Abstract
Sequencing of a cellulosome-integrating gene cluster in Acetivibrio cellulolyticus was completed. The cluster contains four tandem scaffoldin genes (scaA, scaB, scaC, and scaD) bounded upstream and downstream, respectively, by a presumed cellobiose phosphorylase and a nucleotide methylase. The sequences and properties of scaA, scaB, and scaC were reported previously, and those of scaD are reported here. The scaD gene encodes an 852-residue polypeptide that includes a signal peptide, three cohesins, and a C-terminal S-layer homology (SLH) module. The calculated molecular weight of the mature ScaD is 88,960; a 67-residue linker segment separates cohesins 1 and 2, and two ∼30-residue linkers separate cohesin 2 from 3 and cohesin 3 from the SLH module. The presence of an SLH module in ScaD indicates its role as an anchoring protein. The first two ScaD cohesins can be classified as type II, similar to the four cohesins of ScaB. Surprisingly, the third ScaD cohesin belongs to the type I cohesins, like the seven ScaA cohesins. ScaD is the first scaffoldin to be described that contains divergent types of cohesins as integral parts of the polypeptide chain. The recognition properties among selected recombinant cohesins and dockerins from the different scaffoldins of the gene cluster were investigated by affinity blotting. The results indicated that the divergent types of ScaD cohesins also differ in their preference of dockerins. ScaD thus plays a dual role, both as a primary scaffoldin, capable of direct incorporation of a single dockerin-borne enzyme, and as a secondary scaffoldin that anchors the major primary scaffoldin, ScaA and its complement of enzymes to the cell surface.
Cellulosomes are multienzyme complexes that are designed to efficiently degrade cellulose and related plant cell wall polysaccharides (5-10, 18-20, 22, 28, 51, 52). Bacterial cellulosomes are organized by means of a special type of subunit, the scaffoldin, which is comprised of an array of cohesin modules. The cohesin interacts selectively and tenaciously with a complementary type of domain, the dockerin, which is borne by each of the cellulosomal enzyme subunits. The integrity of the complex is thus maintained by the cohesin-dockerin interaction (4).
In Clostridium thermocellum, the first cellulosome to have been described, the scaffoldin gene product (CipA) is located in a gene cluster (24) which also includes a series of genes that encode cohesin-containing anchoring proteins downstream of CipA. These anchoring proteins contain one or more cohesins and a C-terminal S-layer homology (SLH) module that mediates attachment to the cell surface (21). In contrast to the arrangement of the gene cluster in C. thermocellum, for other cellulosome-producing species, such as Clostridium cellulolyticum and Clostridium cellulovorans, gene clusters that comprise a series of genes coding for cellulosomal (dockerin-containing) enzymes instead of anchoring proteins have been described (2, 11, 19, 53). The difference between the scaffoldins of the latter bacteria and those of C. thermocellum is the presence of a distinctive dockerin on CipA, which is lacking on the other scaffoldin species. The CipA dockerin is known to interact with the cohesins of the anchoring proteins, and the complex is thus bound to the cell surface (36, 49, 50).
We have recently described the upstream portion of a gene cluster in the cellulosome-producing, anaerobic mesophile Acetivibrio cellulolyticus. The cluster included a gene coding for a novel scaffoldin, CipV (15), which was subsequently renamed scaA, followed downstream by two tandem genes, scaB and scaC (57). Like CipA of C. thermocellum (25), the A. cellulolyticus scaA scaffoldin contained a C-terminal dockerin domain. This suggested that the gene cluster would resemble the multiple-scaffoldin model observed for C. thermocellum. Indeed, ScaA binds to ScaB, which in turn binds to ScaC via successive cohesin-dockerin interactions, and the multiple cohesins on each of the latter scaffoldins indicates an amplification of the number of enzymes that can be incorporated into the A. cellulolyticus cellulosome (57). Since ScaC contains a resident SLH module, the entire complex would be anchored to the cell surface.
In the present study, we succeeded in completing the sequence of the final gene of the A. cellulolyticus cluster, scaD. The three scaD cohesins are of two different structural types and exhibit divergent specificities. Like scaC, scaD was also found to contain C-terminal segments encoding an SLH module, and hence the respective gene product would presumably act as anchoring protein. The data indicate that the A. cellulolyticus cell surface cellulosome system is characterized by at least two alternative suprastructural arrangements.
MATERIALS AND METHODS
Growth of A. cellulolyticus and isolation of extracellular proteins.
A. cellulolyticus ATCC 33288 was grown on cellobiose as described earlier (15). The cells were used for subsequent protein profiling, preparation of genomic DNA, and scanning electron microscopy. Cell-free culture supernatant fluids from cellobiose-grown cells were absorbed onto amorphous cellulose (29) to isolate extracellular cellulose-binding proteins. These proteins presumably included cellulosome-related components and free enzymes containing a cellulose-binding domain.
Scanning electron microscopy.
Cells were treated with cationized ferritin prior to examination by electron microscopy as described previously (30, 31).
Isolation of genomic DNA and construction of genomic libraries.
A. cellulolyticus genomic libraries were constructed by using the Lambda ZAP II undigested vector kit for an XbaI library and the Uni-ZAP XR vector kit for an EcoRI-XhoI library, according to the manufacturer's instructions (Stratagene Cloning Systems, La Jolla, Calif.).
PCR and subcloning.
PCRs were performed by using a Master Personal device (Eppendorf, Hamburg, Germany) at various annealing temperatures (50 to 60°C). DNA samples were purified by using either a QIAquick PCR purification kit (Qiagen Inc., Valencia, Calif.), or an agarose gel DNA extraction kit (Roche Diagnostics Corp., Indianapolis, Ind.). Plasmids were purified by using the High Puri plasmid isolation kit (Boehringer, Mannheim, Germany). PCR fragments were cloned by using the pGEM-T vector system 1 (Promega Corp., Madison, Wis.). Escherichia coli TG1 or XL-1 strains were used as host cells for transformation.
Library screening.
