Abstract
The dynamic orchestration of gene expression is crucial for the proper differentiation, function, and adaptation of cells. In the brain, transcriptional regulation underlies the incredible diversity of neuronal cell types and contributes to the ability of neurons to adapt their function to the environment. Recently, novel methods for genome and epigenome editing have begun to revolutionize our understanding of gene regulatory mechanisms. In particular, the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system has proven to be a particularly accessible and adaptable technique for genome engineering. Here, we review the use of CRISPR/Cas9 in neurobiology and discuss how these studies have advanced understanding of nervous system development and plasticity. We cover four especially salient applications of CRISPR/Cas9: testing the consequences of enhancer mutations, tagging genes and gene products for visualization in live cells, directly activating or repressing enhancers in vivo, and manipulating the epigenome. In each case, we summarize findings from recent studies and discuss evolving adaptations of the method.
Keywords: CRISPR/Cas9, genome editing, neuron, chromatin, enhancer, transcription
Introduction
The brain presents an ideal frontier into which to deploy the remarkable technical advances in genome biology that emerged from the completion of the Human Genome Project. Although the incredible diversity of cell types in the mammalian brain share a single genomic blueprint, they express highly distinct transcriptional profiles, which neurobiologists have begun to characterize at the single-cell level using the emergence of increasingly sensitive next-generation sequencing technologies. Furthermore, even once they are fully differentiated, neurons experience large-scale dynamic waves of new gene transcription in response to environmental exposures. The evidence that these transcriptional states can be mediated by activity-dependent regulation of chromatin modifying enzymes has driven an explosion in the field of neuroepigenetics [1,2]. This field addresses the tempting hypothesis that upon neuronal activity, biochemical mechanisms of chromatin regulation [3,4] can shift transcriptional output when neuronal circuits are subsequently reactivated. Our goal in this review is to show how the combination of sequencing data with the emerging genome and epigenome editing strategies made easily possible using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology has enabled rapid discoveries that reveal how cooperation between coding and non-coding elements of the genome both establish and adapt neuronal transcriptional programs and brain function.
Functional Relevance of Gene Regulation in the Nervous System
How is the diversity of neuronal cell types established during development? We are only just beginning to understand how different gene expression programs are progressively regulated over the course of development as precursor cells divide, differentiate, and commit to distinct fates [5]. For each gene in the mammalian genome, there exists a cohort of non-coding cis-regulatory elements that enable precise spatiotemporal control over its expression [6,7] (Figure 1). Among these gene regulatory regions, proximal promoters are the best characterized due to their proximity to their target genes. Less is known about the specific functions and regulatory logic of distal regulatory elements, which include the distal enhancers that activate gene expression, as well as insulators that serve to buffer interactions between enhancers and promoters. Improvements in our understanding of the chromatin signatures that mark these gene regulatory elements [8] have enabled genome-wide identification of putative neuronal cis-regulatory elements [9]. However, it remains an ongoing challenge to experimentally test the functional importance of neuronal enhancers in their native genomic context. Existing enhancer reporter assays [10], including lacZ, GFP, and luciferase reporter plasmids, can provide convenient readouts for enhancer activity, but they fail to recapitulate the enhancer’s role in its native genomic context and cannot reveal the specific genes regulated by any given distal element. In contrast, as we discuss below, application of genome editing technologies is beginning to massively accelerate functional annotation of neuronal cis-regulatory elements within the mammalian genome.
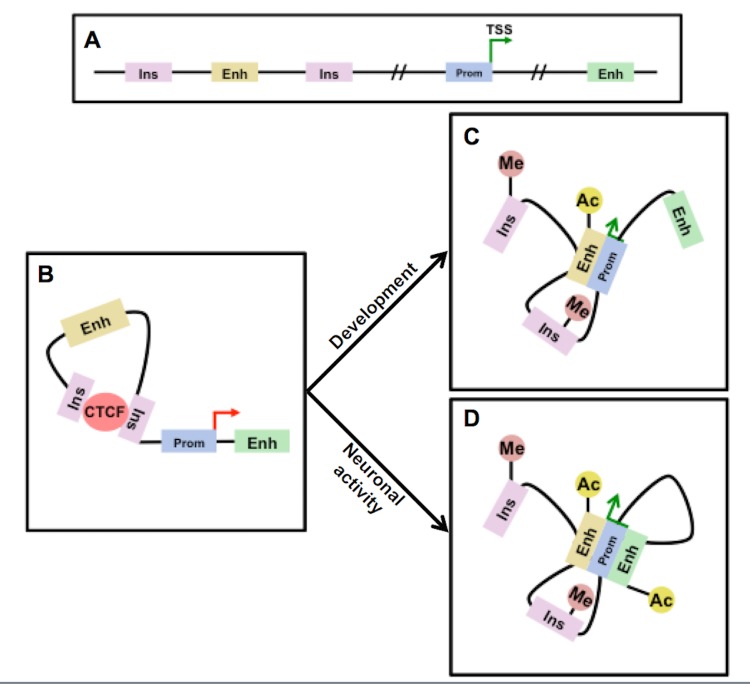
Figure 1.
Cis-regulatory elements and the control of gene transcription. A] Linear diagram of a hypothetical transcribed gene [green arrow] and its promoter [Prom] flanked by two insulator elements [Ins] and two enhancers [Enh]. B] Binding of the chromatin architectural factor CTCF to insulator elements traps the enhancer in a loop away from the gene promoter, inhibiting transcription [red arrow]. C] Following developmental methylation [Me] of insulator elements, which blocks CTCF binding, the enhancer loops over to interact with the promoter, promoting transcription. The active enhancer is marked by H3K27 acetylation [Ac]. D] Upon activity-induced recruitment of transcriptional co-activators, a shadow enhancer is recruited to the gene promoter, further promoting transcription.
How does the environment intersect with the genome to guide activity-dependent brain development? Although each neuron, like all other cells in the body, has a genetic code that provides a blueprint for its development and mature function, the brain is unique in the extreme to which it integrates and responds transcriptionally to a dynamic and diverse set of extrinsic stimuli. Just like in non-neuronal cell types, unique combinations of transcription factors binding to distinct cis-regulatory elements determine the expression of lineage-specific transcriptional programs and ultimately define cellular identities in the central nervous system [11]. However, in addition to these intrinsically defined programs of gene expression, sensory experience during postnatal neural development is capable of eliciting persistent, long-term changes to neuronal structure and function, many of which require the transcription of gene products in an activity-regulated manner that converge from complex signaling pathways [12]. Such neuronal activity-dependent gene expression programs and their regulation have been extensively studied [13,14]. Activity-regulated transcriptional programs are critical for neuronal plasticity because they are fundamentally situated at the interface between the intrinsic genome and the extrinsic environment. The availability of genetic approaches to manipulate specific cellular and progenitor populations in the brain, as well as the technology to interrogate genomic changes with single-cell resolution, makes this an exciting era to apply genome editing technologies to study the genome-environment interactions that define brain development and function.
What is the function of epigenetic regulation in neural plasticity? It is well known that the epigenetic landscape of the genome can influence gene expression in other cell types [15] and it is now better appreciated that the neuronal epigenome imparts dynamic changes that enable specific genes to be activated or repressed with temporal and spatial specificity [16]. This led to the start of the field of neuroepigenetics, which addresses how biochemical features laid on top of the DNA code modulate features of neuronal gene expression and function. These include the biophysical conformation of chromatin, the post-translational modifications of histone proteins that wrap around DNA, and the biochemical alterations to DNA nucleotides. Due to the inducible and often persistent nature of these epigenetic marks [4], it has been suggested that these epigenomic modifications contribute to the ability of a neuron to retain information from prior stimuli to influence how that neuron initiates transcriptional responses to future stimuli. However, it has been challenging to directly test the hypothesis that site-specific epigenomic modifications are actually causative for the neural plasticities that underlie functionally interesting processes in the brain like learning and memory [1,2]. The continued growth of genome editing technologies has expanded in the realm of the epigenome and there now exist methods to probe how each of these epigenomic components regulate transcription and neuronal function in the mammalian brain.
