 | 1.1 | 0.01 ± 0.00 | 1 |
| Pi (2) + Hpt (0.1) | 0.19 ± 0.06 | 1.1 | 0.01 ± 0.00 | 1 |
| Pi (2) + Mpn (0.1) | 0.13 ± 0.02 | NAg | NDf | NAg |
P. stutzeri WM2940.
P. stutzeri WM2033.
The P sources were added to either 2 mM (excess P with a limiting carbon source) or 0.1 mM (P starvation with excess carbon source). Pi, Pt, Hpt, and Mpn are abbreviations for phosphate, phosphite, hypophosphite, and methylphosphate, respectively.
β-Galactosidase activities were determined from duplicate assays of triplicate cultures and are reported in standard units as means ± standard deviations.
Induction relative to the expression observed with growth on 2 mM Pi.
ND, Not detected. The limit of detection was 0.005 U.
NA, Not applicable.
Expression analysis of the htx operon was done in a similar manner. Due to the large size of the htx operon (11.8 kbp), a reporter gene fusion was constructed to measure expression levels at the htx promoter rather than at the distal end of the operon. A chromosomal htxA::lacZ translational fusion was constructed by integration, via homologous recombination at the htx promoter, of a suicide plasmid (pAW41) carrying a 1.0-kbp region directly upstream of the translational start site of htxA (Fig. 2B) (strain WM2940). This allowed for measurements of the expression levels at the plasmid-borne htx promoter without disrupting expression at the native promoter. Strain WM2940 was grown under the conditions described above and its β-galactosidase activity was measured (Table 3). Compared to the expression level during growth on 2 mM Pi (excess Pi), an 11-fold induction of htx expression in response to phosphate starvation (0.1 mM Pi) was observed. The induction of expression was slightly higher for growth on phosphite or hypophosphite as the phosphorus source, resulting in a 13- and 17-fold induction, respectively.
To determine if the presence of the reduced phosphorus compounds that act as phosphorus substrates for P. stutzeri could specifically induce the expression of either the htx or ptx operon in the presence of Pi, we grew the reporter gene fusion strains on 2 mM Pi in addition to 0.1 mM phosphite, hypophosphite, or the organic reduced phosphorus compound methylphosphonate. No induction of expression of either the ptx or htx operon was observed (Table 3).
Identification of P. stutzeri phoBR.
The induction of the htx and ptx operons in response to Pi starvation suggested that htx and ptx might be regulated in a phoBR-dependent manner. This possibility was further supported by the presence of well-conserved putative Pho boxes located within the promoter regions of the htx and ptx operons (Fig. 3). Although the presence of phoBR homologs in P. stutzeri had not been determined previously, homologs of these genes have been identified in the published genome sequences of several pseudomonad species. To examine PhoBR-dependent regulation of the htx and ptx operons in the native host, we identified P. stutzeri phoBR as follows. The phoBR and flanking sequences of six pseudomonads were aligned with ClustalW (28), and degenerate primers were designed from conserved sequences just upstream and downstream of the phoBR operons of these organisms. Using these primers, we amplified a ca. 2.2-kbp PCR fragment, consistent with the predicted size of the phoBR operons from other pseudomonads. The fragment was cloned, and sequence analysis indicated the presence of two open reading frames arranged in a putative operon. Comparisons of the predicted amino acid sequences encoded by the two open reading frames to those in the UniProt database indicated that the first open reading frame, of 690 bp, encodes a protein of 229 amino acids that is 90 to 93% identical on the amino acid level to the PhoB proteins of other pseudomonads and 42% identical to E. coli PhoB. The second open reading frame (located 68 bp downstream of the stop codon of phoB) is 1,299 bp long and encodes a protein of 433 amino acids that shares 69 to 72% amino acid sequence identity with the PhoR proteins from other pseudomonads and 42% identity with the PhoR protein of E. coli. Thus, based on sequence analysis, the cloned fragment encodes a phoBR operon of 2,060 bp from P. stutzeri, in addition to 49 bp directly upstream of the PhoB translational start site and 115 bp directly downstream of the PhoR translational stop codon.
FIG. 3.