Two A. cellulolyticus genomic libraries were screened according to the protocol described in the DIG Application Manual for Filter Hybridization (Roche Molecular Biochemicals). For EcoRI-XhoI library screening, a PCR fragment of 450 bp from primers ACAnF15 and ACAnR13 (Table 1) was labeled and used as a probe. A positive plaque was identified and transferred to the phagemid, and the resulting 6-kb insert was sequenced. This insert contained the terminal portion of scaB, the entire scaC reading frame, and the beginning of a new reading frame (57). Another XbaI library was screened in order to sequence additional portions downstream of scaC. For screening of the XbaI library, a 260-bp PCR product was prepared from primers F-AC-ScaD-1 and R-AC-ScaD-1 (see Table 1 for details). The ∼10-kb insert was obtained from positive plaques, and the 4-kb C-terminal portion was sequenced. The residual 6-kb N-terminal segment was already known from a previous report (57).
TABLE 1.
Primers used in this study
| Primer | Nucleotide sequencea | Location | Comment |
|---|---|---|---|
| ACAn-R13 | CTACTACCATCTACTGGGGC | Coh-1, scaC | Probe for library screening |
| ACAn-F15 | CCCTGTTGAAGAGAAAGAAG | Doc, scaB | Probe for library screening |
| R-AC-SCA9 | CACATCCGCACCTGATAACTTAGCC | SLH-1, scaC | Sequencing |
| R-AC-SCA8 | CAAAACTGTTCCATATTCAAGTAA | Intermediary region be- ween scaC and scaD | Sequencing |
| R-AC-SCA7 | CCATTGTTATATAAGACTCATCGCTTGC | Coh-1, scaD | Sequencing |
| R-AC-SCA6 | CCAACAACAACTGACGTCTTATC | Coh-2, scaD | Sequencing |
| F-AC-ScaD-1 | CAGTTACATATCAATAAAGTTAG | Coh-2, scaD | Probe for library screening |
| R-AC-ScaD-1 | GAATTCCTTTGTCAACATGATTATC | Coh-3, scaD | Probe for library screening |
| F1-SCAD | GGTGATGAAGTAACGGTGCCTGTG | Coh-3, scaD | Sequencing |
| F2-SCAD | AACCTACTGAGGATATACCTGCTGG | Linker between Coh-3 and SLH-1 | Sequencing |
| F3-SCAD | GATGCAAGTACAGGCTTGAG | SLH-2 | Sequencing |
| R1-SCAD | CCAATCATAGCTGCTTCAATCG | C terminal of 10 kb (downstream scaD) | Sequencing |
| R2-SCAD | CTGCTGTTCTAAGCCACAAATTCG | C terminal of 10 kb (downstream scaD) | Sequencing |
| R3-SCAD | CCCGTTAAATAATCTTTGCTC | C terminal of 10 kb (downstream scaD) | Sequencing |
| F-EX-COH1D | ATATCCATGGAGTCTTATATAACAATGGATCTT | Coh-1, scaD | Expression |
| R-EX-COH1D | AATTCTCGAGTACTTTAACCTTAATATCTGAAGG | Coh-1, scaD | Expression (His tag) |
| F-EX-SCA3D | ATACCATGGTAACACCAAATGGATTCCAAGTA | Coh-3, scaD | Expression |
| R-EX-SCA3D | ATACTCGAGTGTTTTTACTGACGTAGTCTTTGT | Coh-3, scaD | Expression (His tag) |
| F-9dxyn-docA | TATAGGTACCAGGTGACATGGCAATAGGCGGTACAC | Doc, cipV (scaA) | Expression, fused to xylanase T6 |
| N-9dxyn-docA2 | TATAGGTACCACCTATACTCATGTGGGCAGGTGAC | Doc, cipV (scaA) | Expression, fused to xylanase T6 |
| R-9dxyn-docA | ATAAGGATCCATAGTCTGAAGATACTTTATTGAAG | Doc, cipV (scaA) | Expression, fused to xylanase T6 |
| F-9dxyn-docB | TATGGTACCGCCTAAATTTATATATGGTGATGTT | Doc, scaB | Expression, fused to xylanase T6 |
| R-9dxyn-Acdoc | TATGGATCCTTCTTCTTTCTCTTCAACAGGG | Doc, scaB | Expression, fused to xylanase T6 |
| F-9dxyn-docE | ATAGGTACCACCTGCACAATACGTATATGGTGAT | Doc, GH9B | Expression, fused to xylanase T6 |
| R-9dxyn-docE | CAAGGATCCCTTTTGTACCGGAAACTTTGAGAT | Doc, GH9B | Expression, fused to xylanase T6 |
| M13/pUC(-21) | AACAGCTATGACCATGATTACG | Plasmid | Sequencing |
| M13/pUC(-20) | TGTAAAACGACGGCCAGT | Plasmid | Sequencing |
| T7 | CGCGCGTAATACGACTCACTATAG | Plasmid | Sequencing |
| SP6 | CCAAGCTATTTAGGTGACAC | Plasmid | Sequencing |
Underlining indicates restriction enzyme cleavage sites.
Sequencing.
DNA sequencing was performed either directly on PCR products or on cloned fragments essentially as described earlier (57).
Cloning and overexpression of recombinant proteins.
Desired genes were subcloned into appropriate expression vectors via PCR (see Fig. 1 for details). Either pET28a or pET9d vectors were used for cloning the PCR products, and their intact sequences were verified by DNA sequencing. Cohesin constructs and xylanase fused with the GH9B dockerin were expressed in E. coli BL21(DE3), and xylanase fused to the ScaA or ScaB dockerin were expressed in E. coli BL21(DE3)(pLyS) (Stratagene) (Table 2). The host cells were grown in the presence of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at either 37 or 16°C. Following growth, the cultures were lysed by sonication as described by Ding et al. (17). The expressed proteins were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10 or 12% polyacrylamide) and stained with Coomassie brilliant blue.
FIG. 1.
