Abstract
Glioblastoma multiforme (GBM) lacks effective therapeutic options leaving patients with a survival time of approximately one year. Recently, the alteration of chromatin modulators has been implicated in the pathogenesis and chemoresistance of numerous cancers; in particular, the Polycomb Group Proteins have been shown to play a role in glioblastoma progression and maintenance [1-5]. In this study, we aimed to identify drug combinations that decrease GBM cell viability by combining small molecule inhibitors against the Polycomb family with two standard chemotherapies. We identified dual inhibition of the CBX chromodomain with doxorubicin as a novel therapeutic strategy. While treatment with chromodomain inhibitor is non-toxic to cells alone, it dramatically increased the toxicity of standard chemotherapy drugs. We further validated an increase in DNA damage resulting in a G2/M block and subsequent apoptosis using the dual inhibitor treatment.
Keywords: CBX, glioblastoma, sensitize, epigenetic, Polycomb, doxorubicin, chromodomain, chemotherapy
Introduction
Glioblastoma multiforme (GBM) is a malignant brain tumor that comprises the majority of all gliomas [6]. Due to GBM’s highly aggressive nature, patients are left with a survival time of approximately twelve months [7]. Current treatments for GBM include surgery, radiation, and chemotherapy; however, numerous challenges exist in eradicating the tumor. Complete resection of the tumor is difficult due to the sticky finger-like morphology. Additionally, small molecules must be capable of permeating the blood brain barrier. Although the development of temozolomide (TMZ) in 2005 initially seemed promising, the five year survival rate for GBM patients has not improved [8,9]. It has been hypothesized that GBM develops resistance through its stem cell-like properties [10,11].
Polycomb group (PcG) proteins have a fundamental role in the development and maintenance of adult stem-cells [12-14]. Known to serve as transcriptional repressors, PcG proteins are classified into two complexes known as Polycomb Repressive Complex 1 and 2 (PRC1/2). PRC2, via its catalytic subunit EZH2, is responsible for the trimethylation of histone H3 lysine 27 (H3K27me3). Consequently, PRC1 is recruited to H3K27me3 via its chromatin-organization modifier domain (chromodomain) where it ubiquitinates H2A lysine 119 resulting in chromatin compaction and gene repression.
The PRC complexes are composed of three subunits (PRC2) and four subunits (PRC1); however, gene duplications have resulted in numerous mutually exclusive paralogs for each subunit [15]. PRC2 is comprised of EZH1 or EZH2, EED, and SUZ12. PRC1, on the other hand, can be much more diverse and is comprised of RING E3 ubiquitin ligase (RING1a/b), Polycomb Group Finger (BMI-1 or MEL18), Polyhomeotic (PHC1-3), and the chromodomain-containing chromobox homolog (CBX2,4,6,7,8).
Numerous PcG proteins have been implicated in GBM progression and maintenance [1,2]. The PRC2 catalytic subunit EZH2 is overexpressed and is important in the development of GBM resistance [1,3,4]. The PRC1 subunit BMI-1 has been demonstrated to play a role in GBM self-renewal and promotes stem cell-like characteristics [2]. The chromobox homolog protein has several misregulated paralogs in GBM [1]; the CBX6 and CBX7 paralogs are downregulated compared to normal tissue, whereas the CBX8 paralog is upregulated [1]. While phenotypical studies have demonstrated the importance of these CBX paralogs [1, unpublished data], the mechanism in which CBX misregulation impacts GBM progression and maintenance is unknown.
Recently, it has been suggested that epigenetic processes may serve as good therapeutic targets [16]. Over the past decade, there has been an emergence of epigenetic inhibitors, including small molecules against the PcG proteins. These inhibitors function in different ways to derepress gene transcription and alter chromatin structure. For example, EZH2 inhibitors block the catalytic methyltransferase domain preventing H3K27 trimethylation. Current inhibitors against PRC1 include RING inhibitors that block histone ubiquitination, CBX inhibitors that prevent chromodomain binding, and inhibitors that block the transcription or incorporation of BMI-1 [17-19]. None of these PcG inhibitors have been tested for efficacy against GBM cell lines.
Because of the limited therapies and low survival time, new therapeutic strategies need to be explored for glioblastoma, particularly to combat chemotherapy resistance. Previous studies have demonstrated that the knockdown of EZH2 and BMI-1 improves response to chemotherapies in a resistant cell population [4,5]. Here, we identify a novel therapeutic strategy to inhibit CBX chromodomain binding to improve GBM response to standard chemotherapy treatment.
Materials and Methods
Cell Culture
U118MG, T98G, A172, and SVGp12 cells from ATCC were maintained in Eagle’s Minimum Essential Medium (Corning) with 10 percent fetal bovine serum (Omega Scientific, Inc.), 1 percent penicillin-streptomycin (Corning), 1 percent non-essential amino acids (Corning), and 1 percent glutamine (Corning) at 37 °C and 5 percent CO2. MDA-MB-231 cells were maintained in Dulbecco’s Modified Essential Medium (Corning) with 10 percent fetal bovine serum (Omega Scientific, Inc.), 1 percent penicillin-streptomycin (Corning), 1 percent sodium pyruvate (Corning), 1 percent glutamine (Corning) at 37 °C and 5 percent CO2. Cells were plated in a 96 well plate at 2x103 (8.9x103 for MDA-MB-231) cells/well 48 hours prior to treatment.
Drug Screen
Cells were treated 48 hours after plating. Cells were dosed in a grid format for every combination of drug at the designated dose: PRT4165 40 µM (Cayman), PTC209 200 nM (Cayman), DZnep 25 µM (Cayman), GSK343 400 nM (Cayman), MS37452 200 µM (Cayman), Doxorubicin 200 nM, temozolomide 50 µM (Cayman), SAHA 1 µM (Cayman). Control cells were treated with 1 percent DMSO or a single drug. Cells were treated for a total of five days but redosed with the same treatments after 48 hours. Following five days of treatment, cells were fixed with 50 percent trichloroacetic acid (TCA) for an hour at 4 °C, washed with water, incubated in sulforhodamine B for 10 minutes, washed with 1 percent acetic acid and dried overnight. Protein was solubilized with 10 mM tris and 515 nm absorbance readings were taken.
