Abstract
Ciliates from the genus Mesodinium are globally distributed in marine and freshwater ecosystems and may possess either heterotrophic or mixotrophic nutritional modes. Members of the Mesodinium major/rubrum species complex photosynthesize by sequestering and maintaining organelles from cryptophyte prey, and under certain conditions form periodic or recurrent blooms (= red tides). Here, we present an analysis of the genetic diversity of Mesodinium and cryptophyte populations from 10 environmental samples (eight globally dispersed habitats including five Mesodinium blooms), using group-specific primers for Mesodinium partial 18S, ITS, and partial 28S rRNA genes as well as cryptophyte large subunit RuBisCO genes (rbcL). In addition, 22 new cryptophyte and four new M. rubrum cultures were used to extract DNA and sequence rbcL and 18S-ITS-28S genes, respectively, in order to provide a stronger phylogenetic context for our environmental sequences. Bloom samples were analyzed from coastal Brazil, Chile, two Northeastern locations in the United States, and the Pribilof Islands within the Bering Sea. Additionally, samples were also analyzed from the Baltic and Barents Seas and coastal California under non-bloom conditions. Most blooms were dominated by a single Mesodinium genotype, with coastal Brazil and Chile blooms composed of M. major and the Eastern USA blooms dominated by M. rubrum variant B. Sequences from all four blooms were dominated by Teleaulax amphioxeia-like cryptophytes. Non-bloom communities revealed more diverse assemblages of Mesodinium spp., including heterotrophic species and the mixotrophic Mesodinium chamaeleon. Similarly, cryptophyte diversity was also higher in non-bloom samples. Our results confirm that Mesodinium blooms may be caused by M. major, as well as multiple variants of M. rubrum, and further implicate T. amphioxeia as the key cryptophyte species linked to these phenomena in temperate and subtropical regions.
Keywords: Mesodinium, Teleaulax, cryptophytes, ciliates, acquired phototrophy, mixotrophy, red tides, ciliate genetic diversity
Introduction
Species belonging to the genus Mesodinium are among the most widely distributed and abundant marine ciliates in coastal and estuarine ecosystems (Leppanen and Bruun, 1986; Sanders, 1995; Bell and Laybourn-Parry, 1999). Red water blooms of M. rubrum-like ciliates (= Myrionecta rubra) have been recorded since Darwin’s journey on the Beagle (Darwin, 1906), and are recurrent features in many coastal ecosystems (Lindholm, 1978; Crawford et al., 1997; Herfort et al., 2011a). While M. rubrum has historically been reported as a single species from numerous global locations (Taylor et al., 1971), cryptic diversity has been suspected within the M. rubrum morphospecies for some time, due primarily to variability in cell size (Leegaard, 1920; Lindholm, 1985; Rychert, 2004). Previously, only one environmental study of the genetic diversity of the Mesodiniidae has been published, and it focused only on the Mesodinium rubrum/major complex within the Columbia River Estuary (Herfort et al., 2011b). Phylogenetic analysis of rRNA genes spanning the internally transcribed spacer region (ITS) have demonstrated that M. rubrum is actually a species complex, composed of at least six major clades (Herfort et al., 2011b; Garcia-Cuetos et al., 2012). One of these clades was described as a new species, M. major, based on molecular and ultrastructural characteristics, and is larger and has more plastids than M. rubrum (Garcia-Cuetos et al., 2012). Additionally, a new mixotrophic species, M. chameleon, was described from primarily benthic habitats, with unique cilia/kinetid structures as well as plastid type and organization of cryptophycean organelles (Moestrup et al., 2012).
Cryptophytes are known to be important components of phytoplankton communities in coastal ocean ecosystems, especially in low light, estuarine, or high latitude environments (Buma et al., 1992; Mallin, 1994; Adolf et al., 2006). While little is known regarding temporal or spatial trends of cryptophyte diversity, Teleaulax, Plagioselmis, and Hemiselmis are commonly encountered in marine environments (Hill et al., 1992; Cerino and Zingone, 2007; Metfies et al., 2010). Despite the well-established connection between Teleaulax-like cryptophytes and growth of M. rubrum (Gustafson et al., 2000; Johnson and Stoecker, 2005; Smith and Hansen, 2007), relatively few studies have documented their relationship in nature. Single cell PCR of M. rubrum from both coastal Japan and the Columbia River Estuary, have revealed predominantly Teleaulax amphioxeia plastids (Nishitani et al., 2010; Herfort et al., 2011b).
All photosynthetic Mesodinium spp. are thought to harbor only cryptophyte organelles, which they acquire through feeding on free-living prey (Gustafson et al., 2000). When acquiring organelles from cryptophyte prey, M. rubrum-like ciliates also retain mitochondria, cytoplasm, and the nucleus (Johnson, 2011). The nucleus, referred to as a kleptokaryon, remains transcriptionally active and appears to facilitate functional control and division of stolen organelles (Johnson et al., 2007; Lasek-Nesselquist et al., 2015). However, relatively few strains of the M. major/rubrum complex have been successfully cultured and studied in detail.
Mesodinium spp. are well-documented to form blooms in numerous coastal regions, particularly in estuaries (Crawford et al., 1997; Herfort et al., 2011a) and coastal upwelling zones (Ryther, 1967; Packard et al., 1978). Blooms of Mesodinium are highly productive (Smith and Barber, 1979) and form dynamic aggregations within the water column (Crawford and Purdie, 1992). M. rubrum-like ciliates can form thin layers in stratified surface waters (Sjöqvist and Lindholm, 2011), concentrate around down-welling frontal regions (Packard et al., 1978), and are capable of self-retention within estuarine systems by vertically migrating to avoid tidal flushing (Crawford and Purdie, 1992). These behaviors are due to M. rubrum’s astounding motility, which allow the ciliate to move at speeds of ∼400 body lengths s-1 through jumping (Fenchel and Hansen, 2006). One of the best-studied recurrent M. rubrum bloom locations is within the Columbia River estuary in the Pacific Northwest of the United States. Blooms in this system have been described to occur annually during late summer, and appear to be caused by only one of the five known genotypes of M. rubrum found in the estuary (Herfort et al., 2011b).
Here we present an analysis of the genetic diversity of Mesodinium and cryptophyte algal communities from both bloom and non-bloom conditions, in order to shed light on which species and variants of each group are associated with the “red tide” phenomenon. We designed new Mesodinium primers are capable of amplifying all known species from the Mesodiniidae family, rather than the M. rubrum/major complex only (Herfort et al., 2011b). We also designed one new cryptophyte rbcL primer in order to better anneal with major marine groups of these flagellates, since previous studies (Hoef-Emden, 2005) were focused on genera dominant in freshwater as well. This study establishes a framework for assessing the biogeography of Mesodiniidae genetic diversity and provides new insights into the cryptic diversity of these ciliates and cryptophyte algae.
Materials and Methods
Collection of Cell Material
Environmental samples analyzed for Mesodinium and cryptophyte diversity were opportunistically gathered from various sources. In most cases (BR, NC, Bar-M4, GF-LL3a, GF-XVI, TV), samples were collected from surface water preserved using 5–10% Lugol’s fixative. However, samples were also collected onto a 0.2 μm SterivexTM filter (EMD Millipore, Billerica, MA USA) (CL) or centrifuged to form a cell pellet (LIS), and kept frozen until analysis. Previous research has shown that Lugol’s preserved material is sufficient for DNA extraction and quantitative (q) PCR assays, and while sensitivity of the assays decreased overtime, positive amplification was still possible for several months of sample storage at room temperature (Bowers et al., 2000). The sample from SGI, however, was unpreserved and shipping of the sample was slow due to the geographical isolation of the collection site. Upon the arrival of the SGI sample it was immediately centrifuged and the pellet was frozen until DNA was extracted.
Cultures Used for Phylogenetic Analysis
Cryptophyte and Mesodinium spp. cultures were grown in the lab for DNA extraction and sequencing of rbcL or rRNA gene fragments for phylogenetic comparison with our environmental data. Cryptophyte cultures including Teleaulax acuta (SCCAP K-1486), T. amphioxeia (CCMP 1170, GCEPO1), Geminigera cryophila (CCMP 2564), Chroomonas sp. (CCMP 270), Hemiselmis pacific (CCMP 706), H. andersenii (CCMP 439), H. rufescens (CCMP 440), Hemiselmis sp. (SUR21-C3), Hemiselmis sp. (NR11), and Hanusia phi (CCMP 325) were grown in the lab in F/2-Si at 18°C on a 14:10 L:D cycle, as were M. rubrum cultures AND-A0711 and CBJR05. Additionally, the M. rubrum cultures were maintained with GCEPO1, by feeding the ciliate weekly with a 1–5% volume addition of log-phase cryptophyte culture. A Falcomonas sp. culture (CCMP 2293) was also grown on F/2 without added Si (F/2-Si), but at 4°C and constant light. All of the above cryptophyte and Mesodinium cultures were harvested for DNA extraction by centrifugation of 10 ml of dense culture within a 15 ml Falcon tube at 4000 RPM for 15 min, the supernatant decanted, and the cell pellet frozen until processed. Cultures of Teleaulax gracilis (Cr6EHU), T. minuta (Cr8EHU), T. cf. merimbula (Cr59EHU), aff. Teleaulax (Cr22EHU), Plagioselmis nannoplanctica (Cr50EHU), P. cf. prolonga (Cr10EHU, Cr127EHU, Cr143EHU, Cr194EHU) and Urgorri complanatus (Cr1EHU) were grown in the same light and temperature conditions described above, on a modified F/2-Si (enriched with soil extract and selenium at 0.006 mM Na2SeO3 final concentration) at a salinity of 30 (P. nannoplanctica strain grown in freshwater), and were preserved in ethanol prior to DNA extraction and analysis of the rbcL gene. A live sample of M. rubrum culture MR-INO200702 and a 5% Lugol’s-preserved sample of culture MR-MAL01 were also acquired for analysis. For all cultures, approximately 10–20 ml of dense stationary-phase culture was collected by centrifugation (4000 RPM, 10 min) and frozen until extraction.