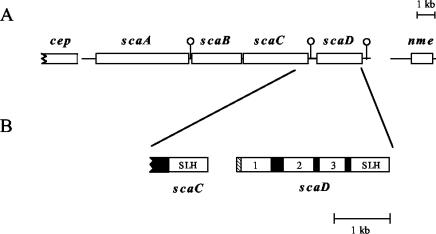
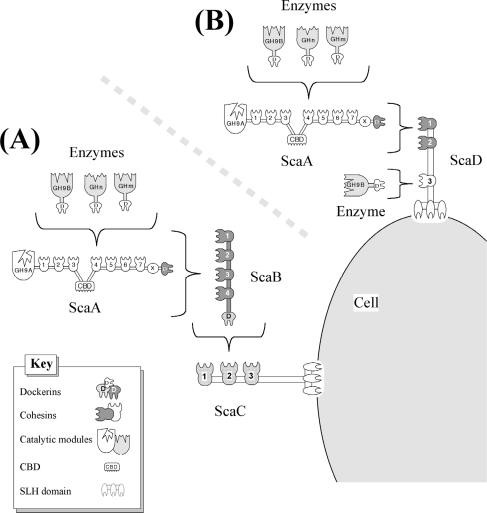
Organization of the A. cellulolyticus cellulosome-integrating cluster. (A) Arrangement of the four scaffoldin genes, bordered upstream by a putative cellobiose phosphorylase gene (cep) and downstream by a putative nucleotide methylase gene (nme). Immediately downstream of scaA, scaC, and scaD are located putative transcription terminators (hairpin loops), designated by lollipop-like symbols. (B) The scaD gene was sequenced in this work. The scaD gene encodes a protein containing a signal peptide (crosshatched), three cohesins (numbered), and a C-terminal SLH module. The modules of ScaD are all separated by well-defined Pro/Thr-rich linker segments (black boxes).
TABLE 2.
Expressed proteins prepared in this study
| Protein | Modular content | Plasmid | Comment |
|---|---|---|---|
| CohD1 | First cohesin of scaD | pET28a | C-terminal His tag |
| CohD3 | Third cohesin of scaD | pET28a | C-terminal His tag |
| Xyn-DocA | Hybrid construct of G. stearothermophilus xylanase T6 harboring the A. cellulolyticus cipV (scaA) dockerin at the C terminus | pET9d | N-terminal His tag |
| Xyn-DocB | Hybrid construct of G. stearothermophilus xylanase T6 harboring the A. cellulolyticus scaB dockerin at the C terminus | pET9d | N-terminal His tag |
| Xyn-DocGH9 | Hybrid construct of G. stearothermophilus xylanase T6 harboring the A. cellulolyticus GH9B dockerin at the C terminus | pET9d | N-terminal His tag |
Immunoblotting.
Proteins were separated by SDS-PAGE, transferred onto nitrocellulose membranes, and treated with peroxidase-conjugated antibody (anti-His[C-terminal]-horseradish peroxidase mouse antibody) as outlined earlier (57).
Protein sequence analysis.
Potential signal sequences were determined with the SignalP version 2.0 program (44). The parameters for molecular weight, theoretical pI, amino acid composition, and extinction coefficient were computed by using the ProtParam Tool (http://www.expasy.org/tools/protparam.html), available via the SWISS-PROT website (3). Identification of protein sequences was performed with the Basic Local Alignment Search Tool (BLAST) (1), available on the website of the National Center for Biotechnology Information (protein-protein BLAST [blastp: http://www.ncbi.nlm.nih.gov/BLAST/]). Multiple-sequence alignments and phylogenetic trees were generated by using the ClustalW program (http://www2.ebi.ac.uk/clustalw/) (26). Sequences for cohesin and SLH modules were obtained from the GenBank website (http://www.ncbi.nlm.nih.gov/) or via the Carbohydrate-Active Enzymes server (CAZy website, http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html), designed by Coutinho and Henrissat (12, 14). The ScaD cohesins were mapped on a background of cohesin modules, as previously described (15, 16, 57). The sequences used to generate the tree for the SLH modules are given in Table 3. Secondary structures of the Thr-rich linker sequences were predicted by using the PredictProtein server at Columbia University (http://cubic.bioc.columbia.edu/predictprotein/).
TABLE 3.
SLH-containing proteins cited in this work.
| Species | Abbreviation | Protein | Description | No. and arrangement of domainsa | Accession no. |
|---|---|---|---|---|---|
| Acetivibrio cellulolyticus | Acece | ScaC | Anchoring protein | 1/2, 2, 1/2 | GenBank AY221113 |
| ScaD | Anchoring protein | 1/2, 2, 1/2 | This work | ||
| Clostridium themocellum | Clotm | OlpA | Anchoring protein | 1/2, 2, 1/2 | SwissProt Q06848 |
| OlpB | Anchoring protein | 1/2, 2, 1/2 | SwissProt Q06852 | ||
| Orf2p | Anchoring protein | 1/2, 2, 1/2 | SwissProt Q06853 | ||
| SdbA | Anchoring protein | 3 | GenBank U49980 | ||
| LicA | Enzyme | 3 | GenBank X89732 | ||
| XynX | Enzyme | 3 | SwissProt P38535 | ||
| Clostridium josui | Clojo | EgA | Enzyme | 1/2, 2 | GenBank D85526 |
| Anaerocellum thermophilum | Anath | ManA | Enzyme | 3 | GenBank Z86105 |
| Bacillus anthracis | Bacan | Eal | S-layer protein | 3 | GenBank X99724 |
| Bacillus sp. | Bacsp | AapT | Enzyme | 3 | GenBank D28467 |
| EgA | Enzyme | 3 | GenBank M27420 | ||
| Bacillus thuringiensis | Bacth | SlpA | S-layer protein | 3 | GenBank AJ249446 |
| Caldicellulosiruptor sp | Calsp | XynB | Enzyme | 2, 1/2 | GenBank AF036923 |
| Cellulomonas fimi | Celfi | Man26A | Enzyme | 1/2, 2 | GenBank AF126471 |
| Thermoanaerobacterium thermosulfurigenes | Theth | PglA | Enzyme | 3 | GenBank U50951 |
See reference 21.
Nucleotide sequence accession numbers.
The DNA sequences for the complete scaD gene and the nme gene have been deposited in the GenBank database under accession numbers AY221114 and AY221111.
RESULTS
Completion of the cellulosome-integrating gene cluster of A. cellulolyticus.
