Abstract
Epigenetics is the study of phenotypic variation arising from developmental and environmental factors regulating gene transcription at molecular, cellular, and physiological levels. A naturally occurring biological process driven by epigenetics is the egg-to-embryo developmental transition when two fully differentiated adult cells – egg and sperm – revert to an early stem cell type with totipotency but subsequently differentiates into pluripotent embryonic stem cells that give rise to any cell type. Transposable elements (TEs) are active in mammalian oocytes and early embryos, and this activity, albeit counterintuitive because TEs can lead to genomic instability in somatic cells, correlates to successful development. TEs bridge genetic and epigenetic landscapes because TEs are genetic elements whose silencing and de-repression are regulated by epigenetic mechanisms that are sensitive to environmental factors. Ultimately, transposition events can change size, content, and function of mammalian genomes. Thus, TEs act beyond mutagenic agents reshuffling the genomes, and epigenetic regulation of TEs may act as a proximate mechanism by which evolutionary forces increase a species’ hidden reserve of epigenetic and phenotypic variability facilitating the adaptation of genomes to their environment.
Keywords: transposons, retrotransposons, endogenous retroviruses, LINE elements, LTR, DNA methylation, histone modification, epigenetics, oogenesis, preimplantation development, variability
Introduction
The realm of epigenetics covers developmental canalization of gene expression and its interaction with the environment, resulting in phenotype variation at cellular, tissue and organismal levels. Research in functional epigenetics endeavors to understand reversible changes in DNA and chromatin function, in contrast to irreversible genotype changes in DNA nucleotide sequence, a realm of genetics, underlying phenotypic variation. Within the context of developmental biology, an utmost example of a biological process driven by epigenetics is the de-differentiation followed by re-differentiation naturally occurring during the egg-to-embryo transition [1]. Two fully differentiated adult cells – egg and sperm – revert to an early developmental stem cell type that retains totipotency, and subsequently differentiates into embryonic stem cells that give rise to any cellular type of the embryonic germ layers, such as muscle cells, neurons, epithelial cells, etc. In mammals, a hallmark of the egg-to-embryo transition is a dramatic, dynamic change in gene expression patterns that associates with the activity of specific transposable elements (TEs) [2-4]. Bridging the realms of genetics and epigenetics, transposons are genetic elements regulated by epigenetic factors at biochemical and molecular levels. First described by Barbara McClintock in maize as controlling units, in eukaryotes transposons propagate throughout genomes, changing their size, structure and function, and may be turned off or on by environmental factors, or developmental checkpoints [5]. Originally alleged as junk or parasitic DNA [6,7], transposons may have broader biological roles in the process of cellular differentiation because in mammalian genomes, experimental evidence supports epigenetic regulation of transposons as being critical to initiate synchronous, temporal expression of genes in germline, early embryos, and stem cells [2,8].
Transposons Background
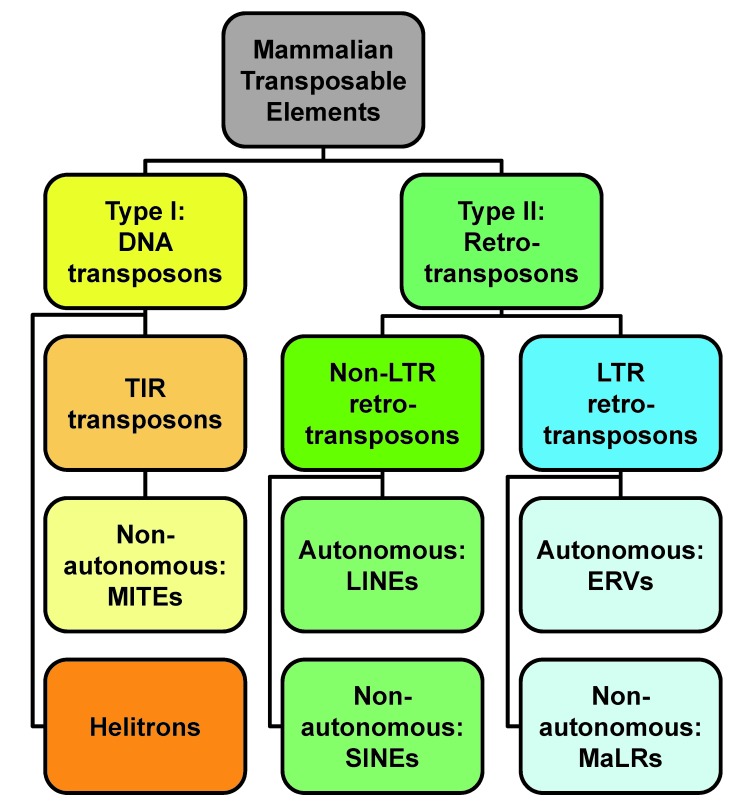
Transposons, defined as a class of genetic elements that can change their position in the genome, are indisputably major contributors to genomic evolution, but recent evidence supports their involvement in major developmental processes as well. Two broad classes of TEs exist, class I DNA transposons and class II retrotransposons [9] (Figure 1). Class I TEs, DNA transposons (Figure 1), do not use an RNA intermediate for replication. The best-studied types of DNA transposons encode transposase protein flanked by terminal inverted repeats (TIRs). Transposase enables these TEs to self-excise and reintegrate into another location in the genome, also known as “cut-and-paste” mechanism. The TIR DNA TEs are further classified in several subgroups, and those in mammals include representatives of Tc1/Mariner [9], piggyBac [10-12] and hAT [13,14] superfamilies. Active TIR DNA transposons appear to be mostly absent from the sequenced mammalian genomes [15,16] although their no-longer-coding remnants are transcriptionally active in the germline, as evidenced from EST libraries [3,4,17]. A notable exception is little brown bat, Myotis lucifugus, whose genome contain intact representatives of all three TIR DNA TE superfamilies [12]. Non-autonomous TIR DNA TEs called Miniature Inverted-repeat Transposable Elements (MITEs) which lack transposase are also found in mammals [14,18,19]; however, they underwent exaptation, at least in humans, and act as microRNA genes [18]. A different class of DNA transposons, the “copy-and-paste” helitrons [20], also have restricted distribution among mammals. Interestingly, while most mammalian species lack helitrons, Vespertilionidae, common bats, are again an exception, and helitrons specific to bats contribute upwards of six percent of genomic sequence in, e.g., Myotis lucifugus [21].
Figure 1.
Classification of mammalian transposons. The figure depicts only the major classes and types of TEs identified to date in mammalian genomes, according to RepBase [9]. Due to complicated phylogenetic relationships, subfamilies of TEs are not depicted here; for complete information, see RepBase online: http://www.girinst.org/repbase/
Class II TEs, retrotransposons, which propagate through an RNA intermediate using reverse transcription and reintegration mechanisms, are sometimes referred to as the “copy-and-paste” transposons. Retrotransposons constitute a large “undocumented” part of mammalian genomes, accumulated over many previous generations. In fact, on average ~40 percent of all mammalian genomic sequences comprise various retrotransposons [15,16], consisting of four major types [9,22]: Long Interspersed Nuclear Elements, Short Interspersed Nuclear Elements, Endogenous Retroviruses, and Mammalian Apparent LTR retrotransposons (Figure 1).
Long Interspersed Nuclear Elements (LINEs), and their remnants, belong to the class of non-Long Terminal Repeat (LTR) retrotransposons. Their activity is evidenced by numerous mutations found in human population, and plausibly is a major contributor to sporadic mutagenesis in humans [23,24]. Short Interspersed Nuclear Elements (SINEs), which resemble tRNAs and other structural RNAs, use LINEs for propagation in the genome. De novo SINE insertions are continuously discovered, highlighting their mutagenic potential [25,26]. SINEs, which do not encode any proteins, are clearly dependent on LINEs for their retrotransposition, also known as trans-mobilization. SINEs subvert LINEs propagation machinery, and particularly L1 open reading frame 2 (ORF2) protein, via expansion of poly(A) tail [27,28].
Endogenous retroviruses (ERVs) are another type of retrotransposable TEs in mammalian genomes. Like retroviruses, ERVs have long terminal repeats (LTRs) at their ends, flanking an internal sequence, with typically two ORFs encoding Group antigen (Gag) and PRO-POL-IN-dUTPase polyprotein [29]. Proteolytically processed PRO-POL-IN-dUTPase allows production and reintegration of a new ERV cDNA copy back into the genome via sequential activities of protease, reverse transcriptase, RNase H, and integrase. On the other hand, unlike retroviruses, ERVs either lack viral envelope (ENV) proteins altogether, or their ORFs are severely truncated and mutated, rendering ERVs non-infectious, and notably incapable of horizontal transposons transfer between individuals. Lastly, an unusual type of LTR retrotransposons has been documented, Mammalian apparent LTR retrotransposons (MaLR) [30]. Members of the MaLR family, which are quite abundant in murine genomes, along with Early (expressing) Transposon (ETn) family [31,32] lack either meaningful or long ORFs. This structure relegates MaLRs in an analogous role to SINEs dependence on LINEs for genomic propagation, and requires ERVs retrotransposition machinery for their spread in genomes [33-35].