DNA Extraction and Gene Fragment Amplification and Sequencing
Nucleic acids of environmental samples were extracted from frozen cells collected by centrifugation (4000 RPM, 10 min) or on SterivexTM filters (EMD Millipore) using either the PowerWater Sterivex DNA Isolation Kit (MO BIO Laboratories, Inc.) or a hot detergent lysis method as described by Gast et al. (2004), modified to exclude zirconia-silica bead disruption. All cultures as well as the North Carolina Bloom sample were extracted using the Qiagen DNeasy Plant Mini Kit. PCR was conducted using GoTaq (Promega) or GoTaq G2 Hot Start mix in 50 mL reactions, with a final concentration of 2.5 mM MgCl2, 200 μM dNTPs, 2.5 U GoTaq Flexi polymerase, and 0.1 μM primers for normal and 0.2 μM for hot start. Primers for Mesodinium spp. (Table 1) were designed to amplify the majority of the SSU and LSU rRNA genes, and the entire ITS region, resulting in a ∼1880 bp amplicon. Gene fragments of the large subunit of the ribulose-1,5-bisphosphate carboxylase oxygenase gene (rbcL) were PCR amplified from DNA extracts using primers targeting cryptophyte plastids. A new primer, crypt_rbcLR2 (5′-CAGTGAATACCACCTGAAGCTA-3′), designed using an alignment of cryptophyte rbcL sequences (See Supplementary Data Sheet 1 for accession numbers) was used in combination with L2F (Hoef-Emden et al., 2005). PCR conditions were: 95°C for 5 min followed by 40 cycles of 95°C for 60 s, 55°C for 60 s, and 72°C for 90 s followed by 72°C for 7 min. The genus-specific primers MESO_245F and MESO_28S_R were used to amplify a combined fragment of the Mesodinium spp. 18S-ITS-28S genes. PCR conditions were: 95°C for 5 min followed by 40 cycles of 95°C for 60 s, 57°C for 60 s, and 72°C for 90 s followed by 72°C for 7 min. PCR products were visualized by agarose gel electrophoresis and later excised and purified from the gels using the Zymoclean Gel DNA Recovery Kit (Zymo Research). Clone libraries were constructed from gel purified fragments using the pGEM-T Easy Vector in the pGEM-T Easy Vector System II cloning kit (Promega Corporation) according to the manufacturer’s protocol. Selected clones were submitted for Sanger sequencing with a single primer to either Beckman Coulter Genomics (Single Pass Sequencing) or the W. M. Keck Ecological and Evolutionary Genetics Facility at the Marine Biological Laboratory (Woods Hole). Full-length Sanger sequencing of select Mesodinium clones were run at Genewiz (Boston) (see Supplementary Data Sheet 1 for accession numbers). Sequences were edited and assembled into contigs using Sequencher (Gene Codes Corporation). We sequenced and analyzed 687 cryptophyte rbcL clones (accession numbers in progress) from environmental samples. For Mesodinium spp., a total of 903 clones were sequenced (accession numbers in progress) from all stations. Using a sequence similarity criterion of 98 and 99% for cryptophytes and Mesodinium, respectively, we constructed independent contigs and generated consensus sequences for use in our global alignment for each sample. This process helped to reduce the number of sequences used in our phylogenetic analyses while maintaining meaningful diversity data.
Table 1.
Primers designed and used for amplifying the SSU, ITS, and LSU rRNA genes of Mesodinium spp. and rbcL gene fragments of cryptophytes in this study.
| Primer | Sequence | Gene: position (bp) |
|---|---|---|
| Mesodinium spp. primers | ||
| MESO245F | CGACTCGACGTCCCG | 18S: 246 |
| MESO580R | CGTCCGTAGTCTGTACGTC | 18S: 585 |
| MESO1200F | ATTCCGGTAACGAACGAGAC | 18S: 1217 |
| MESO1440F | AACTAGGAATGTCTCGTAAGC | 18S: 1446 |
| MESO580R | GACGTACAGACTACGGACG | 18S: 603 |
| MESO865R | ACCTTCGTCCTTTGTCGCA | 18S: 1017 |
| MESO1480R | CTAAACACTCGATCGGTAGG | 18S: 1545 |
| MESO28S_R | AGACTTGGATGACTTTTATCACC | 28S: 298 |
| Cryptophyte primers | ||
| rbcL2F-800 | AGGAGGAAWAYATGTCTCAAT CCG | rbcL: 1 |
| Crypt_rbcLR2 | CAGTGAATACCACCTGAAGCTA | rbcL: 1185 |
Phylogenetic Analysis and Species Assignment
Consensus sequences from assembled contigs were used for separate alignments of Mesodinium spp. and cryptophytes, in combination with available sequences in Genbank and sequences generated from cultures in our laboratory (see above). Alignments were constructed using the Clustal X algorithm (Larkin et al., 2007), and refined by eye using MacClade 4.08a (Maddison and Maddison, 2000). All maximum likelihood (ML) phylogenetic trees were executed with PhyML 3.0 (Guindon et al., 2010) using 100 bootstrap replicates and the general time reversible (GTR) substitution model, while estimating the gamma distribution parameter, proportion of invariable sites, and the transition/transversion ratio. Phylogenetic trees were constructed using TreeDyn 198.3 (Chevenet et al., 2006). Assignment of Mesodinium and cryptophyte species was determined by their nearest neighbor match of a known species within ML phylogenetic trees.
Results
Sample Characteristics
Bloom Samples
The St. George Island (SGI; Alaska) bloom in the Bering Sea was observed within the main harbor, creating dense patches of red water (Table 2). The presence of M. rubrum-like ciliates was confirmed by microscopy at the time of sampling. A massive Mesodinium bloom was sampled from Ilha Bela, São Paulo, Brazil (BR) in the South Atlantic, and microscopic images from samples of the bloom confirmed the presence of large cells (not shown). This bloom stretched for over 100 km of the Brazilian coastline, and was visible from space (Carlowicz et al., 2014). A bloom sample was analyzed from the North Atlantic, in Oyster Bay, Long Island Sound (LIS), and was also documented using satellite imagery (Dierssen et al., 2015). This bloom revealed M. rubrum to be present at ∼1000 cells ml-1. Another North Atlantic red water bloom was sampled from the Outer Banks of North Carolina (NC), revealing M. rubrum at 1750 cells ml-1. Finally, streaks of red water were sampled in the South Pacific, ∼25 km off the Chilean coast (CL) during an oceanographic cruise. However, samples from this bloom were only quickly collected from the surface for nucleic acid extraction, with no additional documentation.
Table 2.
Location and description of samples for analysis of Mesodinium and cryptophyte community diversity.
| Region | Description | Abbrev. | Date | Lat | Long | Sal PSU | Temp °C | Mesob cells ml-1 | Sample notesc |
|---|---|---|---|---|---|---|---|---|---|
| South Atlantic | Ilha Bela, BZ | BR | 2/15/14 | -23.80 | -45.22 | - | 24 | 37.5 | bloom |
| North Atlantic | Long Island Sound | LIS | 9/24/12 | 40.91 | -73.60 | 27.9 | 21.9 | 1003 | bloom; Oyster Bay |
| North Atlantic | Waves, NC | NC | 10/6/08 | 35.56 | -75.44 | 28.63 | 22.5 | 1750 | bloom |
| North Atlantic | Barents Sea; station M4 | Bar-M4 | 6/27/11 | 74.53 | 30.11 | 35.06 | 4.9 | 10 | 60m |
| Baltic Sea | Gulf of Finland; station LL3a | GF-LL3a | 7/8/12 | 60.07 | 26.34 | 5.26a | 17.4a | - | depth integrated |
| Baltic Sea | Gulf of Finland; station XVI | GF-XVI | 7/8/12 | 60.25 | 27.25 | 4.52a | 17.5a | - | depth integrated |
| Baltic Sea | Tvärminne Zoological Station | TV | 7/31/13 | 59.83 | 23.25 | 5.5 | 18.5 | - | Hanko, FI; conc.d |
| South Pacific | Chile Coast | CL | 11/23/10 | -19.97 | -70.72 | 34.8 | 19.5 | - | bloom |
| North Pacific | Bering Sea; St. George Is. | SGI | 9/15/12 | 56.57 | -169.68 | - | 6.6 | - | bloom |
| North Pacific | California Current | CC | 7/5/13 | 36.33 | -123.14 | 33.8 | 12.8 | 0.5 | 2m |
aMeasurements from 1 m, while all others at same depth of sample; bDue to the patchy distribution of M. rubrum and its ability to form thin layers, these concentrations are at best approximation; cSample depths are surface unless indicated otherwise. Depth integrated samples are from 0.5, 3, 5, 7.5, and 10 m; dSample concentrated with a 10 μm net.
Non-bloom Samples
Samples from the Baltic Sea included one coastal Finland (Tvärminne) and two off shore Gulf of Finland samples (Table 2). A non-bloom subsurface sample from the Barents Sea (Bar-M4) was also analyzed and was documented to have 10 Mesodinium ml-1. A sample taken during a research cruise in the North Pacific within the California Current (CC) was also analyzed, and had low levels of M. rubrum present (Table 2).
Mesodiniidae Phylogeny
Using primers designed to amplify a partial region of the 18S, the entire ITS, and a partial region of the 28S ribosomal RNA genes of all known Mesodiniidae taxa, we recovered sequence data from both heterotrophic and plastidic species from eight locations and analyzed their phylogenetic relationships (Figure 1). We also analyzed new sequence data from four cultures of M. rubrum and one culture of Mesodinium pulex (Table 3). As shown previously (Garcia-Cuetos et al., 2012) the Mesodiniidae formed four well-supported and distinct clades, represented by M. pulex, M. pupula, M. chameleon, and the M. major/rubrum complex, respectively, and all comparisons between these major clades revealed the greatest p-distance (>7%) within the dataset (Table 4). Relatively few environmental sequences were recovered from the M. pulex, M. pupula, and M. chameleon clades, and intracladal diversity within these groups was lower than within the M. rubrum/major complex (Figure 1A). Compared to the M. major/rubrum variants, the mixotrophic ciliate Mesodinium chamaeleon shared 89.6–92% similarity, while M. pulex had 83.2–87.7%, and M. pupula only 81.2–86% (Table 4). M. chamaeleon had low similarity to the heterotrophic M. pulex (86%) and M. pupula (82.9%), as did the latter two species to one another (83.8%).
FIGURE 1.
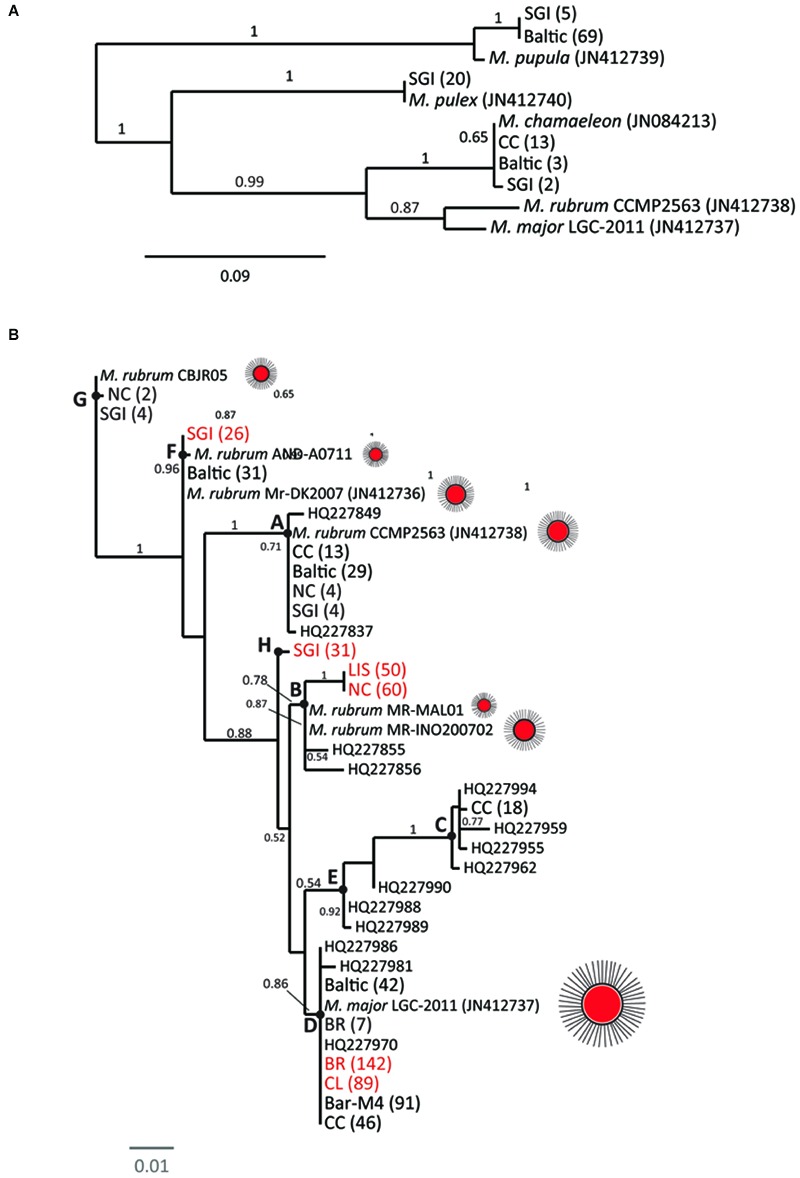
Maximum likelihood phylogenies of the Mesodiniidae using Mesodinium spp. sequences from 10 global locations: the Baltic Sea (stations GF-LL3a, GF-XVI, TV are grouped together in this figure), Barents Sea (Bar-M4), California Current (CC), St George Island (SGI) harbor, coastal Chile (CL), coastal Brazil (BR), Long Island Sound (LIS), and coastal North Carolina (NC). Phylogeny of the Mesodiniide based on a sequence, composed of a partial SSU rRNA, internally transcribed spacer region (ITS), and a partial region of the LSU rRNA gene. (A) Phylogeny of the Mesodiniidae, focusing on community sequences related to M. chamaeleon and heterotrophic Mesodinium spp. (B) Phylogeny of the M. major/rubrum complex. Letters on branches of tree (A–H) refer to variants within the complex, and cartoons of the ciliates (oral view) depict the relative cell size of various isolates (Table 3). Red text is used for genotypes dominating bloom samples and numbers in parentheses refer to the number of clones representing each genotype.