The Genome Engineering Revolution: Why CRISPR is Better
How can protein complexes be recruited to a single site in the genome? Protein engineers recognized the capacity of endogenous transcription factors to design synthetic proteins that target specific sites in the genome. The first such tool fused the DNA binding domain of two different zinc-finger transcription factors to the enzymatic domain of the FokI restriction enzyme [17], demonstrating DNA cleavage in a sequence-specific manner. Cys2-His2 zinc finger domains are about 30bp long and are among the most ubiquitous DNA-binding domains present in transcription factors [18-20]. By linking multiple zinc fingers together in tandem arrays, it is technically possible to design a synthetic protein that can recognize any given DNA sequence, forming the basis of zinc finger nuclease (ZFN) technology that propelled the modern era of genome editing [21]. Similarly, the transcription activator-like effector nucleases (TALENs) are a natural form of DNA-binding proteins produced by plant pathogens that can be stitched together in different permutations to recognize unique DNA sequences [22,23].
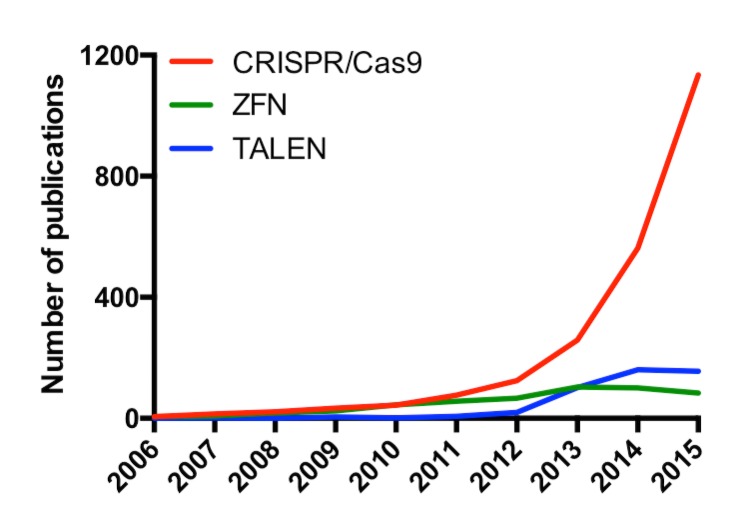
ZFN and TALEN technologies offer what genome engineers want most: high precision, sequence-specific DNA binding coupled with a modular architecture to pair with different effector domains. However, they come at a significant financial and technical cost out of range for most laboratories [24], a likely reason the simpler CRISPR/Cas9 system has been so widely and rapidly adopted. Debates continue on the relative specificity and efficacy of CRISPR/Cas9 versus ZFN or TALEN technologies [25-29]; nevertheless, publication of CRISPR/Cas9 papers has soared in the past three years with no sign of slowing down (Figure 2).
Figure 2.
Number of publications on PubMed per year with CRISPR/Cas9, ZFN, or TALEN in their titles or abstracts. These numbers exclude all review publications.
What makes the CRISPR/Cas9 system more adoptable and adaptable compared with ZFN and TALEN technologies? Instead of a protein with intrinsic sequence-specific DNA binding activity, CRISPR/Cas9 uses an RNA-guided mechanism to recruit the Cas9 effector onto the genome. CRISPR/Cas is a naturally occurring nuclease system from bacteria that recognizes RNA-DNA hybrids, used as an innate form of defense against foreign DNA [30]. The Cas9 nuclease, the core module of the technology, is recruited to DNA upon expression of an RNA (guide RNA or gRNA) that includes a protospacer adjacent motif (PAM) to bind Cas9 [31-34]. Researchers have exploited the fact that different bacterial strains employ unique PAMs to multiplex with different Cas9 molecules and target different effectors to discrete genomic loci [35]. At less than $10 USD to synthesize a new gRNA, CRISPR/Cas9 is the only one of the three genome-editing tools useful for genome-scale screening [36].
What modifications can be elicited by the recruitment of ZFNs, TALENs, or Cas9? ZFN, TALEN, and CRISPR/Cas9 nucleases all elicit double-stranded breaks (DSBs) that result in mutation of the underlying sequence via activation of endogenous DNA repair pathways: non-homologous end joining (NHEJ) and homology-directed repair (HDR) [37]. NHEJ simply ligates the broken ends, often generating insertions or deletions. HDR uses the sequence at the DSB as a template for repair for higher fidelity. When directed at gene regulatory elements, Cas9-induced mutations can reveal the requirement for transcription factor binding sites or other specific regions to activate a target gene.
To use the Cas9 protein as a platform for recruiting transcriptional effectors, the nuclease activity of Cas9 must be inactivated by mutation of specific amino acid residues on Cas9 in the RuvC and HNH domains [38]. The nuclease-dead Cas9 (known as dCas9) [39] serves as a powerful scaffold to target any effector to the genome by leveraging Cas9’s ability to bind to gRNAs that sit down at specific sites of interest.
To turn gene expression on or off, engineers have fused transcriptional activator or repression domains to ZFNs [40], TALENs, and most recently, dCas9 [41], and recruited them to non-coding regions. Recent development of more potent activators, such as dCas9-VPR that combines the VP64, p65, and Rta activators [42], may enable more robust targeting of distal regulatory elements situated up to megabases away from promoters they activate. CRISPR-based activation (CRISPRa) is predominantly used to demonstrate sufficiency of a regulatory element for gene transcription; conversely, CRISPR-mediated repression (CRISPRi) is an alternative for RNA interference, used to demonstrate that an element is necessary for efficient gene expression (Figure 3). Furthermore, to study the function of site-specific modifications to the epigenome, researchers have fused synthetic DNA binding proteins to enzymes that alter the epigenetic landscape by inducing histone [43], DNA, and chromatin modifications. This form of manipulation is most practical for disambiguating whether sequence changes at the DNA level or chromatin state of regulatory elements is the dominant contributor to alterations in gene transcription. Various iterations of chemical- or light-inducible Cas9 are being developed to improve spatiotemporal resolution for activation of Cas9 or dCas9-based effectors [44-46].
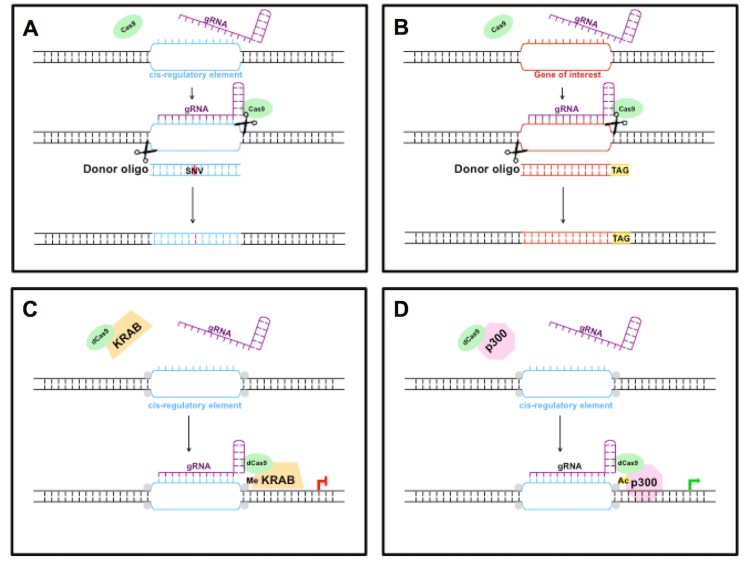
Figure 3.