DNA sequences of promoter regions of htx operon (A) and ptx operon (B). The partial deduced amino acid sequence of each protein is shown below the coding sequence. The boxed sequence represents a putative Pho box and the match to the consensus sequence is shown above it. The predicted ribosomal binding site for each sequence is indicated by a line above the sequence.
Hypophosphite and phosphite oxidation is phoBR dependent in P. stutzeri.
To examine the role of phoBR on the utilization of the reduced phosphorus compounds hypophosphite and phosphite in P. stutzeri, we constructed in-frame deletions in either phoB alone (ΔphoB), phoR alone (ΔphoR), or both (ΔphoBR). To examine the phenotypes of these mutants with respect to the oxidation of reduced phosphorus compounds, we streaked the mutants alongside the wild-type parental strain on glucose-MOPS minimal medium containing one of a variety of phosphorus sources, as described above. The absence of growth on any of these substrates by any of the phoBR mutants contrasted with the robust growth observed for the wild-type strain (WM567) and demonstrated that both hypophosphite and phosphite oxidation is dependent on functional phoBR (data not shown). Similarly, growth on methylphosphonate, a substrate for the two C-P lyase pathways encoded by htxBCDEFGHIJKLMN and phnC-phnP, which are predicted to be Psi operons, was also abolished in the phoBR mutants (data not shown).
Several other interesting phenotypes were observed for the phoBR mutants. A marked decrease in growth on low-Pi solid medium was observed for the ΔphoB, ΔphoR, and ΔphoBR mutants compared to that of the wild-type strain WM567. This phenotype was not observed for medium with excess Pi. To examine the nature of the growth defect in the mutant strains, we performed a growth analysis of the wild type and the mutants in broth cultures. Although the doubling times for the wild type and the mutants on low-Pi medium were similar (ca. 2.4 h), the maximum OD600 reached by the wild-type strain was 0.74 ± 0.02, whereas the maximum OD600 reached by the mutant strains was only 0.33 ± 0.01. This indicates that the decrease in growth observed for the phoBR mutants was due to a decrease in maximum growth yield rather than to an increase in the doubling time.
Expression of htx and ptx operons is phoBR dependent in P. stutzeri.
To examine the mechanism of phoBR regulation of the htx and ptx operons in P. stutzeri, we compared the expression of the ptxE::lacZ and htxA::lacZ fusions in the wild type and the ΔphoB, ΔphoR, and ΔphoBR mutants of P. stutzeri. The appropriate strains were grown under Pi starvation and Pi excess conditions and the β-galactosidase activities were measured as described above. Both the ptx and htx induction levels in the wild-type strains (WM2033 and WM2940, respectively) with low Pi and high Pi were similar to those that were previously observed (Table 3). However, the induction of expression of both the ptx and htx fusions in response to Pi starvation was completely lost in each of the mutants (Table 4). Similar decreases in response to Pi starvation were observed for each of the mutants, indicating that the effects of a null mutation in phoB, phoR, or phoBR are the same. These data provide additional support for a difference in the regulation of the Pho regulons of E. coli and P. stutzeri, as the constitutive expression of ptx or htx was not observed for the ΔphoR mutant. Thus, Pi starvation-dependent expression of the htx and ptx operons in P. stutzeri is dependent on phoBR, and the regulation of these operons occurs at the level of transcription.
TABLE 4.