Table 3.
Origin and cellular dimensions of Mesodinium rubrum variant isolates.
| Variant | Strain | Origin/year | Length (μm) | Width (μm) | L/W | Volumea (μm3) | Cryptophyte prey | Type referenceh | |
|---|---|---|---|---|---|---|---|---|---|
| A | NCMA 2563 | McMurdo Sound, AN 1996 | Mean Range |
23.1 (2.6) 17.3–30.2 |
22.4 (2.7) 15.1–32.5 |
1.1 (0.1) 0.9–1.2 |
5707 (1826) 2139–10,277 |
GCf | 1 |
| B | MR-MAL01 | Gomso Bay, KR 2001 |
Mean Range |
21b | 13 | 1.6 | 1857c | TAg | 2 |
| B | MR-INO200702 | Inokushi Bay, JP 2007 |
Mean Range |
25.8 (3.8) 16.8–32.1 |
21.8 (3.8) 13.1–28.6 |
1.2 (0.1) 0.9–1.6 |
6719 (3008) 1508–13,007 |
TA | 3 |
| F | AND-A0711 | Huelva, ES 2007 |
Mean Range |
16.2 (1.7) 12.6–19.4 |
13.0 (1.0) 10.1–15.5 |
1.3 (0.1) 1.1–1.5 |
1374 (308) 672–1939 |
TA | 4 |
| F | Mr-DK2007 | Frederikssund, DK 2007 |
Mean Range |
31d 25–35 |
21 16–25 |
1.5 | 7154e 3349–11,448 |
TA | 5 |
| G | CBJR05 | James River, US 2011 |
Mean Range |
21.7 (3.0) 15.8–33.7 |
16.0 (1.7) 11–21.7 |
1.4 (0.2) 1.1–2.0 |
2937 (914) 1234–5537 |
TA | Here |
All measurements made here unless otherwise noted. Mean values presented with standard deviation in parentheses.
aVolumes calculated as spheroids using 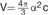 where a is the equatorial radius and c is the polar radius; bestimated from Yih et al. (2004); cre-calculated here and different from reported value (5996 ± 30) in Yih et al. (2004); dfrom Garcia-Cuetos et al. (2012); ecalculated here; fGC: Geminigera cryophila; gTA: Teleaulax amphioxeia; htype references: (1) Gustafson et al. (2000), (2) Yih et al. (2004), (3) Nishitani et al. (2008), (4) Jaén (unpublished), Riobó et al. (2013), (5) Garcia-Cuetos et al. (2012).
where a is the equatorial radius and c is the polar radius; bestimated from Yih et al. (2004); cre-calculated here and different from reported value (5996 ± 30) in Yih et al. (2004); dfrom Garcia-Cuetos et al. (2012); ecalculated here; fGC: Geminigera cryophila; gTA: Teleaulax amphioxeia; htype references: (1) Gustafson et al. (2000), (2) Yih et al. (2004), (3) Nishitani et al. (2008), (4) Jaén (unpublished), Riobó et al. (2013), (5) Garcia-Cuetos et al. (2012).
Table 4.
Genetic p-distance matrix of a partial ‘18S–28S’ rDNA region for variants within the Mesodinium major/rubrum complex and other Mesodinium spp.
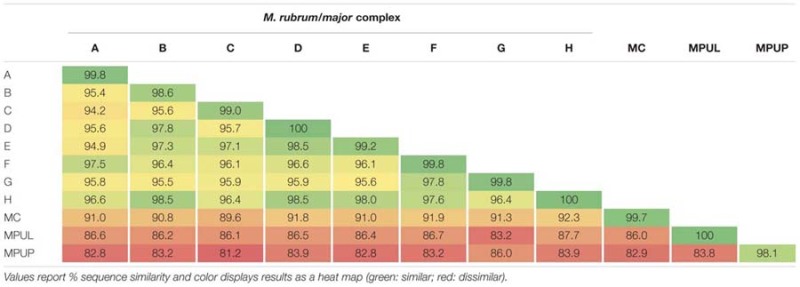 |
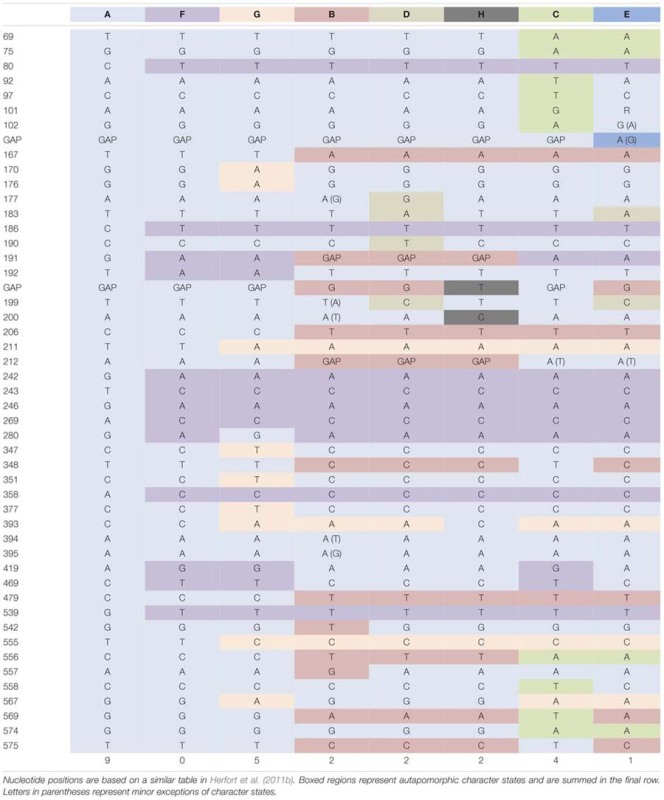
In contrast, the predominantly phototrophic M. major/rubrum species complex formed eight subclades (Figure 1B) of which six have been described previously (Herfort et al., 2011b; Garcia-Cuetos et al., 2012). The M. major/rubrum complex was well-resolved, with four of the clades having 100% bootstrap support within a maximum likelihood phylogeny and three with >75% support (Figure 1B). The least resolved clade was that of E, which had both low bootstrap support (54%) and is comprised of only a few environmental sequences from the Columbia River (Herfort et al., 2011b). One of the new clades, designated G, was closely related to clade F, described originally from the Roskilde Fjord in Denmark, and clade A, described originally from McMurdo Sound, Antarctica. Together these three variants form one subgroup within the M. major/rubrum complex, while clades B, C, D, E, and H form another. Sequences forming a second new variant, H, were recovered from the SGI bloom sample, and shared high sequence similarity (>98%) with variants B and D (Table 4). Lowest intracladal sequence variation was found within variants A, D, F, G, and H (<0.5%), while the greatest variation (>1%) was found within clades C and B (Table 4). Polymorphisms between the different M. major/rubrum variants are shown in Table 5 (modified from Herfort et al., 2011b), revealing the sequence variation that shape clade phylogeny within this group. For instance, several shared polymorphisms are found between clades A, F, and G at bases 167, 206, 479, 556, and 569 while clades C and E share four such polymorphisms (bp: 69, 75, 556, and 574). Clades B, D, and H share polymorphisms with each other at bases 191, 212, and 556, and at additional loci with clades C and E at 167, 206, and 479. The most divergent variants within the M. major/rubrum group were A and C, sharing 95.7 and 95.9% similarity, respectively, with the other six variants, relative to the average among all groups of 96.6%. Of all M. major/rubrum variants, group A had the most autapomorphic characters (10) within the analyzed region, followed by groups G (5) and C (4) (Table 5), and these variants were the only to have 100% boot strap support (Figure 1B).
Table 5.
Details of genetic variation for the Mesodinium major/rubrum complex within the partial ‘18S–28S’ rDNA region for all available sequence data.
 |
Mesodinium Diversity
The most widely encountered variant was D (M. major), found at 6 of the 10 sampling sites, while variants A and F were found at 5 (Figure 2). Sequences from variants C and H, as well as the heterotrophic M. pulex were each only recovered from one station, while no clade E sequences were found in this study. Analysis of samples from red-water events of Mesodinium revealed that the bloom phenomenon within this species complex is not associated with one particular variant. Sequences from subclades B and D dominated (>85%) clones recovered from the four blooms analyzed in this study, and in two cases sequences consisted only of M. major (Figure 2). Variant B dominated two blooms from the eastern United States, and was not found at non-bloom sites (Figure 2). Clones from variants F and H dominated the bloom sample from SGI, however, the sample was not preserved when shipped and thus the relative proportion of M. major/rubrum variants is uncertain. Samples from the Baltic Sea were from coastal and open water stations, and were not associated with bloom events. Collectively, the Baltic Sea had the greatest diversity of Mesodiniidae sequences, with clones recovered from heterotrophic M. pupula, the mixotrophic M. chameleon, as well as from multiple M. rubrum/major clades. The CC sample was also diverse, with three variants of M. major/rubrum as well as M. chamaeleon (Figure 2). In contrast, a non-bloom sample from an off shore station in the Barents Sea was composed entirely of M. major.
FIGURE 2.