Applications of CRISPR/Cas9 in neurons. A] Substitution of cis-regulatory sequences. B] Introduction of small epitope tag to C-terminus of neuronal gene. C] Repression of cis-regulatory element. D] Activation of cis-regulatory element by editing its epigenomic landscape. Histone proteins denoted in gray, histone methylation in pink, and histone acetylation in yellow.
What are the continued technical challenges for CRISPR/Cas9 in neurons? One of the great challenges for any molecular genetic study is disambiguating the cause of phenotypic effects. Cellular phenotypes emerging from globally disrupting gene expression could be a direct effect of a specific gene in a given cell type; alternatively, they are indirect consequences of a disrupted cellular environment or altered development. To address this concern for in vivo studies of the brain, in utero electroporation (IUEs) or viral-based methods are widely used for pre- and post-natal gene manipulations in neurons respectively. In particular, adeno-associated viruses (AAVs) are frequently used for neuronal transgene expression because they exhibit low levels of toxicity and high levels of in vivo neuronal infection. However, AAVs are limited in the size of DNA they can carry and the size of Cas9 from Streptococcus pyogenes (4.2 kb) makes it impossible to package into a single AAV with the gRNA and associated scaffold. By contrast, Staphylococcus aureus Cas9 (saCas9) is only 3.3 kb [35]. This system is compatible with incorporating saCas9, gRNAs, and GFP into a single AAV vector for Cas9-dependent manipulation of the genome in post-mitotic neurons. saCas9 and spCas9 are recruited by different PAM sequences; thus, distinct gRNAs must be engineered for use in the two systems.
The ability to make precise mutations in the neuronal genome with Cas9 is critical to study the effect of discrete changes to gene regulatory elements. Cas9 is a highly active nuclease and it can routinely elicit multiple DSBs at targeted loci, leading to unwanted additional mutations. A recent method called CORRECT aims to overcome this challenge by simultaneously mutating the target sequence and the adjacent PAM to prevent re-targeting by Cas9 [47]. Moreover, the modeling of monogenic disorders caused by heterozygous mutations requires mutational knock-ins at single alleles. It turns out the rate of mutation incorporation is inversely related to the distance between the Cas9 cut site and the desired mutation; this fact is now exploited to elicit heterozygous mutations at a higher frequency by selecting PAM sites at specific distances from the variant to be introduced [47].
Potential off-target effects of CRISPR/Cas9 compound the issues discussed above; in other words, single gRNAs can inadvertently target Cas9 to more than one genomic location. Several experimental strategies are used to validate the specificity of CRISPR/Cas9 action directly with respect to gene regulation. dCas9 ChIP-seq can validate the protein is binding to a single genomic location [48,49]. An RNA-seq comparison between the introduction of Cas9 with and without a gRNA targeting gene regulatory elements ensures that expression of a single gene is altered by the manipulation [50]. Computational tools exist to evaluate efficiencies of genome editing events [51] and criteria for gRNA length and target selection to minimize off-target effects have been examined [52]. Improving the specificity of the CRISPR/Cas9 system is an extensive field of study and it has been reviewed in detail [53,54].
CRISPR Applications in Neurobiology
Using CRISPR/Cas9 to Mutate Coding and Non-coding Elements
One of the principal applications for CRISPR/Cas9-based genome editing is knockout of specific genes of interest for functional studies. In this regard, CRISPR/Cas9 represents a more rapid way to make germline null alleles compared with homologous recombination [55,56]. Variations of the CRISPR/Cas9 method can also be used to make germline knock-in mutations and Cre-dependent expression of Cas9 permits conditional genome editing, though the efficiency of the CRISPR/Cas9 method compared with traditional genetic techniques for the generation of these more complex alleles remains to be fully established. What is most important for neurons, is that CRISPR/Cas9 genome editing represents a new way to achieve gene knockout in post-mitotic neurons [57-59]. By Cre-dependent Cas9 expression paired with transfection or infection of gRNAs, the CRISPR/Cas9 system has the temporal and cell-type specificity of RNA interference with the added benefit of potentially yielding complete knockout, rather than knockdown of gene expression in single cells [57].
However, beyond just knocking out genes by introducing indels into coding sequences, CRISPR/Cas9 genome editing can also be used to introduce function-disrupting mutations into non-coding gene regulatory elements. Because the non-coding genome is far more expansive than the coding genome, functional annotation of noncoding elements is substantially benefitted by the higher throughput nature of CRISPR/Cas9 compared with conventional homologous recombination. This method is particularly useful for interrogating the genomic control of brain development, as manipulating enhancer activity in situ can give insight into the finer mechanisms of genome regulation that give rise to neuronal diversity.
The murine retina is a model system of CNS neuronal diversification that is readily amenable to gene regulation studies. Differentiation is driven by a well-defined complement of transcription factors that regulate binary cell fate decisions [60,61], leading a highly stereotyped structure that facilitates discrimination of cellular phenotypes [62]. One such cell fate decision occurs in retinal progenitor cells that terminally divide to produce two cell types with distinct structures and functions: rod photoreceptors that capture light and bipolar cells that function as interneurons [63]. Blimp1, a transcription factor encoded by the Prdm1 gene, is required for this differentiation; conditional knockout of Prdm1 leads to excess bipolar cells, suggesting that Blimp1 drives photoreceptor fate. Wang et al. [64] identified a putative Prdm1 enhancer that correlates with the developmental time course of Blimp1 protein expression. Fate mapping revealed that the putative enhancer was most active in the progenitors that would ultimately become rods, not in those that differentiated into bipolar cells. The authors used CRISPR/Cas9 to delete the putative Prdm1 enhancer in the mouse retina at P0 to demonstrate its requirement for proper Prdm1 expression and rod photoreceptor fate. Loss of the enhancer resulted in an excess of bipolar cells phenocopying the Prdm1 conditional knockout, suggesting that this distal cis-regulatory element is essential for Prdm1 expression in the retina during early postnatal development. Enhancer deletions have long been accomplished using HR in ES cells [65], though the CRISPR/Cas9 method has potential advantages. It is faster to generate deletions, potentially allowing for screening of multiple putative enhancers in parallel. Furthermore, enhancer mutations can be introduced into a subset of cells at a specific developmental timepoint. For genes that have pleotropic functions during development, the precision of these enhancer mutations can improve temporal resolution on our understanding of gene regulation in development.
Enhancers are robust substrates for evolutionary change that manifests in tissue-specific gene expression changes [66,67]. At the same time, sequence variation at enhancers can also result in CNS cellular dysfunction and disease [68-70] in a cell type-specific manner [71]. It remains a considerable hurdle to gain mechanistic insight into how single nucleotide variants (SNVs) and other genetic variants identified in genome-wide association studies cause disease in the CNS [72]. It is suggested that many of disease-associated SNVs alter local chromatin and histone state, falling within genomic regions that occlude endogenous transcriptional regulators [73,74]. Soldner et al. [75] have used CRISPR/Cas9 to address Parkinson’s disease cases without clear Mendelian inheritance patterns, which accounts for up to 90 percent of patients. Since enhancers are thought to modulate gene expression, they sought genomic regions with active epigenetic signatures in the substantia nigra of post-mortem Parkinson’s patients. They found putative enhancers in an intronic region of SNCA and in the 3’ UTR that are proposed to regulate α-synuclein expression. Soldner et al. [75] recapitulated the disease genotype by deleting the putative enhancer elements by CRISPR/Cas9-mediated genome editing in human ES cells and restoring the regions with known variants from Parkinson’s patients (Figure 3A) before differentiating cells into neural precursors. The resultant neuronal cultures were useful because they enabled the study of allele-specific differences in epigenetic marks indicative of enhancer activity, as well as the disruption of binding of key transcription factors in human cortical function (EMX2, NKX6-1) by enhancer sequence variation observed in Parkinson’s patients. With improved AAVs for introducing CRISPR/Cas9 to the CNS [76,77], there is a promising platform to correct such variants in brain-specific enhancer regions.