Dose Response Curves
U118MG, A172, SVGp12, and MDA-MB-231 cells were treated in a 96 well-plate with varying concentrations of MS37452 (0, 15.6, 31.25, 62.5, 125, 250 µM) in combination with doxorubicin (200 nM) or DMSO for five days or and with varying concentrations of doxorubicin with standard dose of MS37452 (250 µM) or DMSO. After five days, protein levels were measured with the sulforhodamine assay described above. Cells were plated at 1.3x104 cells/well in a 24 well plate and treated with MS37452 (0, 31.25, 250 µM) and doxorubicin (100 nM) or DMSO in a similar format. Additional cells were plated at 2x103 cells/well in a 96 well plate and treated with MS37452 (0, 31.25, 250 µM) and doxorubicin (50 nM) or DMSO. Cells were washed with PBS, trypsinized, harvested and counted.
Western Blot Analysis
Cells were treated as indicated above for five days in a 6 well plate. Cells were harvested and lysed with Buffer A (25 mM HEPES, 5 mM KCl, 25 mM MgCl2, 0.05 mM EDTA, 10 percent glycerol, 0.1 percent NP-40, protease inhibitors) for 15 minutes on ice. Nuclei were pelleted and resuspended in RIPA buffer (50 mM tris, 150 mM NaCl, 0.1 percent SDS, 0.5 percent Na DOC, 1 percent triton-X, protease inhibitors, benzonase (Sigma)) for 15 minutes. Lysates with LDS and BME loading buffer were boiled and ran on an SDS-page 4-12 percent gel (Invitrogen). Gels were transferred to PVDF membranes (Millipore) and exposed to 5 percent BSA in PBSt (0.1 percent tween-20). Membranes were blotted with primary antibodies overnight at 4°C. The blots were washed with PBSt and incubated for an hour at room temperature with goat anti-rabbit or mouse conjugated to IR800CW or IRDye 680 (LI-COR) secondary antibody. Blots were imaged on the LI-COR Odyssey. Antibodies used are rabbit CBX8 (Bethyl, A300-882A), rabbit CBX7 (Abcam, ab21873), rabbit cleaved PARP-1 (Cell Signaling Technology, 9541), mouse H3 (Active Motif, am-61475), and rabbit phospho-histone H2A.X (ser139) (Cell Signaling Technology, 9718).
Peptide Pulldown
Nuclear lysate was made with untreated cells lysed with Buffer A for 15 minutes on ice. Nuclei were pelleted and resuspended in IP buffer (25 mM tris, 300 mM NaCl, 1 mM EDTA, 1 percent NP-40) for 15 minutes. 3 µg of biotinylated [Lys(Me3)27]-Histone H3 (21-44) and Histone H3 (21-44) (Eurogentec) were incubated with 10 µL of equilibrated streptavidin agarose resin (Solulink) (150 mM NaCl, 0.5 mM DTT, 50 mM tris, 1 percent NP-40) [20] at 4°C for an hour. Nuclear lysate was aliquoted and MS37452 was added to the lysate (0, 31.25, 250 µM). Lysate was divided and added to each peptide saturated resin and incubated at 4°C overnight. Resin was washed with equilibration buffer twice for ten minutes at 4°C. LDS with BME was added to the sample, boiled and ran on an SDS-page 4-12 percent gel (Invitrogen) as described above.
Flow Cytometry
U118 cells were plated in a 6 well plate 24 hours prior to drug treatment. Cells were treated with DMSO or CBX7i (250 µM) every 48 hours for four days. On the fifth day of treatment, doxorubicin (100 nM) was added to the cells for 16 hours. Cells were fixed and permeabilized following the Click iT Plus EdU Alexa Fluor 488 Flow cytometry protocol (Life Technologies). FxCycle PI/RNase staining solution (Life Technologies) was used for PI staining following the product manual. Cells were pelleted and resuspended in 300 µL of PBS and loaded on a 96-well plate. Unstained and single color controls were used to adjust the laser intensity of the Guava Easy-Cyte flow cytometer (Guava Technologies). 10,000 events were collected and data was processed and analyzed on FlowJo.
Immunofluorescence
U118MG cells were plated on glass coverslips in a 24-well plate seven days prior to staining. Cells were treated with CBX7i (250 µM) every 48 hours following plating. On day five, cells were treated with 1 µM of doxorubicin for an hour. After an hour, doxorubicin was removed and cells were allowed to recover for 0, 2, 4, 8, or 12 hours. Cells were fixed with 4 percent paraformaldehyde for 20 minutes. Fixed cells were stained with mouse phospho-H2A.X antibody (Millipore) overnight at 4 °C followed by secondary staining with a rabbit anti-mouse Alexa Fluor 594 antibody (Molecular Probes) for an hour at room temperature. The nuclei were stained with DAPI at 5µg/mL for 10 minutes. All cells were imaged at the same light intensity, brightness, and contrast at 40x magnification.
Data Analysis
Replicate absorbances (n = 3) were averaged and normalized to the DMSO control (n = 24) or to a single drug treatment (n = 3) to generate a heat map using RStudio. Red indicating low cell viability and blue indicating high cell viability. The mean corrected total cell fluorescence (CTCF) per cell was calculated using ImageJ and the equation CTCF = Integrated Density–(area of the cell x mean fluorescence background). Student’s t-tests were performed on the 515 nm absorbance data, cell count, flow cytometry data, and mean CTCF for microscopy. Immunoblots were quantitated with ImageJ software. The intensity of each band was measured, normalized to the loading control (H3). Normalized data was used to calculate fold change compared to the DMSO control. Peptide immunoblots were quantitated in a similar manner and normalized to the H3K27me3 0 µM treatment.