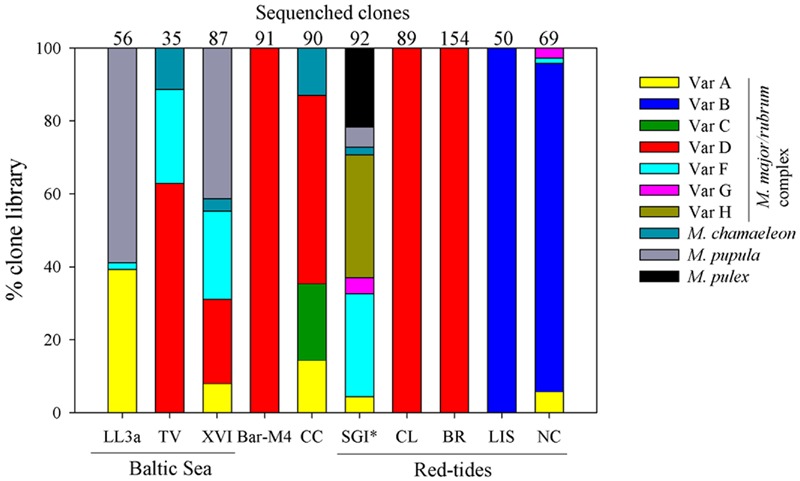
Community genetic diversity of Mesodinium from the Baltic Sea (GF-LL3a, GF-XVI, TV), Barents Sea (Bar-M4), California Current (CC), St George Island (SGI) harbor, coastal Chile (CL), coastal Brazil (BR), Long Island Sound (LIS), and coastal North Carolina (NC). Community diversity of Mesodinium based on a sequence composed of a SSU rRNA gene fragment, the complete internally transcribed spacer region (ITS), and a partial region of the LSU rRNA gene. Categories include seven variants within the M. major/rubrum species complex, the mixotrophic M. chamaeleon, and the heterotrophic species M. pupula and M. pulex. ∗SGI sample was not properly preserved, so the relative proportion sequences within each category may have been compromised.
Cryptophyte rbcL Phylogeny
Our cryptophyte rbcL gene data set included new sequences from 22 cryptophyte cultures, comprised largely of TPG and Hemiselmis species. Analysis of our environmental clones resulted in 39 distinct contigs, at 99% sequence similarity, from the nine sample sites (one Baltic site was not assessed for cryptophyte rbcL diversity). Of these, 21 consensus sequences were determined to be from the Teleaulax/Plagioselmis/Geminigera (TPG) clade (Figure 3). Our maximum likelihood tree had strong support (>90 boot strap) for the cryptophyte clades comprised of TPG, Proteomonas, Rhodomonas-like cryptophytes, Guillardia/Hanusia, Urgorri, and Falcomonas, and Cryptomonas, while the clade comprised of Hemiselmis and Chroomonas had weaker support (78%) (Figure 3). Culture sequences from T. amphioxeia and P. cf. prolonga were polyphyletic, forming a species complex that appears to be in need of taxonomic revision. The position of Proteomonas as sister of TPG was strongly supported. No environmental sequences were recovered from the Cryptomonas or Proteomonas clades. Of all the recovered environmental sequences, those clustering with Falcomonas sp. appeared to represent the most novel lineages of uncharacterized cryptophyte species (Figure 3).
FIGURE 3.
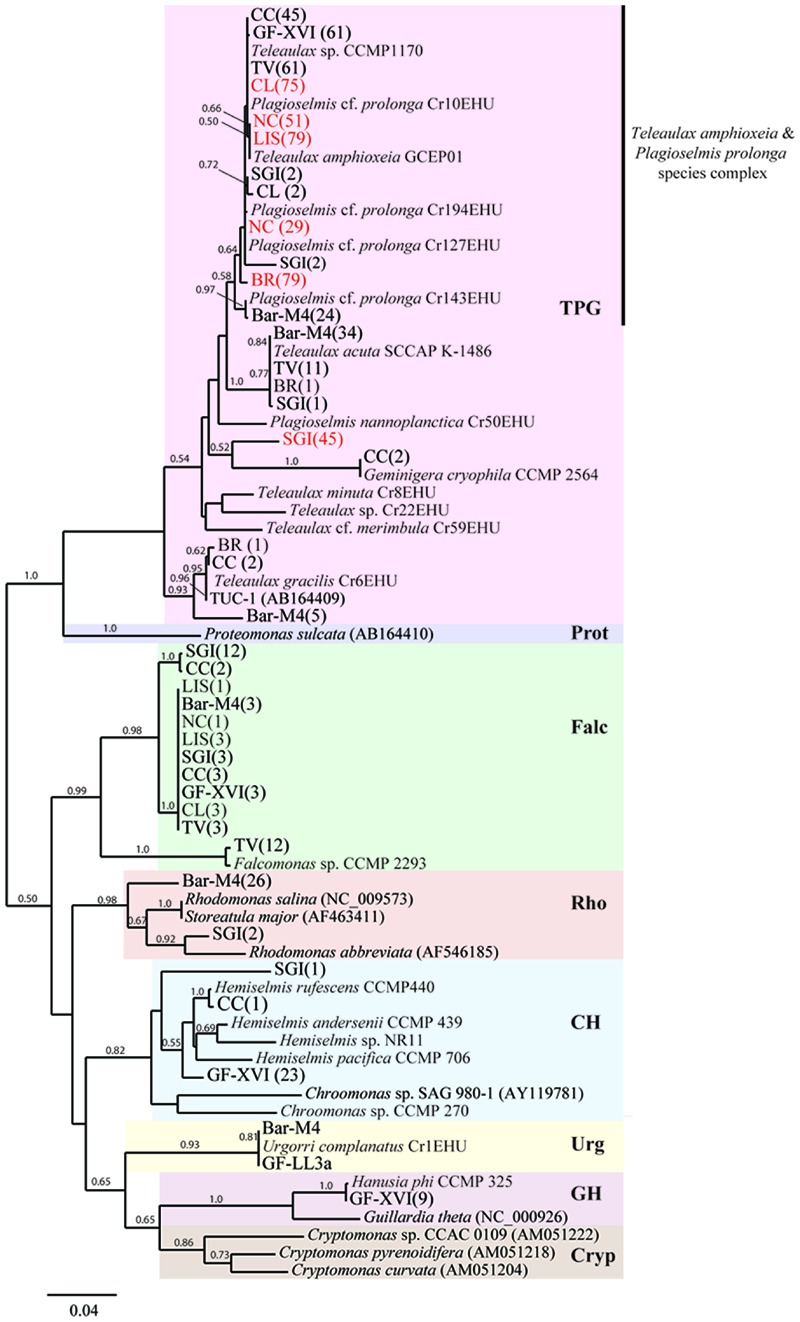
A maximum likelihood phylogeny of cryptophytes using partial plastid LSU RuBisCO gene (rbcL) sequences from 10 global locations: the Baltic Sea (GF-LL3a, GF-XVI, TV), Barents Sea (Bar-M4), California Current (CC), St George Island (SGI) harbor, coastal Chile (CL), coastal Brazil (BR), Long Island Sound (LIS), and coastal North Carolina (NC). Red text refers to genotypes dominating bloom samples and numbers in parentheses refer to the number of clones representing a genotype. Major cryptophyte clades are boxed by color: TPG: Teleaulax/Plagioselmis/Geminigera group; Falc: Falcomonas-like species; CH: Chroomonas/Hemiselmis group; Rho: Rhodomonas-like cryptophytes; GH: Guillardia/Hanusia group; Urg: Urgorri-like cryptophytes.
Cryptophyte Plastid rbcL Diversity
Community plastid rbcL sequences were analyzed for each sample in order to test the hypotheses that M. rubrum blooms are associated with TGP-like cryptophyte sequences and that the number of cryptophyte phylotypes are greater in non-bloom samples. Since these samples are unfractionated community samples, it is not possible to determine the proportion of cryptophyte sequences that are from free-living cells or associated with a particular protist species (e.g., M. rubrum). However, while these samples may not specifically represent the diversity of the free-living cryptophyte community per se, they are ultimately derived from it. Overall 74% of rbcL sequences from all sample sites originated from T. amphioxeia/P. cf. prolonga-like sequences, while a striking 82% were from the TPG clade. Bloom samples were dominated by TGP cryptophytes (92%), with samples from BR, CL, LIS, and NC sites mostly (>90%) comprised of T. amphioxeia/P. cf. prolonga-like sequences. In contrast to other bloom samples, the SGI bloom was dominated by a phylotype branching near G. cryophila. From non-bloom stations (343 sequences), 56% were T. amphioxeia/P. cf. prolonga-like sequences, while 72% belonged to the TPG clade. The Bar-M4 sample was primarily composed of T. amphioxeia/P. cf. prolonga-like sequences, T. acuta, and a Rhodomonas sp., while T. amphioxeia/P. cf. prolonga-like sequences dominated the Baltic Sea and CC samples. One of the Baltic Sea sites, station XVI in the Gulf of Finland, was also rich in a Hemiselmis sp. and Hanusia phi. Sequences from Falcomonas spp. were found to comprise a minor component of cryptophyte genetic diversity from eight of the nine sampling sites. U. complanatus, which was originally described from Atlantic estuaries in southwestern Europe (Laza-Martínez, 2012), was detected in the Baltic and Barents Seas.
Discussion
This study is the first to assess the environmental genetic diversity of both non-photosynthetic and plastidic marine Mesodinium species, across a broad geographical range of sampling sites. Our findings expand upon the known diversity of the M. major/rubrum species complex, and further solidify the relationship of Teleaulax amphioxeia cryptophytes with their bloom events. Further, we show novel diversity of cryptophyte algae, particularly among Falcomonas-like species, and provide evidence for the widespread dominance of the Teleaualx/Plagioselmis/Geminigera (TPG) cryptophyte group.
Phylogeny of Mesodinium
Mesodinium spp. have an unusual 18S rRNA gene with numerous deletions and substitutions within conserved regions compared to other ciliates (Johnson et al., 2004), and they appear to lack a separate 5.8S rDNA region, suggesting that it may be fused to the 28S rDNA gene (Herfort et al., 2011b). This characteristic is also found in the microsporidia and bacteria (Hoef-Emden, 2005). While Mesodinium show typical litostome secondary structure in their V4 region, including reduction of helices 23_1, 23_8, and 23_9 and the absence of 23_5 (Strüder-Kypke et al., 2006), phylogenies based on the SSU rRNA gene consistently result in a novel, deep basal branch within the ciliates (Johnson et al., 2004; Chen et al., 2015a). From ultrastructural observations, only the ciliary transition region supports their placement within the litostomes (Garcia-Cuetos et al., 2012). A recent multigene phylogeny of the Mesodiniidae also suggests a deep basal ciliate branch and strongly implies that they may represent a distinct ciliate class, the Mesodiniea (Gao et al., 2016). However, a phylogenomic analysis of non-model ciliates using transcriptomic data failed to resolve the monophyly of the Mesodiniea (Chen et al., 2015b).