Looking ahead, simultaneous targeting of multiple enhancers will be a useful tool to test the combinatorial nature of enhancer-promoter activation [78]. Moreover, high-throughout CRISPR/Cas9-based enhancer screens [79-81] are emerging to tile tens of kilobases of genomic search space in an unbiased manner for cis-regulatory activity, or for understanding the hierarchy of transcription factor binding sites within these enhancers. The studies highlighted in this section are some of the first to utilize CRISPR/Cas9 technology to dissect both the cis- and trans-regulators of gene expression in neurons and can be paired with improved cell type-specific sorting methods [82,83] in future to learn about the functional diversity of transcriptional regulation across neuronal and glial subtypes of mammalian brain.
Using CRISPR/Cas9 to Watch Gene Expression in Living Cells
The classic model for enhancer activation of gene expression includes the interaction between enhancer and promoter to recruit transcriptional co-activators nearby transcription start sites to initiate gene expression (Figure 1). At the moment, there are limited strategies to monitor these biophysical interactions that result in the formation of so-called chromatin loops. These include chromatin capture to provide an aggregate measure of chromatin interactions across all cells in a population [84], though this often lacks the resolution required to study neuronal subtype-specific enhancers. Alternatively, two-color DNA fluorescence in situ hybridization (FISH) [85] is an approach that enables cell type-specific resolution (co-localization of two fluorophore labeled probes is indicative of a chromosomal interaction between the putative enhancer and promoter being targeted), but for only one set of loci at a time and in fixed cells. Since there is some evidence to suggest that these chromatin loops are transient interactions between enhancer and promoter [86], it would be particularly useful to visualize these interactions in real-time in living cells. To that end, some labs have developed new techniques to recruit dCas9 fused to a variety of fluorophores that are targeted to distinct loci in the genome as an alternative to DNA FISH that is more efficient for multiplexing purposes [87,88].
For cellular localization of RNAs, RNA FISH is a robust method whereby DNA probes fused to fluorophores are allowed to hybridize to complementary RNA sequences of interest for their visualization [89], a technique that has been used for neurons in vitro and in vivo. Yet again, although this technology is sufficient to detect and quantify individual single RNA molecules at cellular resolution [90], they require that neurons be fixed at a given time point, which severely limits temporal resolution on transcriptional analysis. To address this gap, researchers have developed live imaging systems that rely on the complementarity of gRNAs to target RNAs to recruit dCas9 fused with fluorescent proteins [91]. This approach could be highly useful for studying dynamic gene expression programs in the brain, such as neuronal activity-dependent transcription. This process includes a wave of delayed primary response genes that could be interesting to track with CRISPR/dCas9, because they are often cell type-specific and enriched for diverse synaptic functions [92]. Moreover, given the observation that RNA-binding proteins (RBPs) enable the trafficking of specific RNAs to specialized neuronal compartments (e.g. dendrites vs. axons) for local translation [93], CRISPR/dCas9-mediated RNA live imaging could be an important tool for determining the components present in individual RNA granules as they are translocated by RBPs [94].
Lastly, localization of newly synthesized proteins from dynamic neuronal processes in the mammalian brain, such as neuronal activity- or neurotrophin-induced transcription, is relevant because they represent the functional output of gene regulation. Several groups have employed CRISPR/Cas9 gene editing to knock in small epitope tags following the open reading frame for a protein of interest in the nervous system, specifically in the context of the Drosophila olfactory system [95,96]. Within the mammalian brain, generating conditional knock-in mice to tag a protein with a small epitope is a common approach to study cell type-specific functions of proteins. Leveraging the potential of mitotic neural progenitors in mice for HDR, Mikuni et al. [97] recently published a technique known as single-cell labeling of endogenous proteins by CRISPR/Cas9-mediated homology-directed repair (SLENDR). SLENDR permits the knock-in of small epitopes adjacent to neuronal genes of interest in single progenitors by IUE of CRISPR/Cas9 machinery into the embryonic mouse brain (Figure 3B). As proof of principle, Mikuni et al. [97] knocked in an HA tag adjacent to the CaMKIIβ locus to achieve nanometer-scale resolution of CaMKIIβ localization by electron microscopy. SLENDR was also used to localize cytoskeletal proteins (β-actin) within the context of a sparse knockout by CRISPR/Cas9 (of methyl-CpG-binding protein 2 or MeCP2), which allows the study of cell-autonomous protein function of normal and knockout cells in the same tissue.
Overall, these CRISPR/Cas9-based approaches are powerful because they enable both temporal and spatial specificity to the imaging of a protein that is transcribed and translated within cells from its native genomic locus, which could shed light on the function of the protein in its endogenous gene regulatory context. To this end, as described below, several groups have developed CRISPR/Cas9 approaches to localize transcriptional regulators to specific genomic loci, an elegant approach to probe the function of a given cis-regulatory element to gene transcription and downstream functional consequences to neurons.
Activation and Repression of cis-regulatory Elements in their Endogenous Context
The ability of synthetic DNA binding proteins to recognize coordinates in the genome with high specificity can be expanded beyond the editing of DNA sequences. As we describe above, in all three of the genome editing methods, researchers have also co-opted the specificity of protein-DNA or RNA-DNA binding to recruit transcriptional co-activators and co-repressors to specific loci of interest, with the goal of interrogating the function of these regulatory elements in their endogenous chromatin context. This expands the traditional toolbox for neurobiologists to study cis-regulatory elements beyond plasmid-based reporter assays that often fail to faithfully recapitulate an enhancer’s level of activity in its endogenous locus of the neuronal genome.
Recruitment of nuclease-dead dCas9 alone to promoter or enhancer elements by gRNAs has been shown to occlude endogenous transcriptional activators from accessing chromatin, likely by blocking relevant transcription factor binding sites, to reduce the activity of these cis-regulatory elements [98,99]. However, a more powerful strategy to functionally annotate genome regulatory elements is to fuse the synthetic DNA binding protein with a transcriptional regulatory domain. Examples include dCas9 fusion proteins that incorporate activators, such as VP64 [100,101] and p65 [102], which serve to recruit RNA pol II and associated general transcriptional machinery to DNA loci of interest. Studies targeting dCas9-VP64 to promoters [50,103] and even enhancers located distally to target genes [16,104] have been shown to be sufficient to activate expression of relevant target genes outside the context of the nervous system [105]. Importantly, VP64 recruitment is thought to function by the addition of relevant epigenetic signatures associated with active regulatory regions [106]. These specifically mediate increased chromatin accessibility as quantified by DNase-seq or ATAC-seq, as well as increased acetylation of lysine 27 of histone H3 (H3K27ac) and increased methylation of lysine 4 of histone H3 (H3K4me1 and H3K4me3) as determined by ChIP-seq, all indicators of active DNA regulatory elements.
Recent studies have demonstrated the efficacy of this approach by exogenously activating enhancers in specific neuronal populations. Frank et al. [16] characterized genome-wide chromatin accessibility changes and epigenetic marks (DNase-seq and H3K27ac ChIP-seq respectively) to identify thousands of putative regulatory elements that are dynamically turned on and off at discrete stages of neuronal differentiation in the cerebellum. The authors selected specific enhancers to target by dCas9-VP64 based on epigenomic features and transcription factor occupancy. Localizing dCas9-VP64 to two putative enhancers in a gene-dense region demonstrated the selective activation of a single gene (Grin2c) among many situated within this genomic locus. This study is important because it validated the function and gene targets of putative neuronal enhancers in their endogenous chromatin and cellular context. Moreover, it enabled the robust empirical identification of a specific target gene associated with each enhancer, which is otherwise limited to bioinformatics methods [107].