Results
Identifying Drug Combinations Enhancing Cell Response
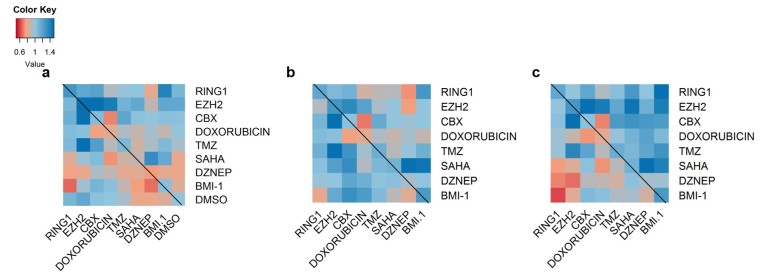
Initially, we performed a drug screen with a wide array of epigenetic inhibitors and chemotherapies to identify therapeutic combinations that decrease GBM cell viability. In a grid format to ensure all possible combinations, we treated GBM U118MG cells with inhibitors against histone deacetylase complexes (SAHA), CBX7 chromodomain (MS3742) [18], BMI-1 (PTC-209) [19], RING1 ubiquitin ligase (PRT4165) [17], and histone methyltransferases (GSK343, specific for EZH2 [21]; DZnep, inhibits global histone methylation [22]). Additionally, we included the chemotherapies, doxorubicin, a topoisomerase II inhibitor, and temozolomide, a DNA alkylating agent [23]. After treatment, bulk protein in the screen was stained with sulforhodamine B and the absorbance at 515 nm was taken and correlated to cell viability.
From our screen, we identified several combinations that resulted in consistently decreased cell viability compared to DMSO treated and single drug treatment: SAHA/TMZ and MS37452/doxorubicin (Figure 1a-c). It was necessary to normalize the absorbance values to both DMSO-treated and a single drug treatment. Normalization to a single drug treatment is important to ensure that the decrease in viability is a result of the combination of drugs, and not just one of the drugs. If it were the result of only one of the two drugs normalized viability would be 100 percent. The SAHA/TMZ combined treatment has been identified prior to our study, and clinical trials examining the effect SAHA and TMZ have together on GBM progression are in progress [24]; however, MS37452, which is a CBX7 chromodomain inhibitor (CBX7i), in combination with doxorubicin is a novel therapeutic strategy.
Figure 1.
Select epigenetic inhibitors sensitize GBM U118MG cells to doxorubicin and temozolomide chemotherapies. a. A heat map demonstrates the effect of a dual drug treatment on cell viability (n = 3, high viability ( > 77 percent), blue; low viability ( < 76 percent), red) compared to DMSO treated cells (n = 18). b,c. Heat maps represent the effect of a dual drug treatment compared to cells treated with a single drug. Each dual treatment was normalized to the single drug treatment indicated on the right column (b) or the drug indicated on the bottom (c). Heat maps in a-c were generated from sulforhodamine 515 nm assays to correlate bulk protein to cell viability.
CBX7 Chromodomain Inhibition with Doxorubicin Decreases Viability
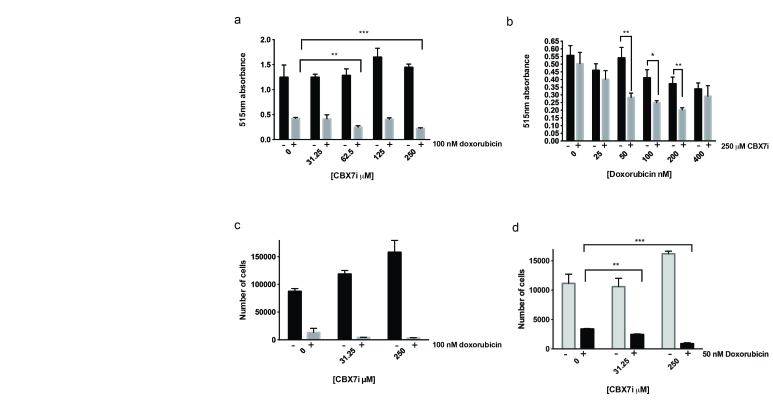
We subjected U118MG cells to varying concentrations of CBX7i while the doxorubicin concentration remained constant (200 nM) for five days. Following the five-day treatment regimen, we used a sulforhodamine assay to measure bulk protein adhered to the plate, which should correlate to cell viability. Interestingly, we observed a slight increase in cell viability with increasing concentrations of CBX7i; however, at high concentrations of CBX7i with doxorubicin, cell viability was significantly decreased compared to CBX7i or doxorubicin only treatments (Figure 2a). Unsurprisingly, when we treated cells with varying concentrations of doxorubicin and kept CBX7i concentration constant (250 µM), we observe a similar response to the combinatorial treatment (Figure 2b).
Figure 2.
CBX7i with doxorubicin decreases cell viability. a. U118MG cells treated with CBX7i in the presence (grey) or absence (black) of 100 nM doxorubicin total protein measured by sulforhodamine B 515 nm absorbance (n = 3) b. U118MG cells treated with doxorubicin in the presence (grey) or absence (black) of 250 µM CBX7i bulk protein measured by sulforhodamine B 515 nm absorbance (n = 3) c. U118MG cells were treated with CBX7i in the presence (grey) or absence (black) of 100 nM doxorubicin and counted (n = 3) d. U118MG cells in a 96 well plate were treated CBX7i in the presence (grey) or absence (black) of 50 nM doxorubicin for five days and counted (n = 3) Data in a-d represented by mean ± SEM, p-values calculated with student’s t-test: (*) < 0.05, p (**) < 0.01, p (***) < 0.001.
The sulforhodamine assay is limited to measuring bulk protein, which can remain adhered to the plate after cell death, so we also measured viability by counting live cells. Under the same conditions, we observe a similar trend to the sulforhodamine assay, confirming our results (Figure 2c). Finally, at minimal doxorubicin (50 nM), we still observe a significant decrease in cell viability (Figure 2d). This suggests that dual therapeutic strategy for GBM will require lower doses of doxorubicin, potentially minimizing the toxic side effects of chemotherapy.