Our phylogenetic analysis of taxa within the Mesodiniidae agree with Garcia-Cuetos et al. (2012), with strong support for four major clades distinguishing the two heterotrophic lineages of M. pulex and M. pupula, the mixotrophic M. chamaeleon, and the predominantly phototrophic M. major/rubrum complex. At least one additional major clade of Mesodinium-like ciliates are found in freshwater habitats, and phylogenies based on the SSU rRNA gene group them as a sister group to M. pulex (Bass et al., 2009). Since we did not sequence samples from freshwater habitats and no cultures of these species were available, comparisons of their full rRNA gene cassette to marine Mesodiniidae were not made here. Our analysis resolved two novel subclades of M. rubrum: (1) variant G was identified from sequence data of a culture from Chesapeake Bay, USA, along with environmental sequences from the North Carolina (NC) and St. George Island (SGI) bloom events, and (2) variant H, which is only represented by environmental sequences from the SGI sample. One sequence from a culture isolated in Spain (Riobó et al., 2013) grouped with a Danish culture from the F subclade (Garcia-Cuetos et al., 2012) as well as clones from the Baltic Sea and SGI. Together, variants F and G grouped strongly with variant A to form one major branch of the M. rubrum/major complex. We found M. rubrum sequences from the Baltic Sea, CC, NC, and SGI that grouped with variant A, which includes a well-studied cultured strain from Antarctica (Gustafson et al., 2000; Johnson et al., 2007) and sequences from coastal Oregon (Herfort et al., 2011b). Cultures isolated from Korea (Yih et al., 2004) and Japan (Nishitani et al., 2008) were found to belong to subclade B, which included sequences from blooms in LIS and NC, as well as the recurrent blooms of the Columbia River Estuary (Herfort et al., 2012). M. major (subclade D) was found in the Baltic Sea, Bar-M4, and in blooms from coastal BR and CL, and has also been found in the Columbia River Estuary (Herfort et al., 2011b) as well as coastal Denmark (Garcia-Cuetos et al., 2012). Two other M. rubrum variants, C and E, remain uncultured and have only been found in the Northeastern Pacific (here and Herfort et al., 2011b). Subclades B, C, D, E, and H formed the second major branch of the M. rubrum/major complex. While C, E, and H have not yet been cultured, subclade B appears to be highly variable in size, both within and between strains (Table 2). A bloom of M. rubrum variant B in LIS was found to contain cells of typical size for M. rubrum cultures (Tables 2 and 3).
Genetic Diversity of Mesodinium
The studies of Herfort et al. (2011b) and Garcia-Cuetos et al. (2012) revealed that “M. rubrum” is a species complex of closely related genetic variants, most of which are morphologically indistinguishable. While cell size clearly distinguishes M. major from M. rubrum variants, cell size within M. rubrum strains appears to be highly variable considerably (Table 2). Furthermore, sequence variation of rRNA genes and the ITS region in M. major relative to M. rubrum, is equal to or lower than variation among M. rubrum variants (Table 4), making the distinction of species vs. variant somewhat ambiguous based on these sequences alone. While we are beginning to identify trends in genotype distribution of M. major/rubrum ciliates and their association with bloom events, we know little about distinctions in the physiological or behavioral diversity among these variants and if they may possess distinct ecological niches.
While our results strongly suggest that heterotrophic species have less intracladal diversity in marine ecosystems, our samples focused on bloom events and are therefore biased toward the mixotrophic species. Stations where heterotrophic species were abundant, such as two Gulf of Finland (Baltic Sea) samples rich in M. pupula (>40% of Mesodinium clones), revealed essentially identical sequences within and between these populations. Furthermore, M. pupula sequences from the Baltic Sea were nearly identical to those from SGI. In addition, M. pulex sequences from SGI and M. chamaeleon sequence data from SGI and the Baltic Sea were remarkably similar to strains isolated from Danish waters.
Bloom Samples
All temperate and subtropical bloom samples were dominated either by variant B or D from the M. rubrum/major complex. The only previous bloom where genetic diversity of Mesodinium spp. were assessed was in the Columbia River Estuary, and these populations are also associated with subclade B (Herfort et al., 2011b). Annual blooms of M. rubrum occur within the Columbia River, developing first near the mouth of the estuary and later within the main channel in coincidence with neap tides and decreased turbulence (Herfort et al., 2011a). While summer Mesodinium populations in the Columbia River are dominated by subclade B, they are largely absent in spring samples, which were instead associated with subclades C and D (Herfort et al., 2011b). Our study only provided “snapshots” of community genetic diversity, and it is likely that diversity at our various sites changes seasonally. Danish cultures of variant F have also been isolated from red-water within islands of the Danish straits (Garcia-Cuetos et al., 2012), however, it is unclear if this clade was primarily responsible for the observed bloom.
Blooms of M. rubrum along the North Carolina coastline have been periodically observed (Hathaway, 2014), but no published studies are available. The North Carolina bloom was the only preserved bloom sample that had more than one subclade of M. rubrum present, with three other subclades comprising ≤5% of the community. Since the bloom sample was collected near Oregon Inlet, which empties the Pamlico Sound into the Atlantic Ocean, some of the strains present may have been washed in from estuarine populations. The sequences of one of these subclades (G) are identical to that of a strain isolated from the James River estuary in nearby Chesapeake Bay, which would support mixing of estuarine populations into this coastal bloom. The Long Island Sound bloom was also dominated by subclade B (Figure 2), and was associated with calm wind speeds (Dierssen et al., 2015). This bloom was detected from the Space Station, using a Hyperspectral Imager for the Coastal Ocean sensor (Dierssen et al., 2015).
Numerous studies have commented on the size variability in natural populations of M. rubrum-like ciliates in diverse geographical locations (Taylor et al., 1971; Lindholm, 1981; Crawford, 1993; Rychert, 2004; Montagnes et al., 2008). Garcia-Cuetos et al. (2012) formally described the largest of plastidic Mesodinium spp. as M. major, which attains cell dimensions of 50 μm × 40 μm (L × W). Particularly large M. rubrum-like cells have been observed in many blooms and coastal ocean communities, particularly those associated with upwelling regions (Packard et al., 1978). Both of our M. major-dominated bloom samples were from coastal blooms in Brazil and Chile, and were likely associated with upwelling. Both regions have a rich history of documented Mesodinium “red tides,” many of which have been described reaching massive proportions. Mesodinium blooms have been previously reported in the coastal region of northern São Paulo, Brazil, where our sample was taken, and have been described as forming thin layers of red water 1–2 m below the surface with a maximum thickness of ∼30 cm (Owen et al., 1992). An image of the bloom sampled near Ilha Bela, Brazil, was captured by NASA’s Aqua satellite using the moderate resolution imaging spectrophotometer (MODIS), and the red water formed an offshore patch of staggering proportions, extending 800 km from Rio De Janeiro to south of Florianópolis (Carlowicz et al., 2014). Mesodinium blooms in southern Brazil have also been documented to occur near the Itajaí-Açu River (Proença, 2004).
Darwin is thought to have documented the first Mesodinium bloom off the Chilean coast while on the H. M. S. Beagle (Darwin, 1906; Hart, 1943), and such occurrences are common in this region during non El Niño periods (Avaria and Muñoz, 1987; Marín et al., 1993). Blooms documented from the Peruvian upwelling zone have reported some of the highest rates of primary production ever measured (Smith and Barber, 1979), and red water patches spanning 100s of square miles (Ryther, 1967). Based on these previous observations and our phylogenetic analysis of the Chilean and Brazilian blooms, it is likely that these recurrent events and perhaps blooms in other upwelling ecosystems are predominantly M. major.
Non-bloom Samples
Mesodinium is abundant in the Baltic Sea (Leppanen and Bruun, 1986) and is known to form occasional blooms in brackish basins around the archipelago region of Åland (Lindholm, 1978). During spring in the northern Baltic Sea, M. rubrum-like ciliates dominate ciliate biomass and are thought to account for ∼10% of all primary production (Leppanen and Bruun, 1986). In the lower Baltic Sea, both large and small M. rubrum-like ciliates dwell in the upper water column during June, with mostly large cells found at greater depths (Rychert, 2004). Within the Åland region, up to three size classes (∼20, 40, 60 μm) of M. rubrum-like ciliates are observed, with the smallest being dominant during autumnal red water events (Lindholm, 1978). Deep (>70 m) layers of pigmented Mesodinium have also been found in the Baltic Sea, below the thermocline (Setälä et al., 2005) and near the anoxic boundary of the Gotland Basin (Weber et al., 2014). Our two samples from the Gulf of Finland (GF) revealed a mixed community of M. rubrum subclades A, D, and F. While subclade D (M. major) is the largest Mesodinium spp., both subclades A and F may have a range of cell sizes, even within a single strain (Table 2). Surprisingly, the largest component of both GF samples was M. pupula, a heterotrophic species that is slightly larger than M. pulex (Garcia-Cuetos et al., 2012). The ecology of M. pupula is poorly known, but has been shown to be one of the most abundant heterotrophic ciliates in coastal Yellow Sea microbial communities (Jiang et al., 2011). While M. pulex has instead been reported as a dominant heterotrophic ciliate species in Baltic communities (Setälä and Kivi, 2003), this species is difficult to discern from M. pupula using traditional light microscopy. The near shore station from Hanko, FI, also had subclades D and F, as well as the mixotrophic M. chamaeleon, which is capable of possessing a variety of cryptophyte plastid types, including those containing phycocyanin (Moestrup et al., 2012). M. chameleon differs from M. rubrum-like ciliates, in the structure of its cirrus, its predominantly benthic niche, and in the organization of sequestered cryptophycean organelles (Moestrup et al., 2012). M. chamaeleon-like ciliates, reported as green-blue in appearance, have been observed in the Åland region (Lindholm, 1985), as well as in a coastal Rhode Island estuary (Hargraves, 1991). Our Baltic sequence of M. chamaeleon was essentially identical to an isolate from coastal Denmark, and both were slightly different from sequences found in SGI.
The CC sample was one of the more surprising due to the unexpected presence of M. chamaeleon (13% of clones) in an offshore site. The presence of this “benthic” ciliate in a pelagic environment underscores how little we know about the ecology of this species. The remaining portion of the community was split among variants A, C, and D. Variants C, E, and H are the least distributed and most poorly studied of the M. major/rubrum complex, as none have yet been cultured and all have only been found at only one or two sites along the North Pacific coast of the US.
Cryptophyte Phylogeny
Taxonomic relationships within the cryptophytes, based on traditional morphological and structural traits of their pigments and cell surface, have recently been tested using gene phylogenies (Fitt et al., 2001; Hoef-Emden et al., 2002). These studies have found that while biliprotein type is congruent with molecular phylogenies, the type of inner periplast is not (Hoef-Emden et al., 2002). These phylogenies revealed seven separate lineages within plastid-bearing cryptophytes, all except Cryptomonas, containing representatives from marine environments. An additional lineage, represented by the brackish-water species U. complanatus, was later discovered (Laza-Martínez, 2012). Previously Hoef-Emden et al. (2005) commented on the potential for accelerated evolutionary rates when constructing rbcL phylogenies of cryptophytes, due to shifts in the number of amino acids showing changes in codon usage among early and late diverging taxa. This study was based only on genus Cryptomonas, an exclusively freshwater group containing some highly divergent taxa. Yet, despite finding high evolutionary rates for rbcL, resulting cryptophyte phylogenies were largely congruent with those of 18S rDNA (Hoef-Emden et al., 2005). Our results, which used the most diverse rbcL dataset to date, are also congruent with previously published phylogenies of nuclear 18S rRNA genes (Hoef-Emden et al., 2002). The eight lineages of plastid-bearing cryptophytes were recovered in the rbcL phylogeny. Moreover, the Proteomonas lineage appeared as the sister group of TPG with high support. While major clades are well-supported in previous phylogenies, the relationships among lineages have not been well-resolved. The clustering of Proteomonas and TPG was only partially supported in nucleomorph, but not nuclear, 18S rRNA genes phylogenies in a clade that also included Guillardia/Hanusia (Hoef-Emden et al., 2002).