In previous work from our laboratory, the scaA (15), scaB, and scaC (57) genes were sequenced and described, thus establishing the presence of a cellulosome-integrating gene cluster in the A. cellulolyticus genome (Fig. 1A). A new 260-bp probe was designed (based on primers F-AC-ScaD-1 and R-AC-ScaD-1 [Table 1]) and used to screen a lambda ZAPII (XbaI) library in order to continue sequencing of the genome downstream of scaC. A phage containing a 10-kb insert was obtained. Direct sequencing of PCR products derived from this insert provided overlapping parts of the puzzle, allowing completion of the scaD gene plus immediate downstream portions of the genome.
A new reading frame was detected downstream of the scaD stop codon; the gene was sequenced in its entirety, and the product was identified as a putative nucleic acid methylase on the basis of a BLAST search. Most (1.6 kb) of the 2.7-kb intergenic segment was sequenced in straightforward fashion without detection of additional reading frames (the residual 1.1-kb segment was considered inconsequential for the purposes of the present report).
At a distance of 3 kb, upstream of the scaA gene, a 600-bp fragment was sequenced and found by a BLAST search to unambiguously (∼85% identity) represent a gene encoding a cellobiose phosphorylase (cep). A segment of only about 150 bp remained unaccountable, on the basis of the size of the highly conserved cep genes, together with the position of its 600-bp fragment and the previously sequenced 780-bp portion immediately upstream of scaA (15). Thus, no new open reading frames would exist between cep and scaA.
Sequence analysis of the scaffoldin gene cluster from A. cellulolyticus.
Sequence analyses of the scaffoldin gene cluster reveal that the genes are organized in three putative transcriptional units. The scaA gene (cipV) is the first transcriptional unit, with a potential −35 region (TTGTTT) and a −10 region (TATTAA) upstream of the gene. Downstream of the scaA gene, two other genes (scaB and scaC) are oriented in the same direction without any obvious transcriptional initiation signals or transcriptional terminators separating them. A potential promoter upstream of the scaB gene includes a −35 region (TTGGAG), separated by 17 bp from a −10 region (CATAAA), with four out of six bases matching the σA consensus sequence. Downstream of the scaC gene there is a palindromic sequence, corresponding to an mRNA hairpin loop, followed by a clear stretch of T nucleotides, typical of a Rho-independent terminator. Thus, it is likely that scaB and scaC together constitute a second transcriptional unit. The third transcriptional unit is probably monocistronic and encodes the scaD gene. Upstream of the gene is a potential promoter region (5′-TTGTCA…16 bp…CTTAAT-3′), and downstream of the scaD stop codon is a potential Rho-independent transcriptional terminator.
Description of ScaD.
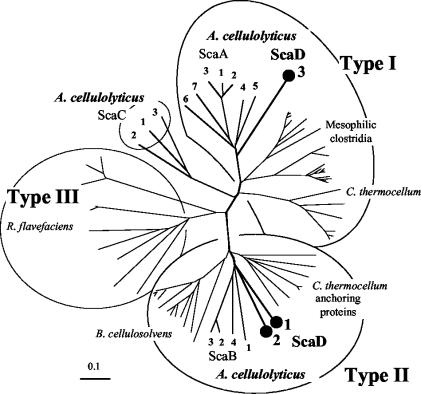
The modular architecture of scaD is shown in Fig. 1B. The scaD gene encodes an 852-residue protein that includes a signal peptide, three cohesins, and a C-terminal SLH module. The predicted scaD signal peptide cleavage site is located between residues 27 and 28 (VNA-SD). The predicted molecular weight of the mature ScaD is 88,960, and the calculated pI of the unfolded polypeptide is 6.59. The three cohesins and SLH module are separated by Pro/Thr-rich linker sequences of 67, 30, and 32 residues, respectively. Like ScaC, ScaD likely serves as an anchoring protein, owing to its SLH module. The nature of the ScaD cohesins is of particular interest. As illustrated in Fig. 2, the first two are similar to the type II cohesins of ScaB (57) and to those of the C. thermocellum anchoring proteins (33, 34, 36, 48) and the Bacteroides cellulosolvens CipBc scaffoldin (16). In contrast, the third ScaD cohesin maps within the type I cohesins, in a position close to those of ScaA (15). ScaD is the first case of a single scaffoldin that carries different types of cohesins.
FIG. 2.
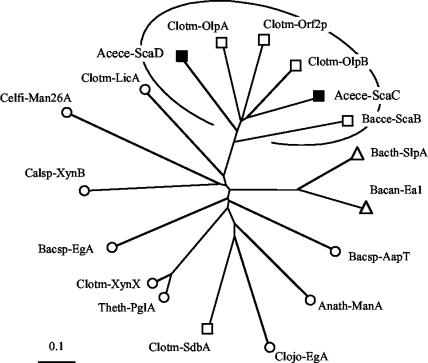
Phylogenetic analysis of the A. cellulolyticus ScaD cohesins. The first two ScaD cohesins map together with the ScaB cohesins on a separate branch of the type II cohesins, radiating from approximately the same bifurcation point. In contrast, the third ScaD cohesin is clearly a member of the type I cohesins, closely aligned to the seven type I cohesins of ScaA. The scale bar indicates the percentage (0.1) of amino acid substitutions. Sources for sequences used in this figure are provided in references 15, 16, 46, and 56.
Specificity of the ScaD cohesins.
The presence of two different types of cohesins in a single scaffoldin polypeptide suggested two different specificities. Since ScaD cohesins 1 and 2 are similar to those of ScaB, the question remained whether they both bind selectively to the same dockerin, i.e., the ScaA dockerin. Likewise, the similarity of ScaD cohesin 3 to those of ScaA raised the question as to whether these cohesins also exhibited the same specificity for the enzyme-borne dockerins. For this purpose, recombinant cohesins (ScaD cohesins 1 and 3) were prepared together with a C-terminal His tag for subsequent isolation and detection (Table 2). The recombinant cohesins were used as probes in affinity blotting and enzyme-linked immunosorbence-based assays.