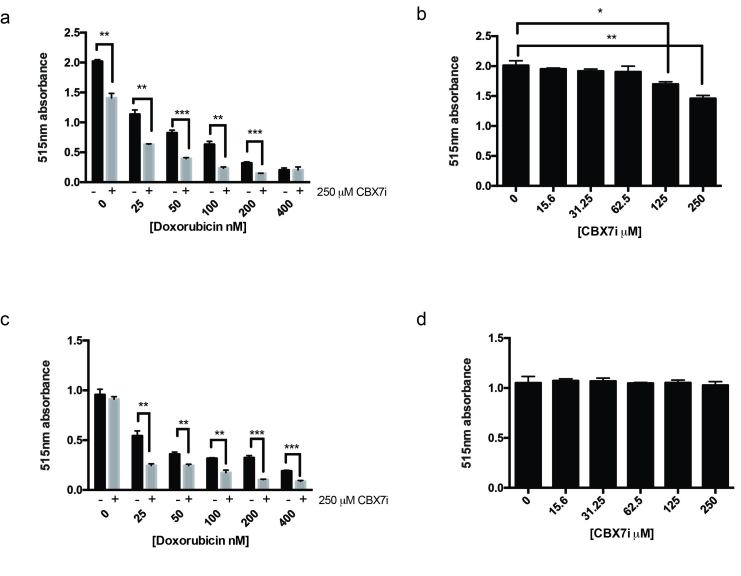
GBM tumors, however, are highly heterogeneous, thus it is important to determine if this phenomenon is observed in other GBM derived cells. To answer this question, we performed our sulforhodamine cell viability assay with the A172 GBM cell line. As expected, the dual treatment increases doxorubicin toxicity in A172 cells (Figure 3a). Interestingly, unlike the U118MG cell lines, the A172 cells displayed sensitivity to the CBX7i (Figure 3b). Because of the observed sensitivity and decrease in cell viability in the presence of only the CBX7i, it is difficult to interpret if the combined effect is a result of increased toxicity to doxorubicin.
Figure 3.
A172 GBM cells are sensitive to CBX7i, but astrocytes are not. a. A172 cells treated with doxorubicin in the presence (grey) or absence (black) of 250 µM CBX7i bulk protein measured by sulforhodamine B 515 nm absorbance (n = 3) b. A172 cells were treated with CBX7i and bulk protein measured by sulforhodamine B 515 nm absorbance (n = 3) c. SVGp12 cells treated with doxorubicin in the presence (grey) or absence (black) of 250 µM CBX7i bulk protein measured by sulforhodamine B 515 nm absorbance (n = 3) d. SVGp12 cells were treated with CBX7i and bulk protein measured by sulforhodamine B 515 nm absorbance (n = 3) Data in a-d represented as mean ± SEM, p-values calculated by student’s t-test: p (*) < 0.05, p (**) < 0.01, p (***) < 0.001.
In glioblastoma, CBX7 is downregulated in over 80 percent of patients compared to normal brain tissue [1]. Thus, we found it important to understand the impact of the CBX7i on non-tumorigenic cells. Using our sulforhodamine assay with the SVGp12 astrocyte cell line, we observed that the astrocytes were not sensitive to the CBX7i alone (Figure 3d). As an intercalator and topoisomerase 2 inhibitor, doxorubicin primarily targets cycling cells. Since the SVGp12 cell line is highly proliferative, doxorubicin was, not surprisingly, toxic to the cells (Figure 3c). Similar to the GBM cell lines, this doxorubicin toxicity was exacerbated in the presence of CBX7i.
The A172 cells’ sensitivity to CBX7i is interesting and could be a result of the heterogeneity of GBM and the diversity of PRC1 composition. While CBX7 expression is generally low across GBM patients, it is possible that A172 cell lines have higher expression of CBX7 and are thus more sensitive to the CBX7i. The response of A172 cells also suggests that CBX7 may serve as a therapeutic target for a subset of glioblastomas that are reliant on CBX7 activity.
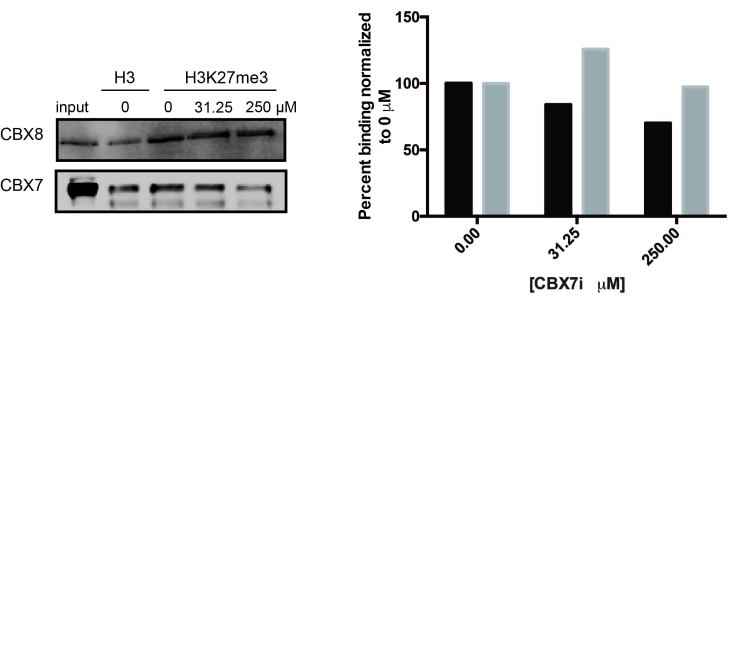
CBX7i is Specific for CBX7
To confirm the specificity of chromodomain inhibition, we performed peptide pulldown studies with unmethylated H3 and tri-methylated H3K27 peptides in presence of the inhibitor. The CBX inhibitor has previously been published to be specific for the CBX7 paralog chromodomain with a Kd of 28.9 µM [18]. As expected, we observed a decrease in CBX7 binding to the methylated peptide in the presence of the inhibitor, particularly at 250 µM (Figure 4a,b). CBX8 binding, however, in the presence of 250 µM was not affected (Figure 4a,b). The pulldown suggests that the effect seen at 250 µM is not due to inhibition of CBX8 chromodomain binding; however, inhibition of CBX4 (Kd of 95.8 µM) cannot be ruled out as our highest concentration used is 1.5 fold higher than the CBX4 Kd.