Similarly, a series of dCas9-based transcriptional repressors have been optimized as a complement to the dCas9-VP64 co-activator. This set of reagents is especially useful in the context of studying enhancers because repression of potentially active cis-regulatory elements enables neurobiologists to quantitatively determine the requirement for any given enhancer to a gene’s proper expression level in neurons. In particular, dCas9 has been fused with the Krupel-associated box (KRAB) domain found in zinc-finger transcription factors [108]. KRAB is thought to mediate repression by binding to DNA and recruiting heterochromatin-forming protein complexes [109] to remodel the local epigenetic state, inducing the methylation of lysine 9 of histone H3 (H3K9me) [110], an epigenetic mark indicative of constitutive heterochromatin. dCas9-KRAB recruitment to gene regulatory regions has been shown to require only a single gRNA for efficient targeting and repression and it has been applied to study both proximal and distal regulatory elements [111].
Recently, Joo et al. [112] employed dCas9-KRAB to interrogate the function of distal enhancers in the activation of Fos, a well-characterized transcription factor [113] that is known to be induced by a variety of extracellular stimuli. Prior studies of Fos in the context of neuronal activity have described a set of five putative activity-regulated enhancers that regulate Fos [3], each of which show induction of the H3K27ac epigenetic mark associated with active regulatory elements upon neuronal stimulation [4]. Joo et al. describe in their study that while the Fos gene is activated in neurons by diverse stimuli, including neuronal activity and neurotrophin signaling, individual Fos enhancers are differentially responsive to each of these activating signals. In particular, they demonstrate by dCas9-KRAB recruitment to each of the five enhancers that they are able to reduce enhancer RNA levels, an epigenetic readout of enhancer activity [114], and Fos transcripts at the mRNA level in a stimulus-specific manner. This method is an illustration of how CRISPR interference (CRISPRi, Figure 3C) is able to dissect the functional contribution of each non-coding cis-regulatory element to the neuron’s ability to initiate transcription of a single gene, which will lend insight into how more complex gene expression programs are regulated by the non-coding genome and its epigenomic state.
CRISPR/dCas9-based Epigenome Editing
In recent years, seminal work in the field of neuroepigenetics has revealed the importance of stimulus-dependent changes in the epigenome, which includes a complex array of DNA modifications and post-translational modifications to histone proteins, for the regulation of neuronal gene expression. Within all cells, there exists an interplay between two broad families of proteins: epigenetic enzymes or effectors, which catalyze reactions that result in biochemical changes, and epigenetic readers, which interpret these biochemical changes and facilitate the active recruitment of transcriptional co-factors to modulate gene expression. The interesting part in neurons is that the expression and function of many of these chromatin regulatory factors is subject to neuronal activity-dependent regulation, leading to correlations in epigenome changes with functionally relevant properties of neurons, such as synaptic plasticity. This biochemical plasticity of chromatin adds an additional layer of complexity to gene regulation beyond cis-regulatory element sequences, a realization that has prompted some seminal work to uncover the epigenomic code of the neuron [115,116] and how these epigenetic changes might alter behavioral output in mouse models [117,118].
Among the biochemical modifications of histone proteins used to identify specific classes of gene regulatory elements, one of the best known is the acetylation of lysine 27 at histone H3 (H3K27ac), shown to be highly correlated with active regulatory elements that are both proximally and distally situated relative to the TSS [119,120]. The histone acetyltransferase p300 has been shown to deposit this specific post-translational modification [121] and is bound widely to active neuronal enhancers [3]. Due to the association of acetylation of particular histone residues at cis-regulatory elements with activation of gene transcription, it is possible that epigenomic editing of the histone acetylation state could be co-opted as a powerful resource for activating genes. Hilton et al. [122] have optimized a dCas9-p300 fusion protein (Figure 3D), which has been targeted to both promoters and distal enhancers to robustly activate genes, often to a greater degree than dCas9-VP64 recruitment. Although other chromatin modifying enzymes have not yet been fused to dCas9, Konermann et al. [123] have introduced a TALE fusion protein linked to SIN3 transcriptional regulator family member A (SIN3A) to neurons, which removes a histone acetylation mark (H3K9ac) that is functionally similar to H3K27ac. When recruited to the Grm2 promoter, TALE-Sin3a significantly reduced H3K9ac and concurrently diminished Grm2 expression in Neuro-2a cells. These histone-specific epigenetic editors provide neurobiologists with a molecular handle to activate and repress genes in neurons, as well as to investigate the epigenomic code and its role neuronal gene regulation.
The most common epigenetic mark associated with DNA is the methylation of cytosines at the C5 position (5mC), which has historically been studied in the context of CpG (CG) dinucleotides and is associated with repression of gene expression. The importance of DNA methylation in the mammalian brain was brought home by the discovery that mutations in the methyl-DNA binding protein MECP2 cause Rett syndrome, an X-linked neurological disorder [124,125]. The major enzymes responsible for depositing methyl groups on cytosine are the de novo methyltransferases 3A (Dnmt3a) and 3B (Dnmt3b) [126]. The level of Dnmt3a expression peaks in the mammalian brain relatively early in postnatal development [127], a period that is critical for proper neuronal circuit formation and refinement. Mouse models with Mecp2 mutations [128,129], as well as conditional Dnmt3a knockouts [130], both display a vast array of neurobiological deficits, underscoring the importance of this epigenetic modification for normal brain development and function. Interestingly, recent studies also show that non-CG methylation is highly enriched in the mammalian brain, is deposited by Dnmt3a [131], and is bound by MeCP2 [132]. Finally, 5-methylcytosine (5mC) can be converted to 5-hydroxymethylcytosine (5hmC) by a series of dioxygenases termed the ten-eleven translocation (TET) family [133]. Studies have shown that 5hmC is an epigenetic mark present in the mammalian brain [134], that 5hmC is dynamically regulated in development [135], and that cell type-specific patterns of 5hmC in the genome are present in active genes [136].
To develop new tools for DNA methylation analysis, Swiech et al. [59] generated adeno-associated viruses for Cas9, as well as for gRNAs targeting Dnmt3a, Dnmt3b, and Dnmt1, the methyltransferase responsible for maintaining DNA methylation patterns over cell division. Dnmt3a has been fused with TALENs [137,138] and Cas9 [139,140] and shown to mediate site-specific regulation of DNA methylation. Finally, there is TALEN technology incorporating the Tet1 catalytic domain [141] and some work has been done to generate a mouse ES cell line deficient in all three TET family members with CRISPR/Cas9 technology [142]. This is an example of where the modular nature of CRISPR/Cas9 could promote rapid development of new tools to probe the function and mechanism of the epigenome in the nervous system (Table 1). In fact, groups have very recently developed and applied new dCas9-Tet1 and dCas9-Dnmt3a fusion proteins to increase our epigenome editing capacity in neurons [143].