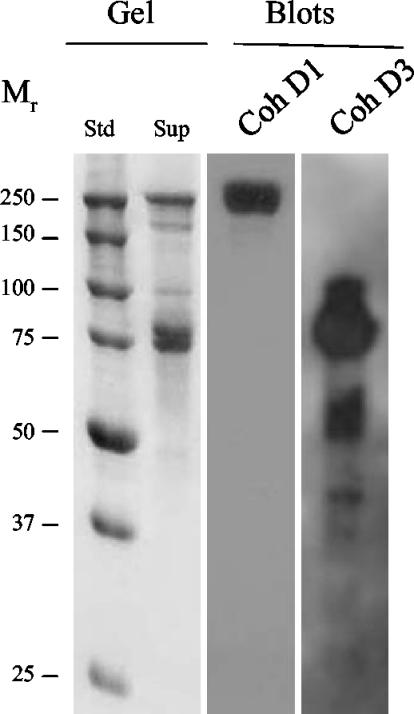
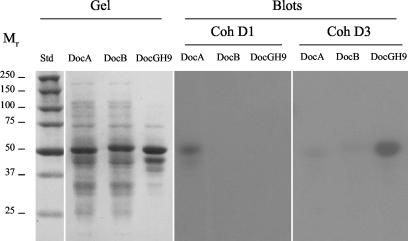
Affinity blotting experiments indicated that the ScaD cohesin 1 probe (CohD1) recognized selectively the high-molecular-mass ScaA band in the supernatant fraction (Fig. 3). The observed labeling appeared to be identical to that determined earlier with ScaB cohesin 1 as a probe (57). The evidence thus suggests that the four ScaB cohesins and the first two ScaD cohesins all recognize the ScaA dockerin. The results were corroborated by using a xylanase-dockerin fusion protein system (A. Mechaly, Y. Barak, T. Handelsman, et al., unpublished results), whereby the CohD1 probe labeled the xylanase hybrid construct containing the dockerin of ScaA but not those of ScaB or the enzyme-borne dockerin (Fig. 4).
FIG. 3.
Affinity blotting of cell-derived proteins, using representative recombinant ScaD cohesins as probes. A. cellulolyticus cells were grown on cellobiose and pelleted, and the supernatant fraction was adsorbed onto phosphoric acid-treated cellulose. Samples were subjected to SDS-PAGE (Gel), and blotted onto nitrocellulose membranes (Blots). Gels were stained with Coomassie brilliant blue. The blots were probed with the designated recombinant protein sample, and labeled bands were detected by chemiluminescence with peroxidase-conjugated, anti-His tag antibody. Probes: CohD1, the first cohesin of ScaD; CohD3, the third cohesin of ScaD. Std, prestained molecular weight markers (in thousands). Sup, original sample of extracellular protein, adsorbed onto cellulose.
FIG. 4.
Affinity blotting of selected dockerin-containing fusion proteins with the two types of recombinant ScaD cohesins. Recombinant fusion proteins, comprising Geobacillus stearothermophilus xylanase T6 and C-terminal dockerins from ScaA (DocA), ScaB (DocB), or the cellulosomal GH9B enzyme (DocGH9), were expressed in an appropriate E. coli host cell system. The ∼50-kDa Xyn-Doc fusion proteins were subjected to SDS-PAGE (Gel), transferred to nitrocellulose membranes (Blots), and probed with the ScaD cohesins (CohD1 and CohD3, respectively) as described in the legend to Fig. 3.
In contrast to CohD1, the ScaD cohesin 3 probe (CohD3) appeared to label several bands, presumably denoting dockerin-bearing enzymes (Fig. 3). The labeling by CohD3 was similar to that reported recently for the ScaA-derived cohesin probe (57). CohD3 appeared to label numerous bands between 40 and 100 kDa; the majority of label concentrated in a band or bands of about 80 kDa. The two Coomassie blue-stained bands in this region were extracted from the gel, the extract was subjected to trypsin digestion, and the tryptic peptides were subjected to mass spectrometry sequence analysis. Two relevant sequences were obtained (TQVYLPSGWTGK and LNNKSGWPAR), which corresponded to segments consistent with family-48 and family-9 glycoside hydrolases, respectively, the latter being distinct in sequence from the corresponding conserved segments from either of the two currently known family-9 enzymes from A. cellulolyticus (i.e., GH9A and GH9B). Indeed, CohD3 selectively labeled the xylanase hybrid construct that harbored the GH9B dockerin domain (Fig. 4). Only negligible labeling of the constructs containing the scaffoldin-derived dockerins (DocA and DocB) was apparent, thus confirming the specificity of the interaction.
Description of the SLH module of ScaD.
The SLH module of A. cellulolyticus ScaD is closely related in sequence to that of ScaC from the same species and to those of other anchoring scaffoldins from C. thermocellum and B. cellulosolvens (Fig. 5). The C. thermocellum anchoring protein, SdbA, represents an exception that maps on the opposite side of the phylogenetic tree among those of the SLH-bearing surface enzymes derived from different gram-positive species. Nevertheless, the similarity in sequence and the resemblance in the number and arrangement of their domains (Table 3) may allude to common structural and/or functional features that are shared by the designated group of anchoring scaffoldins in attaching the respective cellulosome complexes to the cell surface.
FIG. 5.
Comparative phylogenetic analysis of the ScaD-borne SLH module. The A. cellulolyticus SLH modules (filled symbols) map on a separate branch of the phylogenetic tree, together with those of the anchoring proteins of C. thermocellum (OlpA, OlpB, and Orf2p) and B. cellulosolvens ScaB. See Table 3 for sources of sequences used to prepare this figure. Symbols: squares, anchoring proteins; circles, surface-bound enzymes; triangles, surface layer proteins.
DISCUSSION
Two different types of cellulosomal gene clusters are known for cellulosome-producing bacteria (4). The enzyme-linked gene clusters of various mesophilic bacteria, such as C. cellulolyticum and C. cellulovorans, are characterized by a primary scaffoldin gene followed downstream by a series of genes encoding various dockerin-bearing enzymes (11, 19). In contrast, the multiple-scaffoldin gene clusters, such as those observed in C. thermocellum, B. cellulosolvens, and Ruminococcus flavefaciens, comprise two or more genes in tandem that encode scaffoldins, at least one of which appears to anchor the cellulosome onto the bacterial cell surface (17, 23, 46, 47, 56). The cellulosomal gene cluster of A. cellulolyticus is clearly of the latter type. The three initial genes of the complex, scaA, scaB, and scaC, were described earlier (57), and the specificity characteristics of their divergent cohesins and dockerins allowed us to propose a suprastructural model for cellulosome architecture in this bacterium. Owing to the multiple cohesins of the latter scaffoldins, the model implied a dramatic amplification in the number of cellulosomal enzymes that can be incorporated onto the A. cellulolyticus cell surface. According to the model, ScaA acts as the primary scaffoldin, which mediates direct incorporation of the dockerin-bearing enzymes into the complex (Fig. 6A). ScaC serves as an anchoring scaffoldin by virtue of its C-terminal SLH domain. ScaB, on the other hand, interacts with both ScaA and ScaC and thus functions as an adaptor scaffoldin.