Figure 4.
CBX7i disrupts CBX7 binding to H3K27me3. a. Pulldowns with unmethylated H3 or H3K27me3 peptides confirm that increasing concentrations block CBX7 (T98G cells) binding but not CBX8 (U118MG cells). b. Quantitation of immunoblots normalized to H3K27me3 0 µM treatment. CBX7, black; CBX8, grey.
CBX7 Inhibition Increases Doxorubicin-induced DNA Damage and Apoptosis
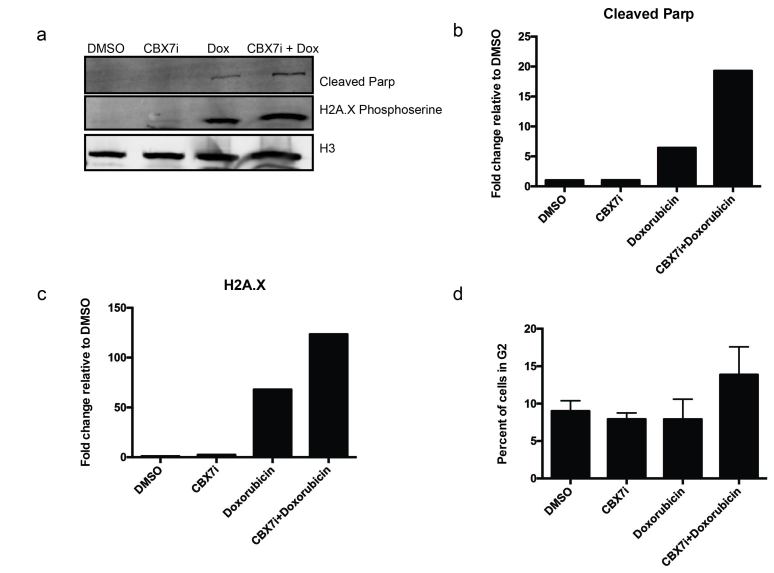
Doxorubicin acts through DNA intercalation and causes double stranded DNA breaks by inhibiting the re-ligation step of topoisomerase II [25]. To understand the role of CBX7i in enhancing doxorubicin response, we examined the protein expression level of DNA damage and apoptosis markers by immunoblot. When cells were treated with doxorubicin only (100 nM), we observe an induction of γH2A.X indicating DNA damage; however, in the presence of 250 µM CBX7i and doxorubicin, we observe a further 1.5-fold increase in γH2A.X induction (Figure 5a,c).
Figure 5.
CBX7i and doxorubicin treatment increases DNA damage and apoptosis. a. Western blot analysis of nuclear lysate staining for H2A.X serine 139 phosphorylation, cleaved PARP-1, and histone H3 (loading control) b. Quantitation of cleaved PARP-1 normalized to H3 loading control, fold change relative to DMSO-treated protein levels c. Quantitation of H2A.X normalized to H3 loading control, fold change relative to DMSO-treated protein levels d. Cell cycle analysis of cells treated with DMSO, CBX7i (250 µM), doxorubicin (100 nM) or CBX7i and doxorubicin (250 µM, 100 nM respectively), percent of cells in G2 plotted as a percent of total gated cells.
If DNA damage is not repaired, cells will undergo apoptosis. In order to determine if the dual treatment augments apoptosis, we examined cleaved PARP-1 levels. When cells undergo apoptosis, caspases cleave PARP-1, rendering it inactive. Our immunoblot analysis reveals that both doxorubicin and the combinatorial treated cells undergo apoptosis. Nevertheless, the CBX7i/doxorubicin combination increases cleaved PARP-1 two-fold compared to doxorubicin only (Figure 5a,b). This suggests that CBX7i enhances the impact of doxorubicin on DNA damage and apoptosis.
CBX7i and Doxorubicin Induce a G2/M Block
We performed flow cytometry to examine the differences in cell cycle amongst the different treatments. Following a four-day treatment of either CBX7i or DMSO, we pulsed cells with 100 nM of doxorubicin for 16 hours and stained cells with Propidium Iodide (PI) for cell cycle analysis. Unsurprisingly, our DMSO and CBX7i treated cells had similar cell cycle phase distributions with a majority of the cell population in G1 and a smaller percent of cells in G2/M (Figure 5d) When we treated cells for 16h with a lower dose of doxorubicin (100 nM), we did not observe an increase in G2/M [26]; however, in accordance with an increase in DNA damage and apoptosis, the dual drug treatment increased the number of cells in G2/M by 50 percent (Figure 5d).
Presence of CBX7i Prevents DNA Damage Repair
There are several potential mechanisms in which CBX inhibition can promote DNA damage in doxorubicin treated cells. Chromatin modulators, like the PcG proteins, are responsible for altering chromatin structure and thus DNA accessibility. It has been suggested that open chromatin is more susceptible to DNA damaging agents [27]. It is possible that inhibiting the CBX proteins allow for more open chromatin, as they can no longer be recruited to histones to compact chromatin and repress transcription. Additionally, previous studies have demonstrated that the recruitment of PcG proteins, including the CBXs, to sites of DNA damage are important for the localization of other DNA damage machinery and subsequent DNA damage repair [28,29].