Expression of the ptx and htx operons in wild-type and ΔphoB, ΔphoR, and ΔphoBR P. stutzeri strains
| Straina | β-Galactosidase activityb
|
Fold inductionc | |
|---|---|---|---|
| 0.1 mM Pi | 2 mM Pi | ||
| ptx expression | |||
| ptxE::lacZ | 0.22 ± 0.02 | 0.01 ± 0.00 | 16 |
| ptxE::lacZ ΔphoB | ND | 0.02 ± 0.00 | NA |
| ptxE::lacZ ΔphoR | ND | 0.02 ± 0.00 | NA |
| ptxE::lacZ ΔphoBR | ND | 0.02 ± 0.01 | NA |
| htx expression | |||
| htxA::lacZ | 1.43 ± 0.14 | 0.08 ± 0.01 | 18 |
| htxA::lacZ ΔphoB | 0.09 ± 0.02 | 0.08 ± 0.01 | 1 |
| htxA::lacZ ΔphoR | 0.06 ± 0.00 | 0.07 ± 0.01 | <1 |
| htxA::lacZ ΔphoBR | 0.08 ± 0.00 | 0.09 ± 0.01 | <1 |
| ptx expression in ΔptxE strain | |||
| ΔptxE::lacZ | 0.20 ± 0.04 | 0.01 ± 0.00 | 20 |
| ΔptxE::lacZ ΔphoB | ND | ND | NA |
| ΔptxE::lacZ ΔphoR | ND | ND | NA |
| ΔptxE::lacZ ΔphoBR | ND | 0.02 ± 0.00 | NA |
The strains used were ptxE::lacZ (WM2033), ptxE::lacZ ΔphoB (WM4268), ptxE::lacZ ΔphoR (WM4261), ptxE::lacZ ΔphoBR (WM4300), htxA::lacZ (WM2940), htxA::lacZ ΔphoB (WM4340), htxA::lacZ ΔphoR (WM4341), htxA::lacZ ΔphoBR (WM4342), ΔptxE::lacZ (WM2106), ΔptxE::lacZ ΔphoB (WM4269), ΔptxE::lacZ ΔphoR (WM4292), and ΔptxE::lacZ ΔphoBR (WM4288).
β-Galactosidase activities were determined from duplicate assays of triplicate cultures and are reported in standard units as means ± standard deviations. ND, not detected. The detection limit was 0.01 U.
Induction relative to the expression observed with growth on 2 mM Pi. NA, not applicable.
ptxE does not play a role in the regulation of the htx or ptx operon in response to Pi starvation.
The ptxE gene encodes a putative transcriptional regulator in the LysR family, suggesting that it might be involved in regulating the expression of the ptx and htx operons. To examine the role of ptxE, we constructed a chromosomal ptxE internal deletion mutant in both the ptxE::lacZ (WM2106) (Fig. 2C) and htxA::lacZ (strain WM3021) fusion backgrounds. Surprisingly, there was no significant change in expression level for either the ptx or htx operon in the ΔptxE strain compared to the wild type after growth on each phosphorus source (data not shown). To determine if a role for ptxE could be observed in the absence of phoBR, we constructed ΔphoB, ΔphoR, and ΔphoBR mutations in the ΔptxE::lacZ fusion background. However, again ΔptxE had no effect on the induction patterns in response to Pi starvation (Table 4). Thus, the role for ptxE in the expression of htx and ptx remains unclear.
DISCUSSION
Our expression analysis of ptxE::lacZ and htxA::lacZ fusions in P. stutzeri clearly demonstrated that both the htx and ptx genes are regulated in response to Pi starvation. Furthermore, an analysis of the htx and ptx reporter gene fusions in the wild type compared to those in ΔphoB, ΔphoR, and ΔphoBR mutants of P. stutzeri confirmed that the regulation of the htx and ptx operons is phoBR dependent. Therefore, the htx and ptx operons, encoding products for the oxidation of the inorganic reduced P compounds hypophosphite and phosphite, are novel members of the Pho regulon of P. stutzeri, thus providing convincing evidence that the physiological role of these genes is Pi acquisition from an alternate phosphorus source.
A growth defect was observed for the ΔphoB, ΔphoR, and ΔphoBR mutants of P. stutzeri on 0.1 mM Pi compared to the growth of the wild type. The mutant phenotype appeared to be due to a decrease in the maximum growth yield rather than to an increase in doubling time. The inability of the mutants to continue growing suggests that a high-affinity Pi transport system required for growth on low levels of Pi is no longer expressed in the absence of PhoBR. Although nothing is known about Pi transport in P. stutzeri, PhoBR-dependent high-affinity Pi transport systems have been characterized for numerous bacteria, including E. coli and several pseudomonads (23, 35, 37). It is reasonable to suspect that P. stutzeri also possesses such a transport system as part of its Pho regulon that would be required for growth on limiting Pi.