Table 1. Summary of genes targeted by diverse CRISPR/Cas9 strategies in mammalian neurons.
| CRISPR manipulation | Gene targets | References |
| Conditional knockout in neurons | Abhd6, Dnmt1, Dnmt3a, Dnmt3b, GRIA2, Grin1, GRIN1, Katnal2, Mecp2, Rbfox3, Top1, 3110043O21Rik | [57-59,150-154] |
| Knockin of variant to neuronal gene | APP, FMR1, PSEN1 | [47,155] |
| Deletion of cis-regulatory element | Prdm1 | [64] |
| Activation of proximal and distal enhancers | Grin2c | [16] |
| Repression of distal enhancers | Fos | [112] |
| Knockin of small epitope tag for visualization | Actb, Arc, Camk2a, Cacna1c, CHAT, Dcx, Fmrp, GAD67, Mecp2, Prkca, Rab11a, Ywhae | [97,156,157] |
Conclusions and Outlook
Neurobiologists have just begun to explore the cellular and molecular mechanisms that enable the epigenomic landscape of cis-regulatory elements to deploy transcriptional programs over brain development and in response to neuronal activity. The innovation of CRISPR/Cas9-based epigenetic effectors will accelerate this process of discovery, as the ability to target multiple regulatory elements with high specificity and efficiency will fuel the functional annotation of the genome. The genome and epigenome editing toolbox made possible by the simple and modular design of the CRISPR/Cas9 system can easily be paired with cutting-edge genetic and microscopy techniques to improve resolution on specific epigenomic features of different neuronal cell types. One of the key frontiers for the future will be using CRISPR/Cas9 methods to push the boundaries of understanding transcriptional regulation at the single-cell level. This experimental capacity is especially important in the brain, where the precise interconnection of multiple cell types is key for the function of neural circuits and where information is stored in distributed networks [144]. In 2015, single-cell methods emerged for quantifying RNA transcripts [145,146], histone epigenetic modifications [147], and chromatin accessibility [148]. An advantage that single-cell technologies provide over previous cell type purification-based methods is the ability to quantify CRISPR/Cas9 effects in a largely unbiased manner. In other words, we now have the ability to profile the transcriptional state before and after CRISPR/Cas9 manipulations of all neurons in a population and we may even discover novel gene regulatory patterns in neurons not defined by a single cell type marker. As we look towards the future of neuroepigenomics, CRISPR/Cas9 developments will continue to expand the possibilities for neurobiologists to understand normal brain development and function, as well as how the chromatin landscape is perturbed in neurologicaldisorders [149].
Acknowledgments
We thank Emmy Li for assistance with figures.
Glossary
- CRISPR
clustered regularly interspaced short palindromic repeats
- DBD
DNA binding domain
- TAD
transcriptional activation domain
- ZFN
zinc finger nuclease
- TALEN
transcription activator-like effector nucleases
- PAM
protospacer adjacent motif
- gRNA
guide RNA
- HDR
homology directed repair
- NEHJ
non-homologous end-joining
- dCas9
nuclease-dead Cas9
- SNP
single nucleotide polymorphism
- FISH
fluorescence in situ hybridization
- RBP
RNA binding protein
- SLENDR
single cell labeling of endogenous proteins by CRISPR-Cas9-mediated homology-directed repair
Author Contributions
Marty Yang and Anne West both conceived of the idea, wrote the paper, edited the paper, and designed the figures. This work was supported by NIH grant DA041878 to A.E.W.
References
- Alarcón JM, Malleret G, Touzani K. et al. Chromatin Acetylation, Memory, and LTP Are Impaired in CBP+/− Mice. Neuron. 2004;42(6):947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP Histone Acetyltransferase Activity Is a Critical Component of Memory Consolidation. Neuron. 2004;42(6):961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-K, Hemberg M, Gray JM. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik AN, Vierbuchen T, Hemberg M. et al. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat Neurosci. 2014;17(10):1–13. doi: 10.1038/nn.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman K, Golonzhka O, Lindtner S. et al. Transcriptional Regulation of Enhancers Active in Protodomains of the Developing Cerebral Cortex. Neuron. 2014;82(5):989–1003. doi: 10.1016/j.neuron.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL. et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21(10):1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch CM, Andrews RM, Flicek P. et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17(6):691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306(5696):636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I. et al. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35(Database issue):D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray PA, Fu H, Luo P. et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306(5705):2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- Lyons MR, West AE. Mechanisms of specificity in neuronal activity-regulated gene transcription. Progress in Neurobiology. 2011;94(3):259–295. doi: 10.1016/j.pneurobio.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011;3(6):a005744. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JM, Kim TK, West AE. et al. Genomic Views of Transcriptional Enhancers: Essential Determinants of Cellular Identity and Activity-Dependent Responses in the CNS. J Neurosci. 2015;35(41):13819–13826. doi: 10.1523/JNEUROSCI.2622-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Cattoglio C, Tjian R. Looping Back to Leap Forward: Transcription Enters a New Era. Cell. 2014;157(1):13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CL, Liu F, Wijayatunge R. et al. Regulation of chromatin accessibility and Zic binding at enhancers in the developing cerebellum. Nature. 2015;18(5):1–13. doi: 10.1038/nn.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S. et al. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93(3):1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapska E, Kaczmarek L. A gene for neuronal plasticity in the mammalian brain: Zif268/Egr-1/NGFI-A/Krox-24/TIS8/ZENK? Progress in Neurobiology. 2004;74(4):183–211. doi: 10.1016/j.pneurobio.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Palm K, Belluardo N, Metsis M. et al. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J Neurosci. 1998;18(4):1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer LD, Barajas B, Tao S. et al. ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes Dev. 2014;28(18):2013–2026. doi: 10.1101/gad.246579.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee Y-L. et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435(7042):646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326(5959):1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326(5959):1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- DeFrancesco L. Move over ZFNs. Nat Biotech. 2011;29(8):681–684. doi: 10.1038/nbt.1935. [DOI] [PubMed] [Google Scholar]
- Naitou A, Kato Y, Nakanishi T. et al. Heterodimeric TALENs induce targeted heritable mutations in the crustacean Daphnia magna. Biol Open. 2015;4(3):BIO20149738-369. doi: 10.1242/bio.20149738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Bae S, Park J. et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Meth. 2015;12(3):237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotech. 2014;33(2):187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Geen H, Henry IM, Bhakta MS. et al. A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic Acids Res. 2015;43(6):gkv137-3404. doi: 10.1093/nar/gkv137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Regan SN, Xia Y. et al. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12(4):393–394. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D. et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M. et al. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507(7490):62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096-6. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Gersbach CA. Genome engineering: the next genomic revolution. Nat Meth. 2014;11(10):1009–1011. doi: 10.1038/nmeth.3113. [DOI] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Sanjana NE, Zheng K. et al. Genome-wide CRISPR Screen in a Mouse Model of Tumor Growth and Metastasis. Cell. 2015;160(6):1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Jenkins BV, O'Connor-Giles KM. et al. A CRISPR view of development. Genes Dev. 2014;28(17):1859–1872. doi: 10.1101/gad.248252.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P. et al. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109(39):E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga-Canut M, Agustín-Pavón C, Herrmann F. et al. Synthetic zinc finger repressors reduce mutant huntingtin expression in the brain of R6/2 mice. Proc Natl Acad Sci USA. 2012;109(45):E3136–E3145. doi: 10.1073/pnas.1206506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Scheiman J, Vora S. et al. Highly efficient Cas9-mediated transcriptional programming. Nat Meth. 2015;12(4):326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden AW, Gregory PD, Case CC. et al. Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr Biol. 2002;12(24):2159–2166. doi: 10.1016/s0960-9822(02)01391-x. [DOI] [PubMed] [Google Scholar]