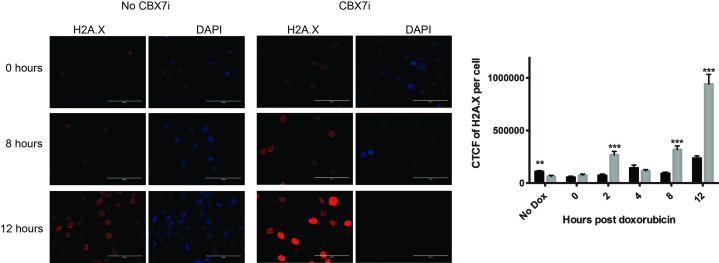
To understand which mechanism may be contributing to the increased sensitivity to doxorubicin with CBX7i, we used H2A.X staining to investigate DNA damage repair after acute doxorubicin treatment (Figure 6). Immediately after doxorubicin treatment there was no difference in DNA damage with or without CBX7i (Figure 6). As cells had time for damage to accumulate and begin recovery, however, we observed a significant increase in DNA damage accumulation in the presence of CBX7i beginning at two hours (Figure 6). This increase in DNA damage was drastically extended as recovery time progressed (Figure 6). Together, our data suggests that treatment of GBM cells with a CBX7 chromodomain inhibitor improves the response to the chemotherapeutic doxorubicin by preventing DNA damage repair, although these results are still preliminary. Based on these data, we hypothesize that CBX7 chromodomain inhibition prevents DNA repair, resulting in a G2/M block and eventual apoptosis. Future studies will be needed to investigate this mechanism further.
Figure 6.
CBX inhibition prevents DNA damage repair. U118MG cells were treated for four days in the presence or absence of CBX7i. On day five, cells were treated with doxorubicin (1 µM) for one hour. After the treatment, doxorubicin was removed and cells were allowed to recovery for either 0, 2, 4, 8, or 12 hours. If cells were treated with CBX7i, they remained on CBX7i throughout the entire process. Following recovery, cells were fixed and stained for phospho-H2A.X and DAPI. The cells were imaged and the mean total corrected cell fluorescence (CTCF) per cell was measured for each treatment type. Mean fluorescence per cell was quantitated and plotted (grey, CBX7i treatment; black, no CBX7i treatment) (n = 77, 111, 87, 64, 66, 64, 65, 80, 43, 78, 72, 88, left to right) Data represented as mean ± SEM, p-values calculated by student’s t-test: p (**) < 0.01, p (***) < 0.001.
CBX7 Inhibition Increases Doxorubicin Toxicity in Breast Cancer
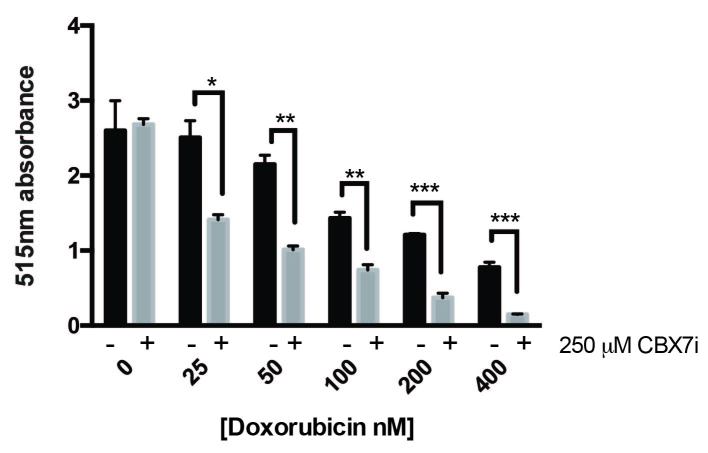
Doxorubicin is not a standard treatment for glioblastoma due to its inability to cross the blood brain barrier. However, it is often used in other cancers such as breast, lung, and leukemia [25]. In order to verify that this dual treatment is applicable to these cancers, we tested the combination of CBX7i and doxorubicin in the breast cancer MDA-MB-231 cell line. Consistent with our findings in GBM, the dual therapy enhances the toxicity of doxorubicin (Figure 7).
Figure 7.
CBX inhibition enhances doxorubicin toxicity in breast cancer. MDA-MB-231 cells were treated with a range of doxorubicin concentrations (0-400 nM) in the presence (grey) or absence (black) of 250 µM of CBX7i; total protein was measured by sulforhodamine B 515 nm absorbance (n = 3) Data represented as mean ± SEM, p-values calculated by student’s t-test: p (*) < 0.05, p (**) < 0.01, p (***) < 0.001.
Discussion
With little improvement in glioblastoma survival over the last decade, it is critical to develop new therapeutic strategies. Although the exact role PcG proteins play in GBM progression and maintenance is unclear, their involvement is necessary [1-5,10,11, unpublished data]. In this study, we identified two therapeutic strategies, inhibition of CBX7 chromodomain binding and histone deacetylase activity, to improve GBM response to traditional chemotherapies. Although we saw a sensitivity to TMZ with the HDAC inhibitor SAHA, there are already currently ongoing clinical trials with the drug combination [24] and previous in vitro studies have investigated potential mechanisms [30-32]. Therefore, we investigated the completely novel approach of inhibiting the CBX7 chromodomain to improve chemotherapeutic response to doxorubicin.
Our findings indicate that inhibition of the CBX7 chromodomain drastically increases DNA damage in response to doxorubicin. Our cell cycle analysis data indicates a G2/M block suggesting that damage cannot be properly repaired without CBX7.