FIG. 6.
Schematic model of the proposed interactions among the A. cellulolyticus cellulosomal components and the two modes of attachment to the cell surface. (A) Dockerin-containing enzymes are incorporated into the ScaA scaffoldin via interaction with the ScaA cohesins. The ScaA dockerin binds to the ScaB type II cohesins, and the ScaB dockerin binds to the ScaC cohesins. The latter complex is anchored to the cell surface by the ScaC SLH module. This arrangement was described by Xu et al. (57). (B) In the additional mechanism of attachment, the enzyme-laden ScaA is bound to the type II cohesins of ScaD, which can also accept a single enzyme via its third type I cohesin. The SLH module of ScaD serves to anchor the alternative complex to the cell surface.
The definition of SLH is somewhat confusing and has been incorrectly employed in the literature. Technically, SLH stands for surface layer homology and refers to homology with a specified sequence that is associated with the anchoring to the cell surface of proteins that carry the SLH module (21, 36, 38, 40). The linkage between the SLH sequence and the cell surface anchoring function has indeed been proven experimentally in several cases (13, 35, 36, 38, 39, 41). While the presence of an SLH module likely implies the anchoring function, the reverse is not necessarily true. Thus, by definition, suspected anchoring proteins with no clear sequence homology with SLH domains should not be referred to as such.
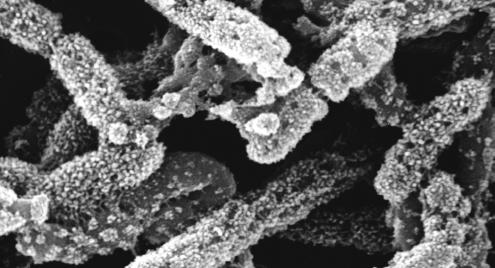
Like ScaC, the ScaD scaffoldin, described in the present report, contains a C-terminal SLH module, which implies that both proteins act as anchoring scaffoldins. However, unlike any other scaffoldin yet described, ScaD contains, in the same polypeptide chain, two different types of cohesin, type I and type II, which exhibit two divergent dockerin-binding specificities. The consequence of this molecular arrangement is that ScaD can integrate two primary scaffoldins via its resident type II cohesins and, additionally, a single dockerin-containing enzyme via the type I cohesin (Fig. 6B). Since each primary scaffoldin represents eight enzymes, the ScaD-anchored cellulosome system of A. cellulolyticus would carry up to 17 enzymes, compared to 96 for the ScaC-anchored system. The particularly elaborate surface topography of A. cellulolyticus (Fig. 7) would appear to reflect the architectural diversity of its cellulosome systems.
FIG. 7.
Scanning electron micrograph of cationized ferritin-treated cells of A. cellulolyticus.
It is interesting to speculate whether the single type I cohesin of ScaD binds a particular type of enzyme in a relatively selective manner. The ScaD cohesin 3 probe appears to bind to an 80-kDa enzyme, consistent perhaps with an unidentified Cel48 enzyme, which has been demonstrated to be a dominant enzyme in several cellulosomal systems (27, 32, 37, 42, 43, 45, 54-56). The latter enzyme has yet to be sequenced in A. cellulolyticus, although a conserved peptide sequence consistent with this type of enzyme has been detected in this work.
The physiological basis for two alternative modes of anchoring cellulases to the cell surface is currently unknown. The presence of the three putative transcriptional units (scaA, scaB-scaC, and scaD) (Fig. 1) would presumably be in line with the two alternative modes of cellulosome architecture as illustrated in Fig. 6. According to this model, there should be three molecules of ScaB per molecule of ScaC, which could correspond with the upstream position of scaB relative to scaC, taking into account possible polarity effects in translation of dicistronic transcriptional units. In fact, the model illustrated in Fig. 6 is but tentative and can be modified in the event of the discovery of additional scaffoldins. In this context, we have previously noted an unidentified 170-kDa scaffoldin that interacted specifically with a ScaC cohesin probe (see Fig. 4 of reference 57). Matrix-assisted laser desorption ionization-time-of-flight analysis of peptides derived from this band (Q. Xu, unpublished data) indicated a close (but nonidentical) sequence similarity to known cohesin sequences from this bacterium, thus implying the presence of at least one additional dockerin-bearing scaffoldin that would bind to ScaC. This unidentified scaffoldin would necessarily be located at an alternative site in the genome, unrelated to the scaffoldin gene cluster described here, and would thus constitute yet another transcriptional unit. The stoichiometry of ScaB versus ScaC would be altered accordingly, and the A. cellulolyticus cellulosome system may well be more intricate than hitherto demonstrated. Future studies will include efforts to clone and characterize further the putative 170-kDa scaffoldin.