The traditionally used morphological trait of periplast structure, as a continuous sheet or polygonal, has been shown to vary with life stage in the genera Cryptomonas and Proteomonas and is thus phylogenetically uninformative (Hill and Wetherbee, 1986; Hoef-Emden and Melkonian, 2003). Similar observations of periplast dimorphism have also been made within the Teleaulax group (Garcia-Cuetos, 2011), and this trait has traditionally been used to distinguish this genus from Plagioselmis. Furthermore, the midventral band, a morphological trait used to distinguish Teleaulax (thought to be absent) and Plagioselmis, was recently shown to be polyphyletic within this group (Laza-Martínez et al., 2012). The finding of strains with the T. amphioxeia and P. prolonga morphology sharing nearly identical rbcL sequences can be interpreted from the perspective of a dimorphic species, making both names synonym. The Teleaulax/Plagioselmis dimorphism can also account for incongruences between microscope-based morphological identifications and environmental DNA sequence identities, as in Bazin et al. (2014), where only T. acuta sequences were retrieved from a P. prolonga red tide sample. These observations, combined with the polyphyletic phylogeny of Teleaulax shown here and previously using 18S rRNA (Deane et al., 2002; Rial et al., 2015), suggest that these genera are in need of taxonomic revision.
Cryptophyte Diversity and its Role in Mesodinium Blooms
About half of our samples were from Mesodinium blooms, and these samples (excluding SGI) were dominated (>95%) by T. amphioxeia, as expected from previous studies (Nishitani et al., 2010, #874; Herfort et al., 2011b, #893). However, clone libraries from non-bloom samples were also largely comprised of TPG sequences, particularly T. amphioxeia (Figure 4). Exceptions were the high latitude BS and SGI samples, which were largely composed of T. acuta and G. cryophila-like sequences, respectively. G. cryophila appears to primarily be a cold-water species, dominating cryptophyte assemblages in Antarctic Dry Valley Lakes (Bielewicz et al., 2011), and was co-isolated with an Antarctic M. rubrum strain (Gustafson et al., 2000). T. gracilis and T. minuta have also been shown to sustain in vitro M. rubrum (clade F) growth (Rial et al., 2015). However, only few sequences of the former and none of the latter were recovered. These results strongly suggest that TPG species may frequently dominate cryptophyte communities in coastal ecosystems, and play critical roles in supporting the productivity of M. major/rubrum.
FIGURE 4.
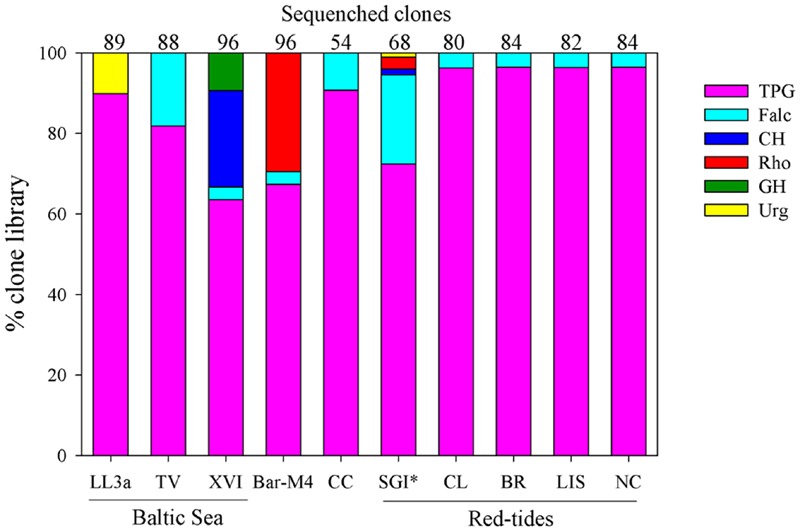
Community genetic diversity of cryptophyte algal plastids from the Baltic Sea (GF-LL3a, GF-XVI, TV), Barents Sea (Bar-M4), California Current (CC), St George Island (SGI) harbor, coastal Chile (CL), coastal Brazil (BR), Long Island Sound (LIS), and coastal North Carolina (NC). Community diversity of cryptophytes based on partial sequences of the plastid LSU RuBisCO gene. Categories include the Teleaulax/Plagioselmis/Geminigera (TPG) group, Falcomonas-like species (Falc), the Chroomonas/Hemiselmis group (CH), Rhodomonas-like cryptophytes (Rho), the Guillardia/Hanusia group (GH), and Urgorri-like cryptophytes (Urg). ∗SGI sample was not properly preserved, so the relative proportion sequences within each category may have been compromised.
Baltic Sea planktonic communities have been shown to be rich in TPG cryptophytes, and are also known to have Hemiselmis and Rhodomonas spp. (Hill et al., 1992; Carstensen and Heiskanen, 2007; Hajdu et al., 2007). While none of our Baltic Sea samples were associated with a Mesodinium bloom, Mesodinium cell counts were not available and thus it is unclear to what degree their plastids may be contaminating the cryptophyte rbcL community signal. In addition to TPG species, we also found Falcomonas, Hemiselmis, and U. complanatus sequences in the Gulf of Finland. U. companatus was originally described from French, Spanish, and Portuguese estuaries and has a maximum growth rate at a salinity of 10 psu (Laza-Martínez, 2012); thus its presence within these brackish waters is not surprising. A single clone of U. complanatus was also recovered from our BS sample. While the BS was dominated by T. amphioxeia, M. major was found in this sample at ∼10 cells/ml, which would equate to about 400–600 T. amphioxeia plastids ml-1 based on plastid numbers reported by Garcia-Cuetos et al. (2012). Since our community samples were not size fractioned, in this case our clone libraries may have been highly enriched with plastids from this species since it is the preferred prey of the M. major/rubrum complex (Yih et al., 2004; Park et al., 2007; Myung et al., 2011; Hansen et al., 2012).
Peaks in cryptophyte abundance have been found to both proceed (Johnson et al., 2013) and co-occur (Kim et al., 2007; Weber et al., 2014) with high levels of M. rubrum-like ciliates in coastal ecosystems. However, most historical accounts of Mesodinium blooms did not note the abundance or composition of co-occurring cryptophyte communities. While certain aspects of M. major/rubrum blooms have been well-studied within a variety of ecosystems, such as the Southampton estuary (Crawford et al., 1997), upwelling zones from Ecuador to Chile (Ryther, 1967; Avaria, 1976; Jimenez and Intriago, 1987), Baltic Sea Fjords (Lindholm, 1978; Lindholm and Mörk, 1990; Sjöqvist and Lindholm, 2011), and portions of the Gulf of California (Gárate-Lizárraga et al., 2002; Bulit et al., 2004), only within the Columbia River estuary (Herfort et al., 2011a) have studies focused on cryptophyte-Mesodinium dynamics. Within this system, cryptophytes are high in abundance both prior to and during early M. rubrum blooms, but decline as ciliate concentrations increased (Peterson et al., 2013). Observations of M. rubrum cells from these communities revealed what appears to be a novel feeding strategy on cryptophyte algae, with numerous prey cells attached to either the cirri and/or feeding tentacles of the ciliates (Peterson et al., 2013). These observations suggest that some M. rubrum populations may be able to act opportunistically and quickly consume a surplus of cryptophytes in order to sustain their productivity. In the Columbia River, M. rubrum cells from blooms were analyzed for 16S rRNA gene diversity during two consecutive years and found to exclusively possess T. amphioxeia-like plastids (Herfort et al., 2011b). In contrast, surface clone libraries from the estuary were nearly devoid of cryptophyte sequences, suggesting they primarily reside at depth (see Peterson et al., 2013).
Conclusion
Our results solidify the link between Mesodinium blooms and T. amphioxeia-like cryptophytes in both temperate and subtropical Atlantic and Pacific ecosystems. While multiple variants of the M. major/rubrum complex are linked to estuarine and near-shore blooms, our results suggest that variant B is the most common agent in temperate and subtropical regions, while variants A and F may be involved in higher latitude blooms. The largest variant, M. major (D), was shown to be associated with shelf-water and offshore blooms in coastal Pacific regions and is likely responsible for the largest and most productive Mesodinium blooms (e.g., Ryther, 1967; Packard et al., 1978; Smith and Barber, 1979) associated with upwelling. Even non-bloom samples in our study were dominated by TPG cryptophyte sequences, implicating this group as playing a critical role in marine microbial foodwebs. Their widespread distribution and abundance, may explain why Mesodinium ciliates have adapted to exploit them for fueling their acquired phototrophy niche. We recommend that future studies of Mesodinium genetic diversity using targeted PCR should use the primers presented here in order to gain further insights into the diversity of the entire genus.
Author Contributions
MJ contributed to sampling, performed data analysis, and wrote the manuscript. DB performed all of the laboratory work for PCR, cloning, and sequencing preparation and contributed to writing the manuscript. AL-M provided cultures, helped interpret data, and contributed to writing the manuscript. SD, EF, SL, AM, MP, and OS provided field samples and helped edit the manuscript. DS contributed to data interpretation and writing the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Drs Wonho Yih and David Jaen for kindly providing the M. rubrum cultures MR-MAL01 and AND-A0711, respectively, for analysis. We also thank Drs Don Anderson and Mengmeng Tong for kindly providing T. amphioxeia culture GCEPO1.