PcG Proteins in DNA Damage Response
Roles for PcG proteins in DNA damage response (DDR) have previously been identified. The majority of PcG proteins, including EZH2 and the CBXs, localize to regions of damaged chromatin in order to recruit DNA damage repair machinery [28,29]. However, efforts to inhibit PRC1 subunits with small molecule inhibitors, particularly with a focus on DNA damage, have been limited. We hypothesized that the inhibition of CBX proteins prevents PcG and other DNA damage repair machinery recruitment to DNA damage. Consequently, DNA damage accumulates and the cells undergo apoptosis. Initial treatment of doxorubicin in both the presence and absence of CBX7i induced similar levels of H2A.X, suggesting that CBX7i is not improving response to chemotherapy by altering DNA accessibility. However, as the cells are allowed to recover from the initial DNA damage, the total DNA damage drastically accumulates, whereas cells with only doxorubicin have significantly less DNA damage. Our data suggests that inhibiting CBX enhances chemotherapeutic response by preventing DNA damage repair, allowing massive accumulation of DNA damage. Although CBX7 has been shown to localize to sites of DNA damage, its role in the process is unknown [28]. CBX4, however, has been studied and shown to be an important part of the DNA damage response [33,34]. These studies have demonstrated that CBX4 is recruited early to sites of DNA damage, and loss of CBX4 extenuates the presence of DNA damage [31,32]. While we used an inhibitor specific for CBX7, the concentration used in our experiments exceeded the Kd for CBX4 [18]. Thus, it is still a possibility that the drastic increase in DNA damage in the presence of CBX7i and doxorubicin is a result of CBX4 inhibition. In addition, we cannot eliminate the possibility that inhibition of CBX7 is playing a transcriptional role in improving the response to chemotherapy. Further studies are necessary to fully dissect the mechanism.
Future Potential of CBX Inhibitors
CBX7i is a first generation chromodomain small molecule inhibitor [18]. Although the CBX7i is not a potent small molecule for the clinic, it is does demonstrate very useful properties. It serves as a useful tool to study the biochemistry of the CBX proteins, particularly CBX7. Additional studies with the recently developed second generation CBX7 inhibitor, as well as the recently developed CBX4/7 inhibitor will be interesting to further understand if the effect observed in these studies can be extended to different CBX inhibitors [35,36].
While we identified a novel therapeutic strategy that results in cell death in vitro, doxorubicin is not currently a viable chemotherapy for glioblastoma as it does not cross the blood brain barrier; however, understanding how inhibition of the CBX proteins can improve response to DNA damaging agents is an important area of research. DNA damaging agents are still the most clinically used chemotherapies, and we have demonstrated that CBX inhibition presents itself as a promising strategy for other cancers such as breast cancer where doxorubicin is a first line of treatment [25]. Finally, there is ongoing research to develop drug carriers that will improve blood-brain barrier penetrance, so that eventually drugs, like doxorubicin can be used in glioblastoma treatment [37,38].
Identifying therapeutic strategies as described in this paper not only will provide us new drug targets for cancers but will also allow clinicians to reduce the amount of chemotherapy necessary to kill cancer cells. This will reduce some of the toxic side effects of the anthracyclines. Though many studies need to be completed, the idea of targeting CBX proteins in combination with current treatments is novel and has great potential.
Acknowledgments
This publication was made possible with partial support from Grant # UL1TR001108 (A. Shekhar, PI) from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award. The authors thank Dr. Wells Brown and the Wendt lab for their assistance with the flow cytometer, the immunofluorescence secondary antibody, and troubleshooting immunofluorescence, the Hu lab for the H2A.X antibody for immunofluorescence, the Weake lab for helpful discussion, and Mr. Joseph Ruhl and Jefferson High School for the opportunity to work with local high school students.
Abbreviations
- PRC1
Polycomb Repressive Complex 1
- CBX
chromobox
- GBM
glioblastoma multiforme
- TMZ
temozolomide
- PcG
Polycomb Group
- PRC2
Polycomb Repressive Complex 2
- H3K27me3
histone H3 lysine 27 trimethylation
- dox
doxorubicin
Author Contributions
KEC and ECD designed experiments. KEC and ECM performed experiments, KEC and ECD wrote the manuscript.
References
- Li G, Warden C, Zou Z. et al. Altered expression of polycomb group genes in glioblastoma multiforme. PLoS One. 2013;8(1):e80970. doi: 10.1371/journal.pone.0080970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdouh M, Facchino S, Chatoo W. et al. BMI1 sustains human glioblastoma multiforme stem cell renewal. J Neurosci. 2009;29(28):8884–8896. doi: 10.1523/JNEUROSCI.0968-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-H, Joshi K, Ezhilarasan R. et al. EZH2 protects glioma stem cells from radiation-induced cell death in a MELK/FOXM1-dependent manner. Stem cell reports. 2015;4(2):226–238. doi: 10.1016/j.stemcr.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan T-Y, Wang H, Xiang P. et al. Inhibition of EZH2 reverses chemotherapeutic drug TMZ chemosensitivity in glioblastoma. Int J Clin Exp Pathol. 2014;7(10):6662–6670. [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Shang C, Xue Y. et al. Silencing of Bmi-1 gene enhances chemotherapy sensitivity in human glioblastoma cells. Med Sci Monit. 2015;21:1002–1007. doi: 10.12659/MSM.893754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109(1):93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- Urbańska K, Sokołowska J, Szmidt M. et al. Glioblastoma multiforme - an overview. Contemp Oncol (Pozn) 2014;18(5):307–312. doi: 10.5114/wo.2014.40559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarica FB, Cekinmez M, Tufan K. et al. Five-year follow-up results for patients diagnosed with anaplastic astrocytoma and effectiveness of concomitant therapy with temozolomide for recurrent anaplastic astrocytoma. Asian J Neurosurg. 2012;7(4):181–190. doi: 10.4103/1793-5482.106650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DR, O’Neill BP. Glioblastoma survival in the United States before and during the temozolomide era. J Neurooncol. 2012;107(2):359–364. doi: 10.1007/s11060-011-0749-4. [DOI] [PubMed] [Google Scholar]