Acknowledgments
This research was supported by the Israel Science Foundation (grants 394/03, 771/01, and 446/01) and by a grant from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel. Additional support was provided by the Otto Meyerhof Center for Biotechnology, established by the Minerva Foundation (Munich, Germany).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bagnara-Tardif, C., C. Gaudin, A. Belaich, P. Hoest, T. Citard, and J.-P. Belaich. 1992. Sequence analysis of a gene cluster encoding cellulases from Clostridium cellulolyticum. Gene 119:17-28. [DOI] [PubMed] [Google Scholar]
- 3.Bairoch, A., and R. Apweiler. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28:45-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayer, E. A., J.-P. Belaich, Y. Shoham, and R. Lamed. 2000. The cellulosomes: multi-enzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58:521-554. [DOI] [PubMed] [Google Scholar]
- 5.Bayer, E. A., H. Chanzy, R. Lamed, and Y. Shoham. 1998. Cellulose, cellulases and cellulosomes. Curr. Opin. Struct. Biol. 8:548-557. [DOI] [PubMed] [Google Scholar]
- 6.Bayer, E. A., E. Morag, and R. Lamed. 1994. The cellulosome—a treasure-trove for biotechnology. Trends Biotechnol. 12:378-386. [DOI] [PubMed] [Google Scholar]
- 7.Bayer, E. A., L. J. W. Shimon, R. Lamed, and Y. Shoham. 1998. Cellulosomes: structure and ultrastructure. J. Struct. Biol. 124:221-234. [DOI] [PubMed] [Google Scholar]
- 8.Bayer, E. A., Y. Shoham, and R. Lamed. 2000. The cellulosome—an exocellular organelle for degrading plant cell wall polysaccharides, p. 387-439. In R. J. Doyle (ed.), Glycomicrobiology. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 9.Béguin, P., and M. Lemaire. 1996. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit. Rev. Biochem. Mol. Biol. 31:201-236. [DOI] [PubMed] [Google Scholar]
- 10.Belaich, J.-P., A. Belaich, H.-P. Fierobe, L. Gal, C. Gaudin, S. Pagès, C. Reverbel-Leroy, and C. Tardif. 1999. The cellulolytic system of Clostridium cellulolyticum, p. 479-487. In K. Ohmiya, K. Hayashi, K. Sakka, Y. Kobayashi, S. Karita, and T. Kimura (ed.), Genetics, biochemistry and ecology of cellulose degradation. Uni Publishers Co., Tokyo, Japan.10581040
- 11.Belaich, J. P., C. Tardif, A. Belaich, and C. Gaudin. 1997. The cellulolytic system of Clostridium cellulolyticum. J. Biotechnol. 57:3-14. [DOI] [PubMed] [Google Scholar]
- 12.Bourne, Y., and B. Henrissat. 2001. Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr. Opin. Struct. Biol. 11:593-600. [DOI] [PubMed] [Google Scholar]
- 13.Chauvaux, S., M. Matuschek, and P. Béguin. 1999. Distinct affinity of binding sites for S-layer homologous domains in Clostridium thermocellum and Bacillus anthracis cell envelopes. J. Bacteriol. 181:2455-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes: an integrated database approach, p. 3-12. In H. J. Gilbert, G. J. Davies, B. Henrissat, and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 15.Ding, S.-Y., E. A. Bayer, D. Steiner, Y. Shoham, and R. Lamed. 1999. A novel cellulosomal scaffoldin from Acetivibrio cellulolyticus that contains a family-9 glycosyl hydrolase. J. Bacteriol. 181:6720-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding, S.-Y., E. A. Bayer, D. Steiner, Y. Shoham, and R. Lamed. 2000. A scaffoldin of the Bacteroides cellulosolvens cellulosome that contains 11 type II cohesins. J. Bacteriol. 182:4915-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding, S.-Y., M. T. Rincon, R. Lamed, J. C. Martin, S. I. McCrae, V. Aurilia, Y. Shoham, E. A. Bayer, and H. J. Flint. 2001. Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J. Bacteriol. 183:1945-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doi, R. H., M. Goldstein, S. Hashida, J. S. Park, and M. Takagi. 1994. The Clostridium cellulovorans cellulosome. Crit. Rev. Microbiol. 20:87-93. [DOI] [PubMed] [Google Scholar]
- 19.Doi, R. H., A. Kosugi, K. Murashima, Y. Tamaru, and S. O. Han. 2003. Cellulosomes from mesophilic bacteria. J. Bacteriol. 185:5907-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doi, R. H., and Y. Tamura. 2001. The Clostridium cellulovorans cellulosome: an enzyme complex with plant cell wall degrading activity. Chem. Rec. 1:24-32. [DOI] [PubMed] [Google Scholar]
- 21.Engelhardt, H., and J. Peters. 1998. Structural research on surface layers: a focus on stability, surface layer homology domains, and surface layer-cell wall interactions. J. Struct. Biol. 124:276-302. [DOI] [PubMed] [Google Scholar]
- 22.Felix, C. R., and L. G. Ljungdahl. 1993. The cellulosome—the exocellular organelle of Clostridium. Annu. Rev. Microbiol. 47:791-819. [DOI] [PubMed] [Google Scholar]
- 23.Fujino, T., P. Béguin, and J.-P. Aubert. 1992. Cloning of a Clostridium thermocellum DNA fragment encoding polypeptides that bind the catalytic components of the cellulosome. FEMS Microbiol. Lett. 94:165-170. [DOI] [PubMed] [Google Scholar]
- 24.Fujino, T., P. Béguin, and J.-P. Aubert. 1993. Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in attachment of the cellulosome to the cell surface. J. Bacteriol. 175:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerngross, U. T., M. P. M. Romaniec, T. Kobayashi, N. S. Huskisson, and A. L. Demain. 1993. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol. Microbiol. 8:325-334. [DOI] [PubMed] [Google Scholar]
- 26.Higgins, D., J. Thompson, T. Gibson, J. D. Thompson, D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kakiuchi, M., A. Isui, K. Suzuki, T. Fujino, E. Fujino, T. Kimura, S. Karita, K. Sakka, and K. Ohmiya. 1998. Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J. Bacteriol. 180:4303-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamed, R., and E. A. Bayer. 1988. The cellulosome of Clostridium thermocellum. Adv. Appl. Microbiol. 33:1-46. [Google Scholar]
- 29.Lamed, R., R. Kenig, E. Setter, and E. A. Bayer. 1985. Major characteristics of the cellulolytic system of Clostridium thermocellum coincide with those of the purified cellulosome. Enzyme Microb. Technol. 7:37-41. [Google Scholar]
- 30.Lamed, R., J. Naimark, E. Morgenstern, and E. A. Bayer. 1987. Scanning electron microscopic delineation of bacterial surface topology using cationized ferritin. J. Microbiol. Methods 7:233-240. [Google Scholar]