Footnotes
Funding. MJ thanks the funding support of the National Science Foundations Grants NSF-OCE 1031718 and NSF-IOS 1326228. Project IT-699-13 from the Basque Government allows for maintenance of the microalgae culture collection at the University of the Basque Country (UPV/EHU).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.02017/full#supplementary-material
References
- Adolf J., Yeager C. L., Miller W. D., Mallonee M., Harding L., Jr. (2006). Environmental forcing of phytoplankton floral composition, biomass, and primary productivity in Chesapeake Bay, USA. Estuar. Coastal Shelf Sci. 67 108–122. 10.1016/j.ecss.2005.11.030 [DOI] [Google Scholar]
- Avaria S. (1976). Marea roja en la costa central de Chile. Rev. Biol. Mar. Valparaíso 16 96–111. [Google Scholar]
- Avaria S., Muñoz P. (1987). Effects of the 1982-1983 El Niño on the marine phytoplankton off northern Chile. J. Geophys. Res. 92 14369–14382. 10.1029/JC092iC13p14369 [DOI] [Google Scholar]
- Bass D., Brown N., Mackenzie-Dodds J., Dyal P., Nierzwicki-Bauer S. A., Vepritskiy A. A., et al. (2009). A Molecular perspective on ecological differentiation and biogeography of cyclotrichiid ciliates. J. Eukaryot. Microbiol. 56 559–567. 10.1111/j.1550-7408.2009.00434.x [DOI] [PubMed] [Google Scholar]
- Bazin P., Jouenne F., Deton-Cabanillas A.-F., Pérez-Ruzafa Á, Véron B. (2014). Complex patterns in phytoplankton and microeukaryote diversity along the estuarine continuum. Hydrobiologia 726 155–178. 10.1007/s10750-013-1761-9 [DOI] [Google Scholar]
- Bell E. M., Laybourn-Parry J. (1999). Annual plankton dynamics in an Antarctic saline lake. Freshw. Biol. 41 507–519. 10.1046/j.1365-2427.1999.00396.x [DOI] [Google Scholar]
- Bielewicz S., Bell E., Kong W., Friedberg I., Priscu J. C., Morgan-Kiss R. M. (2011). Protist diversity in a permanently ice-covered Antarctic Lake during the polar night transition. ISME J. 5 1559–1564. 10.1038/ismej.2011.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers H. A., Tengs T., Glasgow H. B., Burkholder J. M., Rublee P. A., Oldach D. W. (2000). Development of real-time PCR assays for rapid detection of Pfiesteria piscicida and related Dinoflagellates. Appl. Environ. Microbiol. 66 4641–4648. 10.1128/aem.66.11.4641-4648.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulit C., Diaz-Avalos C., Montagnes D. J. S. (2004). Assessing spatial and temporal patchiness of the autotrophic ciliate Myrionecta rubra: a case study in a coastal lagoon. Mar. Ecol. Prog. Ser. 268 55–67. 10.3354/meps268055 [DOI] [Google Scholar]
- Buma A., Gieskes W., Thomsen H. A. (1992). Abundance of Cryptophyceae and chlorophyll b-containing organisms in the Weddell-scotia confluence area in the spring of 1988. Polar Biol. 12 43–52. 10.1007/BF00239964 [DOI] [Google Scholar]
- Carlowicz M., Ciotti A. M., Kuring N. (2014). A Dark Bloom in the South Atlantic Greenbelt: NASA. Available at: http://earthobservatory.nasa.gov/IOTD/view.php?id=82968 [Google Scholar]
- Carstensen J., Heiskanen A.-S. (2007). Phytoplankton responses to nutrient status: application of a screening method to the northern Baltic Sea. Mar. Ecol. Prog. Ser. 336 29–42. 10.3354/meps336029 [DOI] [Google Scholar]
- Cerino F., Zingone A. (2007). “Decrypting cryptomonads: a challenge for molecular taxonomy,” in Unravelling the Algae: The Past, Present, and Future of Algal Systematics eds Brodie J., Lewis J. (Boca Raton, FL: CRC Press; ) 197–214. [Google Scholar]
- Chen X., Ma H.-G., Al-Rasheid K. A., Miao M. (2015a). Molecular data suggests the ciliate Mesodinium (Protista: Ciliophora) might represent an undescribed taxon at class level. Zool. Syst. 40 31–40. 10.11865/zs.20150102 [DOI] [Google Scholar]
- Chen X., Zhao X., Liu X., Warren A., Zhao F., Miao M. (2015b). Phylogenomics of non-model ciliates based on transcriptomic analyses. Protein Cell 6 373–385. 10.1007/s13238-015-0147-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevenet F., Brun C., Bañuls A.-L., Jacq B., Christen R. (2006). TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7:439 10.1186/1471-2105-7-439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford D. W. (1993). Some observations on morphological variation in the red-water ciliate Mesodinium rubrum. J. Mar. Biol. Assoc. U. K. 73 975–978. 10.1017/S002531540003486X [DOI] [Google Scholar]
- Crawford D. W., Purdie D. A. (1992). Evidence for avoidance of flushing from an estuary by a plankton, phototrophic ciliate. Mar. Ecol. Prog. Ser. 79 259–265. 10.3354/meps079259 [DOI] [Google Scholar]
- Crawford D. W., Purdie D. A., Lockwood A. P. M., Weissman P. (1997). Recurrent red-tides in the Southampton Water estuary caused by the phototrophic ciliate Mesodinium rubrum. Estuar. Coast. Shelf Sci. 45 799–812. 10.1006/ecss.1997.0242 [DOI] [Google Scholar]
- Darwin C. (1906). The Voyage of the Beagle. London: JM Dent & sons. [Google Scholar]
- Deane J. A., Strachan I. M., Saunders G. W., Hill D. R., McFadden G. I. (2002). Cryptomonad evolution: nuclear 18S rDNA phylogeny versus cell morphology and pigmentation. J. Phycol. 38 1236–1244. 10.1046/j.1529-8817.2002.01250.x [DOI] [Google Scholar]
- Dierssen H., McManus G. B., Chlus A., Qiu D., Gao B.-C., Lin S. (2015). Space station image captures a red tide ciliate bloom at high spectral and spatial resolution. Proc. Natl. Acad. Sci. U.S.A. 112 14783–14787. 10.1073/pnas.1512538112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenchel T., Hansen P. J. (2006). Motile behaviour of the bloom-forming ciliate Mesodinium rubrum. Mar. Biol. Res. 2 33–40. 10.1080/17451000600571044 [DOI] [Google Scholar]
- Fitt W. K., Brown B. E., Warner M. E., Dunne R. P. (2001). Coral bleaching: interpretation of thermal tolerance limits and thermal thresholds in tropical corals. Coral Reefs 20 51–65. 10.1007/s003380100146 [DOI] [Google Scholar]
- Gao F., Warren A., Zhang Q., Gong J., Miao M., Sun P., et al. (2016). The All-Data-Based evolutionary hypothesis of ciliated protists with a revised classification of the Phylum Ciliophora (Eukaryota, Alveolata). Sci. Rep. 6:24874 10.1038/srep24874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gárate-Lizárraga I., Band-Schmidt C., Cervantes-Duarte R., Urias D. E. (2002). Mesodinium rubrum (Lohmann) Hamburger y Buddenbrock red tides in the Gulf of California (Winter, 1998). Hídrobiológica 12 15–20. [Google Scholar]
- Garcia-Cuetos L. (2011). Photosynthetic Organelle Along a Food Chain. Ph.D. thesis, University of Copenhagen; Copenhagen. [Google Scholar]
- Garcia-Cuetos L., Moestrup O., Hansen P. J. (2012). Studies on the genus mesodinium II. ultrastructural and molecular investigations of five marine species help clarifying the taxonomy. J. Eukaryot. Microbiol. 59 374–400. 10.1111/j.1550-7408.2012.00630.x [DOI] [PubMed] [Google Scholar]
- Gast R. J., Dennett M. R., Caron D. A. (2004). Characterization of protistan assemblages in the Ross Sea, Antarctica, by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70 2028–2037. 10.1128/AEM.70.4.2028-2037.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Gustafson D. E., Stoecker D. K., Johnson M. D., Van Heukelem W. F., Sneider K. (2000). Cryptophyte algae are robbed of their organelles by the marine ciliate Mesodinium rubrum. Nature 405 1049–1052. 10.1038/35016570 [DOI] [PubMed] [Google Scholar]
- Hajdu S., Höglander H., Larsson U. (2007). Phytoplankton vertical distributions and composition in Baltic Sea cyanobacterial blooms. Harmful Algae 6 189–205. 10.1016/j.hal.2006.07.006 [DOI] [Google Scholar]
- Hansen P. J., Moldrup M., Tarangkoon W., Garcia-Cuetos L., Moestrup Ø. (2012). Direct evidence for symbiont sequestration in the marine red tide ciliate Mesodinium rubrum. Aquat. Microb. Ecol. 66 63–75. 10.3354/ame01559 [DOI] [Google Scholar]
- Hargraves P. (1991). Narrow river phytoplankton. Maritimes 35 6–7. [Google Scholar]
- Hart T. J. (1943). Darwin and ’water-bloom’. Nature 152 661–662. 10.1038/152661b0 [DOI] [Google Scholar]
- Hathaway T. K. (2014). FFHS Phytofinders to the Rescue Sea Grant, North Carolina. Available at: https://ncseagrant.ncsu.edu/blog/2014/11/06/ffhs-phytofinders-to-the-rescue/ (accessed June 11, 2014). [Google Scholar]
- Herfort L., Peterson T. D., Campbell V., Futrell S., Zuber P. (2011a). Myrionecta rubra (Mesodinium rubrum) bloom initiation in the Columbia River estuary. Estuar. Coast. Shelf Sci. 95 440–446. 10.1016/j.ecss.2011.10.015 [DOI] [Google Scholar]
- Herfort L., Peterson T. D., McCue L. A., Crump B. C., Prahl F. G., Baptista A. M., et al. (2011b). Myrionecta rubra population genetic diversity and its cryptophyte chloroplast specificity in recurrent red tides in the Columbia River estuary. Aquat. Microb. Ecol. 62 85–97. 10.3354/ame01460 [DOI] [Google Scholar]
- Herfort L., Peterson T. D., Prahl F. G., McCue L. A., Needoba J. A., Crump B. C., et al. (2012). Red waters of Myrionecta rubra are biogeochemical hotspots for the Columbia River Estuary with impacts on primary/secondary productions and nutrient cycles. Estuaries Coast. 35 878–891. 10.1007/s12237-012-9485-z [DOI] [Google Scholar]
- Hill D., Moestrup Ø, Vørs N. (1992). “Rekylalger (Cryptophyceae),” in Plankton i de Indre Danske Farvande ed. Thomsen H. A. (Copenhagen: Havforskning fra Miljøstyrelsen; ). [Google Scholar]
- Hill D. R. A., Wetherbee R. (1986). Proteomonas sulcata gen. et sp. nov. (Cryptophyceae), a cryptomonad with two morphologically distinct and alternating forms. Phycologia 25 521–543. 10.2216/i0031-8884-25-4-521.1 [DOI] [Google Scholar]
- Hoef-Emden K. (2005). Multiple independent losses of photosynthesis and differing evolutionaryrates in the genus Cryptomonas (Cryptophyceae): combined phylogenetic analyses of DNA sequences of the nuclear and the nucleomorph ribosomal operons. J. Mol. Evol. 60 183–195. 10.1007/s00239-004-0089-5 [DOI] [PubMed] [Google Scholar]
- Hoef-Emden K., Marin B., Melkonian M. (2002). Nuclear and nucleomorph SSU rDNA phylogeny in the Cryptophyta and the evolution of cryptophyte diversity. J. Mol. Evol. 55 161–179. 10.1007/s00239-002-2313-5 [DOI] [PubMed] [Google Scholar]
- Hoef-Emden K., Melkonian M. (2003). Revision of the genus Cryptomonas (Cryptophyceae): a combination of molecular phylogeny and morphology provides insights into a long-hidden dimorphism. Protist 154 371–409. 10.1078/143446103322454130 [DOI] [PubMed] [Google Scholar]