- Nguyen DP, Miyaoka Y, Gilbert LA. et al. Ligand-binding domains of nuclear receptors facilitate tight control of split CRISPR activity. Nat Commun. 2016;7:1–10. doi: 10.1038/ncomms12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Volz SE, Zhang F. et al. split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol. 2015;33(2):139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol. 2015;11(3):198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet D, Kwart D, Chen A. et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533(7601):125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ. et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32(7):1–9. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C, Arslan S, Singh R. et al. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32(7):677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM. et al. RNA-guided gene activation by CRISPR-Cas9–based transcription factors. Nat Meth. 2013;10(10):973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boel A, Steyaert W, De Rocker N. et al. BATCH-GE: Batch analysis of Next-Generation Sequencing data for genome editing assessment. Sci Rep. 2016;6:30330–30339. doi: 10.1038/srep30330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J-P, Li X-L, Neises A. et al. Different Effects of sgRNA Length on CRISPR-mediated Gene Knockout Efficiency. Sci Rep. 2016;6:28566. doi: 10.1038/srep28566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella S, Montoya G. The genome editing revolution: A CRISPR-Cas TALE off-target story. BioEssays. 2016;38(S1):S4–S13. doi: 10.1002/bies.201670903. [DOI] [PubMed] [Google Scholar]
- Kanchiswamy CN, Maffei M, Malnoy M. et al. Fine-Tuning Next-Generation Genome Editing Tools. Trends Biotechnol. 2016;34(7):562–574. doi: 10.1016/j.tibtech.2016.03.007. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Qiu Z, Shao Y. et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol. 2013;31(8):681–683. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- Straub C, Granger AJ, Saulnier JL. et al. CRISPR/Cas9-mediated gene knock-down in post-mitotic neurons. PLoS ONe. 2014;9(8):e105584. doi: 10.1371/journal.pone.0105584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incontro S, Asensio CS, Edwards RH. et al. Efficient, complete deletion of synaptic proteins using CRISPR. Neuron. 2014;83(5):1051–1057. doi: 10.1016/j.neuron.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiech L, Heidenreich M, Banerjee A. et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33(1):102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JA, Lamba DA, Reh TA. et al. Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development. 2010;137(4):619–629. doi: 10.1242/dev.043968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Oh ECT, Ng L. et al. Retinoid-related orphan nuclear receptor RORbeta is an early-acting factor in rod photoreceptor development. Proc Natl Acad Sci USA. 2009;106(41):17534–17539. doi: 10.1073/pnas.0902425106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry TJ, Wang S, Bormuth I. et al. NeuroD factors regulate cell fate and neurite stratification in the developing retina. J Neurosci. 2011;31(20):7365–7379. doi: 10.1523/JNEUROSCI.2555-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DL, Cepko CL. A common progenitor for neurons and glia persists in rat retina late in development. Nature. 1987;328(6126):131–136. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- Wang S, Sengel C, Emerson MM. et al. A gene regulatory network controls the binary fate decision of rod and bipolar cells in the vertebrate retina. Dev Cell. 2014;30(5):513–527. doi: 10.1016/j.devcel.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagai T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132(4):797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- Prescott SL, Srinivasan R, Marchetto MC. et al. Enhancer Divergence and cis-Regulatory Evolution in the Human and Chimp Neural Crest. Cell. 2015;163(1):68–83. doi: 10.1016/j.cell.2015.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Berthelot C, Aldridge S. et al. Enhancer Evolution across 20 Mammalian Species. Cell. 2015;160(3):554–566. doi: 10.1016/j.cell.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. Enhancer biology and enhanceropathies. Nat Struct Mol Biol. 2014;21(3):210–219. doi: 10.1038/nsmb.2784. [DOI] [PubMed] [Google Scholar]
- Ghiasvand NM, Rudolph DD, Mashayekhi M. et al. Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nature. 2011;14(5):578–586. doi: 10.1038/nn.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y, Leskow FC, El-Jaick K. et al. Regulation of a remote Shh forebrain enhancer by the Six3 homeoprotein. Nat Genet. 2008;40(11):1348–1353. doi: 10.1038/ng.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Daniels S, Sweeney E. et al. Enhancer-adoption as a mechanism of human developmental disease. Hum Mutat. 2011;32(12):1492–1499. doi: 10.1002/humu.21615. [DOI] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H, Waszak SM, Gschwind AR. et al. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science. 2013;342(6159):744–747. doi: 10.1126/science.1242463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G, van de Geijn B, Degner JF. et al. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342(6159):747–749. doi: 10.1126/science.1242429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Stelzer Y, Shivalila CS. et al. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016;533(7601):95–99. doi: 10.1038/nature17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murlidharan G, Sakamoto K, Rao L. et al. CNS-restricted Transduction and CRISPR/Cas9-mediated Gene Deletion with an Engineered AAV Vector. Mol Ther Nucleic Acids. 2016;5(7):e338. doi: 10.1038/mtna.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung SSC, Chrysostomou V, Li F. et al. AAV-Mediated CRISPR/Cas Gene Editing of Retinal Cells In Vivo. Invest Ophthalmol Vis Sci. 2016;57(7):3470–3476. doi: 10.1167/iovs.16-19316. [DOI] [PubMed] [Google Scholar]
- Meyer MB, Benkusky NA, Pike JW. Selective Distal Enhancer Control of the Mmp13 Gene Identified through Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) Genomic Deletions. J Biol Chem. 2015;290(17):11093–11107. doi: 10.1074/jbc.M115.648394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Smith EC, Sher F. et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527(7577):192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao Y, Li B, Meng Z. et al. A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9-mediated genetic screening. Genome Res. 2016;26(3):397–405. doi: 10.1101/gr.197152.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmaz G, Lopes R, Ugalde AP. et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol. 2016;34(2):192–198. doi: 10.1038/nbt.3450. [DOI] [PubMed] [Google Scholar]
- Hrvatin S, Deng F, O'Donnell CW. et al. MARIS: Method for Analyzing RNA following Intracellular Sorting. PLoS ONE. 2014;9(3):e89459-6. doi: 10.1371/journal.pone.0089459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat Protoc. 2010;6(1):56–68. doi: 10.1038/nprot.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belton J-M, McCord RP, Gibcus JH. et al. Hi-C: A comprehensive technique to capture the conformation of genomes. Methods. 2012;58(3):268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolland DJ, King MR, Reik W. et al. Robust 3D DNA FISH using directly labeled probes. J Vis Exp. 2013;78:e50587-7. doi: 10.3791/50587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartman CR, Hsu SC, Hsiung CCS. et al. Enhancer Regulation of Transcriptional Bursting Parameters Revealed by Forced Chromatin Looping. Mol Cell. 2016;62(2):237–247. doi: 10.1016/j.molcel.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Zhang W, Hu H. et al. Long-term dual-color tracking of genomic loci by modified sgRNAs of the CRISPR/Cas9 system. Nucleic Acids Res. 2016;44(9):e86. doi: 10.1093/nar/gkw066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Tu L-C, Naseri A. et al. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat Biotechnol. 2016;34(5):528–530. doi: 10.1038/nbt.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K. et al. Visualization of single RNA transcripts in situ. Science. 1998;280(5363):585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Mueller F, Senecal A, Tantale K. et al. FISH-quant: automatic counting of transcripts in 3D FISH images. Nat Methods. 2013;10(4):277–278. doi: 10.1038/nmeth.2406. [DOI] [PubMed] [Google Scholar]