- 31.Lamed, R., J. Naimark, E. Morgenstern, and E. A. Bayer. 1987. Specialized cell surface structures in cellulolytic bacteria. J. Bacteriol. 169:3792-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamed, R., E. Setter, and E. A. Bayer. 1983. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 156:828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leibovitz, E., and P. Béguin. 1996. A new type of cohesin domain that specifically binds the dockerin domain of the Clostridium thermocellum cellulosome-integrating protein CipA. J. Bacteriol. 178:3077-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leibovitz, E., H. Ohayon, P. Gounon, and P. Béguin. 1997. Characterization and subcellular localization of the Clostridium thermocellum scaffoldin dockerin binding protein SdbA. J. Bacteriol. 179:2519-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemaire, M., I. Miras, P. Gounon, and P. Béguin. 1998. Identification of a region responsible for binding to the cell wall within the S-layer protein of Clostridium thermocellum. Microbiology 144:211-217. [DOI] [PubMed] [Google Scholar]
- 36.Lemaire, M., H. Ohayon, P. Gounon, T. Fujino, and P. Béguin. 1995. OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J. Bacteriol. 177:2451-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, C. C., and R. H. Doi. 1998. Properties of exgS, a gene for a major subunit of the Clostridium cellulovorans cellulosome. Gene 211:39-47. [DOI] [PubMed] [Google Scholar]
- 38.Lupas, A., H. Engelhardt, J. Peters, U. Santarius, S. Volker, and W. Baumeister. 1994. Domain structure of the Acetogenium kivui surface layer revealed by electron crystallography and sequence analysis. J. Bacteriol. 176:1224-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mesnage, S., T. Fontaine, T. Mignot, M. Delepierre, M. Mock, and A. Fouet. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19:4473-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mesnage, S., M. Haustant, and A. Fouet. 2001. A general strategy for identification of S-layer genes in the Bacillus cereus group: molecular characterization of such a gene in Bacillus thuringiensis subsp. galleriae NRRL 4045. Microbiology 147:1343-1351. [DOI] [PubMed] [Google Scholar]
- 41.Mesnage, S., E. Tosi-Couture, and A. Fouet. 1999. Production and cell surface anchoring of functional fusions between the SLH motifs of the Bacillus anthracis S-layer proteins and the Bacillus subtilis levansucrase. Mol. Microbiol. 31:927-936. [DOI] [PubMed] [Google Scholar]
- 42.Morag, E., E. A. Bayer, G. P. Hazlewood, H. J. Gilbert, and R. Lamed. 1993. Cellulase SS (CelS) is synonymous with the major cellobiohydrolase (subunit S8) from the cellulosome of Clostridium thermocellum. Appl. Biochem. Biotechnol. 43:147-151. [DOI] [PubMed] [Google Scholar]
- 43.Morag, E., I. Halevy, E. A. Bayer, and R. Lamed. 1991. Isolation and properties of a major cellobiohydrolase from the cellulosome of Clostridium thermocellum. J. Bacteriol. 173:4155-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nielsen, H., S. Brunak, and G. von Heijne. 1999. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng. 12:3-9. [DOI] [PubMed] [Google Scholar]
- 45.Reverbel-Leroy, C., S. Pagés, A. Belaich, J.-P. Belaich, and C. Tardif. 1997. The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J. Bacteriol. 179:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rincon, M. T., S.-Y. Ding, S. I. McCrae, J. C. Martin, V. Aurilia, R. Lamed, Y. Shoham, E. A. Bayer, and H. J. Flint. 2003. Novel organization and divergent dockerin specificities in the cellulosome system of Ruminococcus flavefaciens. J. Bacteriol. 185:703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rincon, M. T., J. C. Martin, V. Aurilia, S. I. McCrae, G. Rucklidge, M. Reid, E. A. Bayer, R. Lamed, and H. J. Flint. 2004. ScaC, an adaptor protein carrying a novel cohesin that expands the dockerin-binding repertoire of the Ruminococcus flavefaciens 17 cellulosome. J. Bacteriol. 186:2576-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salamitou, S., M. Lemaire, T. Fujino, H. Ohayon, P. Gounon, P. Béguin, and J.-P. Aubert. 1994. Subcellular localization of Clostridium thermocellum ORF3p, a protein carrying a receptor for the docking sequence borne by the catalytic components of the cellulosome. J. Bacteriol. 176:2828-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salamitou, S., O. Raynaud, M. Lemaire, M. Coughlan, P. Béguin, and J.-P. Aubert. 1994. Recognition specificity of the duplicated segments present in Clostridium thermocellum endoglucanase CelD and in the cellulosome-integrating protein CipA. J. Bacteriol. 176:2822-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salamitou, S., K. Tokatlidis, P. Béguin, and J.-P. Aubert. 1992. Involvement of separate domains of the cellulosomal protein S1 of Clostridium thermocellum in binding to cellulose and in anchoring of catalytic subunits to the cellulosome. FEBS Lett. 304:89-92. [DOI] [PubMed] [Google Scholar]
- 51.Schwarz, W. H. 2001. The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56:634-649. [DOI] [PubMed] [Google Scholar]
- 52.Shoham, Y., R. Lamed, and E. A. Bayer. 1999. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 7:275-281. [DOI] [PubMed] [Google Scholar]
- 53.Tamaru, Y., S. Karita, A. Ibrahim, H. Chan, and R. H. Doi. 2000. A large gene cluster for the Clostridium cellulovorans cellulosome. J. Bacteriol. 182:5906-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, W. K., K. Kruus, and J. H. D. Wu. 1993. Cloning and DNA sequence of the gene coding for Clostridium thermocellum cellulase SS (CelS), a major cellulosome component. J. Bacteriol. 175:1293-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, W. K., and J. H. Wu. 1993. Structural features of the Clostridium thermocellum cellulase SS gene. Appl. Biochem. Biotechnol. 40:149-158. [DOI] [PubMed] [Google Scholar]
- 56.Xu, Q., E. A. Bayer, M. Goldman, R. Kenig, Y. Shoham, and R. Lamed. 2004. Architecture of the Bacteroides cellulosolvens cellulosome: description of a cell-surface anchoring scaffoldin and a family-48 cellulase. J. Bacteriol. 186:968-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu, Q., R. Kenig, Y. Shoham, E. A. Bayer, and R. Lamed. 2003. The cellulosome system of Acetivibrio cellulolyticus includes a novel type of adaptor protein and a cell-surface anchoring protein. J. Bacteriol. 185:4548-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]