- Cheng L, Bao S, Rich JN. Potential therapeutic implications of cancer stem cells in glioblastoma. Biochem Pharmacol. 2010;80(5):654–665. doi: 10.1016/j.bcp.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Hassiotou F, Nowak A. Glioblastoma stem-like cells: at the root of tumor recurrence and a therapeutic target. Carcinogenesis. 2014;36(2):177–185. doi: 10.1093/carcin/bgu243. [DOI] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7(3):299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA. et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125(2):301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J, Bernard D, Peters G. Role of polycomb group proteins in stem cell self-renewal and cancer. DNA Cell Biol. 2005;24(2):117–125. doi: 10.1089/dna.2005.24.117. [DOI] [PubMed] [Google Scholar]
- Sowpati DT, Senthilkumar R, Mishra RK. et al. Expansion of the polycomb system and evolution of complexity. Mech Dev. 2015;138(Pt. 2):97–112. doi: 10.1016/j.mod.2015.07.013. [DOI] [PubMed] [Google Scholar]
- Clarke J, Penas C, Pastori C. et al. Epigenetic pathways and glioblastoma treatment. Epigenetics. 2013;8(8):785–795. doi: 10.4161/epi.25440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail IH, McDonald D, Strickfaden H. et al. A small molecule inhibitor of polycomb repressive complex 1 inhibits ubiquitin signaling at DNA double-strand breaks. J Biol Chem. 2013;288(37):26944–26954. doi: 10.1074/jbc.M113.461699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Morohashi K, Plotnikov AN. et al. Small-Molecule Modulators of Methyl-Lysine Binding for the CBX7 Chromodomain. Chem Biol. 2015;22(2):161–168. doi: 10.1016/j.chembiol.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreso A, van Galen P, Pedley NM. et al. Self-renewal as a therapeutic target in human colorectal cancer. Nat Med. 2014;20(1):29–36. doi: 10.1038/nm.3418. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F. et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142(6):967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Verma SK, Tian X, LaFrance LV. et al. Identification of Potent, Selective, Cell-Active Inhibitors of the Histone Lysine Methyltransferase EZH2. ACS Med Chem Lett. 2012;3(12):1091–1096. doi: 10.1021/ml3003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda TB, Cortez CC, Yoo CB. et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther. 2009;8(6):1579–1588. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Stevens MFG, Bradshaw TD. et al. Temozolomide: mechanisms of action, repair and resistance. Curr Mol Pharmacol. 2012;5(1):102–114. doi: 10.2174/1874467211205010102. [DOI] [PubMed] [Google Scholar]
- Lee EQ, Puduvalli VK, Reid JM. et al. Phase I study of vorinostat in combination with temozolomide in patients with high-grade gliomas: North American Brain Tumor Consortium Study 04-03. Clin Cancer Res. 2012;18(21):6032–6039. doi: 10.1158/1078-0432.CCR-12-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacar O, Sriamornsak P, Dass CR. et al. Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems. J Pharm Pharmacol. 2013;65(2):157–170. doi: 10.1111/j.2042-7158.2012.01567.x. [DOI] [PubMed] [Google Scholar]
- Ling YH, el-Naggar AK, Priebe W. et al. Cell cycle-dependent cytotoxicity, G2/M phase arrest, and disruption of p34cdc2/cyclin B1 activity induced by doxorubicin in synchronized P388 cells. Mol Pharmacol. 1996;49(5):832–841. [PubMed] [Google Scholar]
- Zhou Y, Feng X, Koh DW. et al. Enhanced DNA Accessibility and Increased DNA Damage Induced by the Absence of Poly(ADP-ribose) Hydrolysis. Biochemistry. 2010;49(34):7360–7366. doi: 10.1021/bi100979j. [DOI] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE. et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci. 2010;107(43):18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieni RS, Ismail IH, Campbell S. et al. Polycomb group proteins in the DNA damage response: A link between radiation resistance and “stemness.”. Cell Cycle. 2011;10(6):883–894. doi: 10.4161/cc.10.6.14907. [DOI] [PubMed] [Google Scholar]
- Ryu CH, Yoon WS, Park KY. et al. Valproic Acid Downregulates the Expression of MGMT and Sensitizes Temozolomide-Resistant Glioma Cells. J Biomed Biotechnol. 2012;2012:1–9. doi: 10.1155/2012/987495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitange GJ, Mladek AC, Carlson BL. et al. Inhibition of histone deacetylation potentiates the evolution of acquired temozolomide resistance linked to MGMT upregulation in glioblastoma xenografts. Clin Cancer Res. 2012;18(15):4070–4079. doi: 10.1158/1078-0432.CCR-12-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker CA, Bishop AJ, Chang M. et al. Valproic acid use during radiation therapy for glioblastoma associated with improved survival. Int J Radiat Oncol Biol Phys. 2013;86(3):504–509. doi: 10.1016/j.ijrobp.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail IH, Gagné J-P, Caron M-C. et al. CBX4-mediated SUMO modification regulates BMI1 recruitment at sites of DNA damage. Nucleic Acids Res. 2012;40(12):5497–5410. doi: 10.1093/nar/gks222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisch F, Pozzi B, Risso G. et al. DNA Damage-induced Heterogeneous Nuclear Ribonucleoprotein K SUMOylation Regulates p53 Transcriptional Activation. J Biol Chem. 2012;287(36):30789–30799. doi: 10.1074/jbc.M112.390120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Smith SG, Yap K. et al. Structure-Guided Discovery of Selective Antagonists for the Chromodomain of Polycomb Repressive Protein CBX7. ACS Med Chem Lett. 2016;7(6):601–605. doi: 10.1021/acsmedchemlett.6b00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckey JI, Dickson BM, Cheng N. et al. A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1. Nat Chem Biol. 2016;12(3):180–187. doi: 10.1038/nchembio.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Chen H, Zhang Q. et al. Liposome formulated with TAT-modified cholesterol for improving brain delivery and therapeutic efficacy on brain glioma in animals. Int J Pharm. 2011;420(2):304–312. doi: 10.1016/j.ijpharm.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Sardi I, la Marca G, Cardellicchio S. et al. Pharmacological modulation of blood-brain barrier increases permeability of doxorubicin into the rat brain. Am J Cancer Res. 2013;3(4):424–432. [PMC free article] [PubMed] [Google Scholar]