- Hoef-Emden K., Tran H.-D., Melkonian M. (2005). Lineage-specific variations of congruent evolution among DNA sequences from three genomes, and relaxed selective constraints on rbcL in Cryptomonas (Cryptophyceae). BMC Evol. Biol. 5:1 10.1186/1471-2148-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Xu H., Al-Rasheid K. A., Warren A., Hu X., Song W. (2011). Planktonic ciliate communities in a semi-enclosed bay of Yellow Sea, northern China: annual cycle. J. Mar. Biol. Assoc. U. K. 91 97–105. 10.1017/S002531541000175X [DOI] [Google Scholar]
- Jimenez R., Intriago P. (1987). “Observations on blooms of Mesodinium rubrum in the upwelling area off Ecuador,” in Proceedings of the International Symposium on Vertical Motion in the Equatorial Upper Ocean and its Effects Upon Living Resources and the Atmosphere: Oceanologica Acta Paris. [Google Scholar]
- Johnson M. D. (2011). Acquired phototrophy in ciliates: a review of cellular interactions and structural adaptations1. J. Eukaryot. Microbiol. 58 185–195. 10.1111/j.1550-7408.2011.00545.x [DOI] [PubMed] [Google Scholar]
- Johnson M. D., Oldach D., Delwiche C. F., Stoecker D. K. (2007). Retention of transcriptionally active cryptophyte nuclei by the ciliate Myrionecta rubra. Nature 445 426–428. 10.1038/nature05496 [DOI] [PubMed] [Google Scholar]
- Johnson M. D., Stoecker D. K. (2005). Role of feeding in growth and photophysiology of Myrionecta rubra. Aquat. Microb. Ecol. 39 303–312. 10.3354/ame039303 [DOI] [Google Scholar]
- Johnson M. D., Stoecker D. K., Marshall H. G. (2013). Seasonal dynamics of Mesodinium rubrum in Chesapeake Bay. J. Plankton Res. 35 877–893. 10.1093/plankt/fbt028 [DOI] [Google Scholar]
- Johnson M. D., Tengs T., Oldach D. W., Delwiche C. F., Stoecker D. K. (2004). Highly divergent SSU rRNA genes found in the marine ciliates Myrionecta rubra and Mesodinium pulex. Protist 155 347–359. 10.1078/1434461041844222 [DOI] [PubMed] [Google Scholar]
- Kim S., Park M. G., Moon C., Shin K., Chang M. (2007). Seasonal variations in phytoplankton growth and microzooplankton grazing in a temperate coastal embayment, Korea. Estuar. Coast. Shelf Sci. 71 159–169. 10.1016/j.ecss.2006.07.011 [DOI] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lasek-Nesselquist E., Wisceaver J. H., Hackett J. D., Johnson M. D. (2015). Insights into transcriptional changes that accompany organelle sequestration from the stolen nucleus of Mesodinium rubrum. BMC Genomics 16:806 10.1186/s12864-015-2052-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laza-Martínez A. (2012). Urgorri complanatus gen. et sp. Nov. (cryptophyceae), a red-tide-forming species in brackish waters. J. Phycol. 48 423–435. 10.1111/j.1529-8817.2012.01130.x [DOI] [PubMed] [Google Scholar]
- Laza-Martínez A., Arluzea J., Miguel I., Orive E. (2012). Morphological and molecular characterization of Teleaulax gracilis sp. nov. and T. minuta sp. nov.(Cryptophyceae). Phycologia 51 649–661. 10.2216/11-044.1 [DOI] [Google Scholar]
- Leegaard C. (1920). Microplankton from the Finnish waters during the month of May 1912. Acta Soc. Fenn. 48 1–41. [Google Scholar]
- Leppanen J. M., Bruun J. E. (1986). The role of pelagic ciliates including the autotrophic Mesodinium rubrum during the spring bloom of 1982 in the open northern Baltic proper. Ophelia 4 147–157. [Google Scholar]
- Lindholm T. (1978). Autumnal mass development of the “red water” ciliate Mesodinium rubrum in the Aland archipelago. Memo. Soc. Fauna Flora Fenn. 54 1–5. [Google Scholar]
- Lindholm T. (1981). On the ecology of Mesodinium rubrum (Lohmann) (Ciliata) in a stagnant brackish basin on Åland, SW Finland. Kiel. Meeresforsch. Sonderh. 5 117–123. [Google Scholar]
- Lindholm T. (1985). Mesodinium rubrum– a unique photosynthetic ciliate. Adv. Aquat. Microb. 3 1–48. [Google Scholar]
- Lindholm T., Mörk A. C. (1990). Depth maxima of mesodinium rubrum (Lohmann) hamburger and buddenbrock - examples from a stratified Baltic Sea Inlet. Sarsia 75 53–64. 10.1080/00364827.1990.10413441 [DOI] [Google Scholar]
- Maddison D. R., Maddison W. P. (2000). MacClade 4. Sunderland, MA: Sinauer. [Google Scholar]
- Mallin M. (1994). Phytoplankton ecology of North Carolina estuaries. Estuaries and Coasts 17 561–574. 10.2307/1352404 [DOI] [Google Scholar]
- Marín V., Rodríguez L., Vallejo L., Fuenteseca J., Oyarce E. (1993). Efectos de la surgencia costera sobre la productividad primaria primaveral de Bahía Mejillones del Sur (Antofagasta, Chile). Rev. Chil. Hist. Nat. 66 479–491. [Google Scholar]
- Metfies K., Gescher C., Frickenhaus S., Niestroy R., Wichels A., Gerdts G., et al. (2010). Contribution of the class cryptophyceae to phytoplankton structure in the German Bight. J. Phycol. 46 1152–1160. 10.1111/j.1529-8817.2010.00902.x [DOI] [Google Scholar]
- Moestrup Ø, Garcia-Cuetos L., Hansen P. J., Fenchel T. (2012). Studies on the genus Mesodinium I: ultrastructure and description of Mesodinium chamaeleon n. sp., a benthic marine species with green or red chloroplasts. J. Eukaryot. Microbiol. 59 20–39. 10.1111/j.1550-7408.2011.00593.x [DOI] [PubMed] [Google Scholar]
- Montagnes D. J. S., Allen J., Brown L., Bulit C., Davidson R., Diaz-Avalos C., et al. (2008). Factors controlling the abundance and size distribution of the phototrophic ciliate Myrionecta rubra in open waters of the North Atlantic. J. Eukaryot. Microbiol. 55 457–465. 10.1111/j.1550-7408.2008.00344.x [DOI] [PubMed] [Google Scholar]
- Myung G., Kim H. S., Park J. S., Park M. G., Yih W. (2011). Population growth and plastid type of Myrionecta rubra depend on the kinds of available cryptomonad prey. Harmful Algae 10 536–541. 10.1016/j.hal.2011.04.005 [DOI] [Google Scholar]
- Nishitani G., Nagai S., Baba K., Kiyokawa S., Kosaka Y., Miyamura K., et al. (2010). High-level congruence of Myrionecta rubra prey and Dinophysis species plastid identities as revealed by genetic analyses of isolates from japanese coastal waters. Appl. Environ. Microbiol. 76 2791–2798. 10.1128/AEM.02566-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani G., Nagai S., Takano Y., Sakiyama S., Baba K., Kamiyama T. (2008). Growth characteristics and phylogenetic analysis of the marine dinoflagellate Dinophysis infundibulus (Dinophyceae). Aquat. Microb. Ecol. 52 209–221. 10.3354/ame01233 [DOI] [Google Scholar]
- Owen R. W., Gianesella-Galvão S. F., Kutner M. B. B. (1992). Discrete, subsurface layers of the autotrophic ciliate Mesodinium rubrum off Brazil. J. Plankton Res. 14 97–105. 10.1093/plankt/14.1.97 [DOI] [Google Scholar]
- Packard T., Blasco D., Barber R. (1978). “Mesodinium rubrum in the Baja California upwelling system,” in Upwelling Systems ed. Tomczak M. (Berlin: Springer-Verlag; ) 73–89. [Google Scholar]
- Park J. S., Myung G., Kim H. S., Cho B. C., Yih W. (2007). Growth responses of the marine photosynthetic ciliate Myrionecta rubra to different cryptomonad strains. Aquat. Microb. Ecol. 48 83–90. 10.3354/ame048083 [DOI] [Google Scholar]
- Peterson T. D., Golda R. L., Garcia M. L., Li B., Maier M. A., Needoba J. A., et al. (2013). Associations between Mesodinium rubrum and cryptophyte algae in the Columbia River estuary. Aquat. Microb. Ecol. 68 117–130. 10.3354/ame01598 [DOI] [Google Scholar]
- Proença L. A. O. (2004). A red water caused by Mesodinium rubrum on the coast of Santa Catarina, Southern Brasil. Braz. J. Oceanogr. 52 153–161. [Google Scholar]
- Rial P., Laza-Martínez A., Reguera B., Raho N., Rodríguez F. (2015). Origin of cryptophyte plastids in Dinophysis from Galician waters: results from field and culture experiments. Aquat. Microb. Ecol. 76 163–174. 10.3354/ame01774 [DOI] [Google Scholar]
- Riobó P., Reguera B., Franco J. M., Rodríguez F. (2013). First report of the toxin profile of Dinophysis sacculus stein from LC-MS analysis of laboratory cultures. Toxicon 76 221–224. 10.1016/j.toxicon.2013.10.012 [DOI] [PubMed] [Google Scholar]
- Rychert K. (2004). The size structure of the Mesodinium rubrum population in the Gdañsk Basin. Oceanologia 46 439–444. [Google Scholar]
- Ryther J. H. (1967). Occurrence of red water off Peru. Nature 214 1318–1319. 10.1038/2141318a0 [DOI] [Google Scholar]
- Sanders R. W. (1995). Seasonal distributions of the photosynthesizing ciliates Laboea strobila and Myrionecta rubra ( = Mesodinium rubrum) in an estuary of the Gulf of Maine. Aquat. Microb. Ecol. 9 237–242. 10.3354/ame009237 [DOI] [Google Scholar]
- Setälä O., Autio R., Kuosa H., Rintala J., Ylöstalo P. (2005). Survival and photosynthetic activity of different Dinophysis acuminata populations in the northern Baltic Sea. Harmful Algae 4 337–350. 10.1016/j.hal.2004.06.017 [DOI] [Google Scholar]
- Setälä O., Kivi K. (2003). Planktonic ciliates in the Baltic Sea in summer: distribution, species association and estimated grazing impact. Aquat. Microb. Ecol. 32 287–297. 10.3354/ame032287 [DOI] [Google Scholar]
- Sjöqvist C. O., Lindholm T. J. (2011). Natural Co-occurrence of Dinophysis acuminata (Dinoflagellata) and Mesodinium rubrum (Ciliophora) in Thin Layers in a Coastal Inlet. J. Eukaryot. Microbiol. 58 365–372. 10.1111/j.1550-7408.2011.00559.x [DOI] [PubMed] [Google Scholar]
- Smith M., Hansen P. J. (2007). Interaction between Mesodinium rubrum and its prey: importance of prey concentration, irradiance and pH. Mar. Ecol. Prog. Ser. 338 61–70. 10.3354/meps338061 [DOI] [Google Scholar]
- Smith W. O., Jr., Barber R. (1979). A carbon budget for the autotrophic ciliate Mesodinium rubrum. J. Phycol. 15 27–33. 10.1111/j.0022-3646.1979.00027.x [DOI] [Google Scholar]
- Strüder-Kypke M. C., Wright A.-D. G., Foissner W., Chatzinotas A., Lynn D. H. (2006). Molecular phylogeny of litostome ciliates (Ciliophora, Litostomatea) with emphasis on free-living haptorian genera. Protist 157 261–278. 10.1016/j.protis.2006.03.003 [DOI] [PubMed] [Google Scholar]
- Taylor F. J. R., Blackboun D. J., Blackboun J. (1971). Red-water ciliate Mesodinium rubrum and its incomplete symbionts–review including new ultrastructural observations. J. Fish. Res. Bd. Canada 28 391–407. 10.1139/f71-052 [DOI] [Google Scholar]
- Weber F., Anderson R., Foissner W., Mylnikov A. P., Jürgens K. (2014). Morphological and molecular approaches reveal highly stratified protist communities along Baltic Sea pelagic redox gradients. Aquat. Microb. Ecol. 73 1–16. 10.3354/ame01702 [DOI] [Google Scholar]
- Yih W., Kim H. S., Jeong H. A., Myung G., Kim Y. G. (2004). Ingestion of cryptophyte cells by the marine photosynthetic ciliate Mesodinium rubrum. Aquat. Microb. Ecol. 36 165–170. 10.3354/ame036165 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