The sequence similarity between ptxE and other regulatory proteins of the LysR family (32% amino acid sequence identity to CbbR of Rhizobium meliloti), in addition to the presence of a conserved helix-turn-helix motif for DNA binding (9), suggests that PtxE might act as a regulator of the htx or ptx genes. Despite these properties, PtxE has no apparent role in the regulation of the htx or ptx genes in response to Pi starvation, as seen by the absence of a measurable effect on the expression levels of these genes in the wild-type and ΔptxE strains in the presence or absence of phoBR. Perhaps this observation should not be surprising considering that no genes of the Pho regulon have yet been found to be under individual regulatory control in addition to the regulatory effects exerted by phoBR (30).
The data presented in this report, in addition to the large numbers of bacterial species reported to grow on hypophosphite and phosphite as sole sources of phosphorus, provide strong evidence for both the presence of these reduced phosphorus compounds in the environment and the significant role that they play as alternate phosphorus sources for environmental organisms.
Acknowledgments
We are grateful to Barry Wanner for generously providing strains and plasmids and to Marlena Wilson, Adam Guss, and Shannon Neuhaus for their efforts in transposon mutagenesis.
This work was supported by grant GM59334 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Adams, F., and J. P. Conrad. 1953. Transition of phosphite to phosphate in soils. Soil Sci. 75:361-371. [Google Scholar]
- 2.Argast, M., and W. Boos. 1980. Coregulation in Escherichia coli of a novel transport system for sn-glycerol-3-phosphate and outer membrane protein Ic (e, E) with alkaline phosphatase and phosphate-binding protein. J. Bacteriol. 143:142-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Current protocols in molecular biology, vol. 1 and 2. John Wiley & Sons, New York, N.Y.
- 4.Brunner, U., T. G. Chasteen, P. Ferloni, and R. Bachofen. 1995. Chromatographic determination of phosphine (PH3) and hydrogen sulfide (H2S) in the headspace of anaerobic bacterial enrichments using flame photometric detection. Chromatographia 40:399-403. [Google Scholar]
- 5.Costas, A. M., A. K. White, and W. W. Metcalf. 2001. Purification and characterization of a novel phosphorus-oxidizing enzyme from Pseudomonas stutzeri WM88. J. Biol. Chem. 276:17429-17436. [DOI] [PubMed] [Google Scholar]
- 6.Foster, T. L., L. Winans, Jr., and S. J. Helms. 1978. Anaerobic utilization of phosphite and hypophosphite by Bacillus sp. Appl. Environ. Microbiol. 35:937-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gassman, G., and F. Schorn. 1993. Phosphine from harbor surface sediments. Naturwissenschaften 80:78-80. [Google Scholar]
- 8.Haldimann, A., and B. L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henikoff, S., G. W. Haughn, J. M. Calvo, and J. C. Wallace. 1988. A large family of bacterial activator proteins. Proc. Natl. Acad. Sci. USA 85:6602-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imazu, K. 1998. Enhanced utilization of phosphonate and phosphite by Klebsiella aerogenes. Appl. Environ. Microbiol. 64:3754-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsen, R. A., M. M. Wilson, A. M. Guss, and W. W. Metcalf. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193-201. [DOI] [PubMed] [Google Scholar]
- 12.Lauwers, A. M., and W. Heinen. 1977. Alterations of alkaline phosphatase activity during adaptation of Escherichia coli to phosphite and hypophosphite. Arch. Microbiol. 112:103-107. [DOI] [PubMed] [Google Scholar]
- 13.Makino, K. 1986. Nucleotide sequence of the phoB gene, the positive regulatory gene for the phosphate regulon of Escherichia coli K-12. J. Mol. Biol. 190:37-44. [DOI] [PubMed] [Google Scholar]
- 14.Makino, K., H. Shinagawa, M. Amemura, T. Kawamoto, M. Yamada, and A. Nakata. 1989. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J. Mol. Biol. 210:551-559. [DOI] [PubMed] [Google Scholar]
- 15.Makino, K., H. Shinagawa, M. Amemura, S. Kimura, A. Nakata, and A. Ishihama. 1988. Regulation of the phosphate regulon of Escherichia coli. Activation of pstS transcription by PhoB protein in vitro. J. Mol. Biol. 203:85-95. [DOI] [PubMed] [Google Scholar]