- Nelles DA, Fang MY, O'Connell MR. et al. Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell. 2016;165(2):488–496. doi: 10.1016/j.cell.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinly AR, Spiegel I, Patrizi A. et al. Sensory experience regulates cortical inhibition by inducing IGF1 in VIP neurons. Nature. 2016;531(7594):371–375. doi: 10.1038/nature17187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosker KE, Fenstermacher SJ, Pazyra-Murphy MF. et al. The RNA-binding protein SFPQ orchestrates an RNA regulon to promote axon viability. Nat Neurosci. 2016;19(5):690–696. doi: 10.1038/nn.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Su J-H, Zhang F. et al. An RNA-aptamer-based two-color CRISPR labeling system. Sci Rep. 2016;6:1–7. doi: 10.1038/srep26857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Barish S, Okuwa S. et al. Examination of Endogenous Rotund Expression and Function in Developing Drosophila Olfactory System Using CRISPR-Cas9-Mediated Protein Tagging. G3 (Bethesda) 2015;5(12):2809–2016. doi: 10.1534/g3.115.021857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koles K, Yeh AR, Rodal AA. et al. Tissue-specific tagging of endogenous loci in Drosophila melanogaster. Biol Open. 2015;5(1):83–89. doi: 10.1242/bio.016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikuni T, Nishiyama J, Sun Y. et al. High-Throughput, High-Resolution Mapping of Protein Localization in Mammalian Brain by In Vivo Genome Editing. Cell. 2016;165(7):1803–1817. doi: 10.1016/j.cell.2016.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Gilbert LA, Wang X. et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8(11):2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM. et al. CRISPR RNA-guided activation of endogenous human genes. Nat Meth. 2013;10(10):977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotech. 2013;31(9):833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517(7536):583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saayman SM, Lazar DC, Scott TA. et al. Potent and Targeted Activation of Latent HIV-1 Using the CRISPR/dCas9 Activator Complex. Mol Ther. 2015;24(3):488–498. doi: 10.1038/mt.2015.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Tsang JCH, Gaba F. et al. Comparison of TALE designer transcription factors and the CRISPR/dCas9 in regulation of gene expression by targeting enhancers. Nucleic Acids Res. 2014;42(20):e155. doi: 10.1093/nar/gku836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Tuttle M, Pruitt BW. et al. Comparison of Cas9 activators in multiple species. Nat Meth. 2016;13(7):563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein LR, Perez-Pinera P, Kocak DD. et al. Genome-wide specificity of DNA binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome Res. 2015;25(8):1158–1159. doi: 10.1101/gr.179044.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):nbt.1630–1639. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore PI, Black JB, Hilton JB. et al. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Meth. 2016;13(2):127–137. doi: 10.1038/nmeth.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groner AC, Meylan S, Ciuffi A. et al. KRAB-Zinc Finger Proteins and KAP1 Can Mediate Long-Range Transcriptional Repression through Heterochromatin Spreading. PLoS Genet. 2010;6(3):e1000869-14. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D. et al. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16(8):919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B. et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159(3):647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo J-Y, Schaukowitch K, Farbiak L. et al. Stimulus-specific combinatorial functionality of neuronal c-fos enhancers. Nat Neurosci. 2016;19(1):75–83. doi: 10.1038/nn.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ME, Ziff EB. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Schaukowitch K, Joo J-Y, Liu X. et al. Enhancer RNA Facilitates NELF Release from Immediate Early Genes. Mol Cell. 2014;56(1):29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP. et al. Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron. 2015;86(6):1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Luo C, vDavis FP. et al. Epigenomic landscapes of retinal rods and cones. eLife. 2016;5:e11613-29. doi: 10.7554/eLife.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V. et al. Histone Deacetylase 5 Epigenetically Controls Behavioral Adaptations to Chronic Emotional Stimuli. Neuron. 2007;56(3):517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Renthal W, Carle TL, Maze I. et al. Delta FosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. J Neurosci. 2008;28(29):7344–7349. doi: 10.1523/JNEUROSCI.1043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T. et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V. et al. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87(5):953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Hilton IB, D'Ippolito AM, Vockley CM. et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33(5):1–10. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino A. et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500(7463):472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M. et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Meehan RR, Lewis JD, Bird AP. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20(19):5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA. et al. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR. et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341(6146):1237905-5. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Gabel HW, Hemberg M. et al. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron. 2011;72(1):72–85. doi: 10.1016/j.neuron.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JV, Wan Y, Wang X. et al. MeCP2 Phosphorylation Limits Psychostimulant-Induced Behavioral and Neuronal Plasticity. J Neurosci. 2014;34(13):4519–4527. doi: 10.1523/JNEUROSCI.2821-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL. et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13(4):423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Shin JH. et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci. 2013;17(2):215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel HW, Kinde B, Stroud H. et al. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015;522(7554):1–21. doi: 10.1038/nature14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y. et al. 5-hmC–mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14(12):1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellén M, Ayata P, Dewell S. et al. MeCP2 Binds to 5hmC Enriched within Active Genes and Accessible Chromatin in the Nervous System. Cell. 2012;151(7):1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivenbark AG, Stolzenburg S, Beltran AS. et al. Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics. 2014;7(4):350–360. doi: 10.4161/epi.19507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique AN, Nunna S, Rajavelu A. et al. Targeted Methylation and Gene Silencing of VEGF-A in Human Cells by Using a Designed Dnmt3a–Dnmt3L Single-Chain Fusion Protein with Increased DNA Methylation Activity. J Mol Biol. 2013;425(3):479–491. doi: 10.1016/j.jmb.2012.11.038. [DOI] [PubMed] [Google Scholar]
- Vojta A, Dobrinić P, Tadić V. et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44(12):5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JI, Celik H, Rois LE. et al. Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biol Open. 2016;5(6):866–874. doi: 10.1242/bio.019067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Angstman JF, Richardson ME. et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotech. 2013;31(12):1137–1142. doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Liu Y, Jiang L. et al. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 2014;28(19):2103–2119. doi: 10.1101/gad.248005.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Wu H, Ji X. et al. Editing DNA Methylation in the Mammalian Genome. Cell. 2016;167(1):233–247. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josselyn SA, Köhler S, Frankland PW. et al. Finding the engram. Nat Rev Neurosci. 2015;16(9):521–534. doi: 10.1038/nrn4000. [DOI] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I. et al. Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell. 2015;161(5):1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R. et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A, Ram O, Shoresh N. et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol. 2015;33(11):1–11. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523(7561):486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP. et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MR, Fricano-Kugler CJ, Getz SA. et al. A Retroviral CRISPR-Cas9 System for Cellular Autism-Associated Phenotype Discovery in Developing Neurons. Sci Rep. 2016;6:1–11. doi: 10.1038/srep25611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabb AM, Simon JM, King IF. et al. Topoisomerase 1 Regulates Gene Expression in Neurons through Cleavage Complex-Dependent and -Independent Mechanisms. PLoS ONE. 2016;11(5):e0156439-18. doi: 10.1371/journal.pone.0156439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Zhang J, Jia M. et al. α/β-Hydrolase domain-containing 6 (ABHD6) negatively regulates the surface delivery and synaptic function of AMPA receptors. Proc Natl Acad Sci USA. 2016;113(19):E2695-704. doi: 10.1073/pnas.1524589113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharjan N, Künzli C, Buthey K. et al. C9ORF72 Regulates Stress Granule Formation and Its Deficiency Impairs Stress Granule Assembly, Hypersensitizing Cells to Stress. Mol Neurobiol. 2016:1–16. doi: 10.1007/s12035-016-9850-1. [DOI] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y. et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C-Y, Halevy T, Lee DR. et al. Reversion of FMR1 Methylation and Silencing by Editing the Triplet Repeats in Fragile X iPSC-Derived Neurons. Cell Rep. 2015;13(2):234–241. doi: 10.1016/j.celrep.2015.08.084. [DOI] [PubMed] [Google Scholar]
- Zhou J, Wang C, Zhang K. et al. Generation of Human Embryonic Stem Cell Line Expressing zsGreen in Cholinergic Neurons Using CRISPR/Cas9 System. Neurochem Res. 2016:1–10. doi: 10.1007/s11064-016-1918-9. [DOI] [PubMed] [Google Scholar]
- Liu J, Gao C, Chen W. et al. CRISPR/Cas9 facilitates investigation of neural circuit disease using human iPSCs: mechanism of epilepsy caused by an SCN1A loss-of-function mutation. Transl Psychiatry. 2016;6(1):e703–e709. doi: 10.1038/tp.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]