- 16.Makino, K., H. Shinagawa, M. Amemura, and A. Nakata. 1986. Nucleotide sequence of the phoR gene, a regulatory gene for the phosphate regulon of Escherichia coli. J. Mol. Biol. 192:549-556. [DOI] [PubMed] [Google Scholar]
- 17.Malacinski, G., and W. A. Konetzka. 1966. Bacterial oxidation of orthophosphite. J. Bacteriol. 91:578-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metcalf, W. W., W. Jiang, L. L. Daniels, S. K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 19.Metcalf, W. W., and B. L. Wanner. 1991. Involvement of the Escherichia coli phn (psiD) gene cluster in assimilation of phosphorus in the form of phosphonates, phosphite, Pi esters, and Pi. J. Bacteriol. 173:587-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metcalf, W. W., and R. S. Wolfe. 1998. Molecular genetic analysis of phosphite and hypophosphite oxidation by Pseudomonas stutzeri WM88. J. Bacteriol. 180:5547-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morton, S. C., D. Glindemann, and M. A. Edwards. 2003. Phosphates, phosphites, and phosphides in environmental samples. Environ. Sci. Technol. 37:1169-1174. [DOI] [PubMed] [Google Scholar]
- 23.Nikata, T., Y. Sakai, K. Shibat, J. Kato, A. Kuroda, and H. Ohtake. 1996. Molecular analysis of the phosphate-specific transport (pst) operon of Pseudomonas aeruginosa. Mol. Gen. Genet. 250:692-698. [DOI] [PubMed] [Google Scholar]
- 24.Qi, Y., Y. Kobayashi, and F. M. Hulett. 1997. The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the pho regulon. J. Bacteriol. 179:2534-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 26.Steed, P. M., and B. L. Wanner. 1993. Use of the rep technique for allele replacement to construct mutants with deletions of the pstSCAB-phoU operon: evidence of a new role for the PhoU protein in the phosphate regulon. J. Bacteriol. 175:6797-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Surin, B. P., H. Rosenberg, and G. B. Cox. 1985. Phosphate-specific transport system of Escherichia coli: nucleotide sequence and gene-polypeptide relationships. J. Bacteriol. 161:189-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsubota, G. 1959. Phosphate reduction in the paddy field I. Soil Plant Food 5:10-15. [Google Scholar]
- 30.Wanner, B. L. 1993. Gene regulation by phosphate in enteric bacteria. J. Cell Biochem. 51:47-54. [DOI] [PubMed] [Google Scholar]
- 31.Wanner, B. L. 1992. Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J. Bacteriol. 174:2053-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wanner, B. L. 1986. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J. Mol. Biol. 191:39-58. [DOI] [PubMed] [Google Scholar]
- 33.Wanner, B. L., and J. A. Boline. 1990. Mapping and molecular cloning of the phn (psiD) locus for phosphonate utilization in Escherichia coli. J. Bacteriol. 172:1186-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White, A. K., and W. W. Metcalf. 2002. Isolation and biochemical characterization of hypophosphite/2-oxoglutarate dioxygenase. A novel phosphorus-oxidizing enzyme from Pseudomonas stutzeri WM88. J. Biol. Chem. 277:38262-38271. [DOI] [PubMed] [Google Scholar]
- 34a.White, A. K., and W. W. Metcalf. 2004. Two C-P Lyase Operons in Pseudomonas stutzeri and their roles in the oxidation of phosphonates, phosphite, and hypophosphite. J. Bacteriol. 186:4730-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willsky, G. R., and M. H. Malamy. 1980. Characterization of two genetically separable inorganic phosphate transport systems in Escherichia coli. J. Bacteriol. 144:356-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willsky, G. R., and M. H. Malamy. 1976. Control of the synthesis of alkaline phosphatase and the phosphate-binding protein in Escherichia coli. J. Bacteriol. 127:595-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, H., H. Kosaka, J. Kato, A. Kuroda, T. Ikeda, N. Takiguchi, and H. Ohtake. 1999. Cloning and characterization of Pseudomonas putida genes encoding the phosphate-specific transport system. J. Biosci. Bioeng. 87:273-279. [DOI] [PubMed] [Google Scholar]
- 38.Yakovleva, G. M., S. K. Kim, and B. L. Wanner. 1998. Phosphate-independent expression of the carbon-phosphorus lyase activity of Escherichia coli. Appl. Microbiol. Biotechnol. 49:573-578. [DOI] [PubMed] [Google Scholar]