Transposons Expression in Mammalian Oocytes and Early Embryos
Traditionally, transposon expression is viewed as a threat to genomic stability [6,7]. The basic notion is quite simple: if transposition-competent TEs are constitutively active, their exponential amplification will drive random genome instability in situ, which results in different cells harboring different mutations [36,37], as seen, for example in advanced stages of cancer [38]. To avoid this, living organisms developed and continue reinventing mechanisms for TE silencing, mostly via epigenetic mechanisms, which spares removal of TE loci from their genome, discussed in further detail below. This tug-of-war hypothesis implies rapid co-evolution of genomic “host defense” and TE “suppression avoidance”. The true picture, however, is complicated because TE fitness eventually depends upon the fitness of the host genomes, and thus TEs are under selection pressure to maintain or increase genome fitness; importantly, if certain TEs do increase species’ fitness, the ultimate result is for selection pressure not to eliminate TEs from host genomes. More importantly, we postulate that fitness of TEs is tightly linked to the fitness of host germline, as it is only germline transmission that ensures successful transmission and overall increase in TE copies.
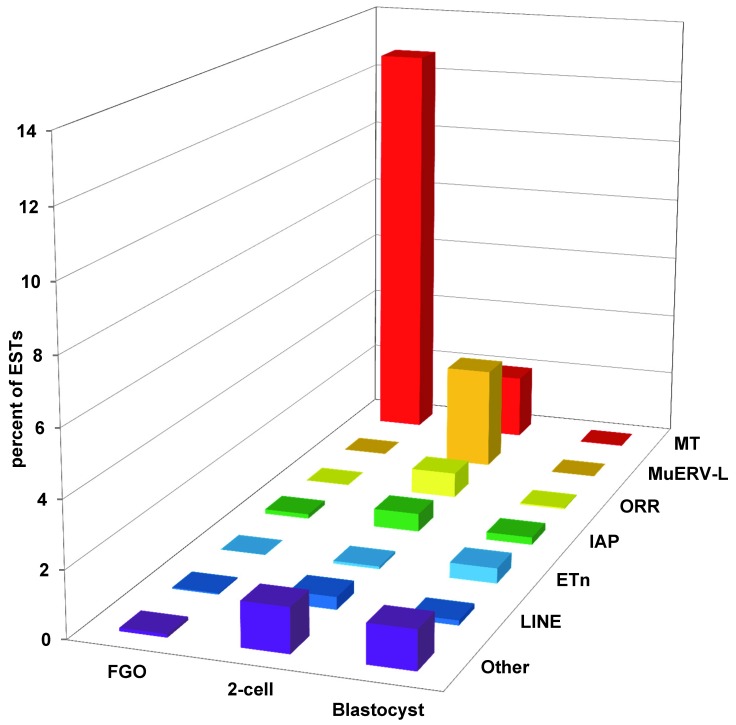
Germline expression of TEs in mammals has been known for many decades. As early as late 1960s, presence of virus-like “intracisternal A particles,” now designated as IAP LTR retrotransposons, was observed in the cytoplasm of mouse oocytes and preimplantation embryos [39,40]. However, some of these electron microscope-detected particles are likely a product of another ERV, MuERV-L [41]. An inverse relationship between IAP expression and the amount of DNA methylation was noted [42] and experimentally verified using 5-azacytidine, an inhibitor of cytosine methylation [43]. The major breakthrough in large-scale unbiased identification of TEs expressed in mammalian germline and early development came with large-scale analysis of transcriptomes via sequencing of cDNA libraries introduced in late 1970s [44]. These analyses revealed overwhelming expression of SINE B1 and B2 elements in the first cDNA libraries constructed from mouse cleavage-stage embryos [45], prompting obligatory RNA size-selection for future cDNA libraries [46]. Consequently, analysis of size-restricted libraries constructed from mouse oocytes and preimplantation embryos prepared in the Davor Solter, Barbara Knowles, and John Eppig laboratories revealed abundant expression of TEs other than SINEs, particularly LTR retrotransposons [2-4] (Figure 2). The first surprising finding is a dynamic change in TE expression across the stages of early development: the expression profiles of LTRs were dramatically different during oogenesis as seen at the fully-grown oocyte stage, compared to nascent embryonic genome activation (EGA) at the 2-cell stage, and finally just before implantation at the blastocyst stage (Figure 2). These waves of differential TE expression correlate to dramatic increases in transcriptional activity during mammalian oogenesis and early development, revealing three distinct bursts of transcription associated with accumulation of maternal RNAs, during embryonic genome activation, and at initial differentiation events [47]. Another surprise is the vast abundance of TE transcripts in the transcriptome – ranging from 2 percent to 13 percent of all ESTs in the fully-grown oocyte, 2-cell-stage embryo and blastocyst libraries, elevating TEs, on the grand scale, as one of the highest-expressed categories of transcripts among all long poly(A) RNAs. For instance, and to illustrate the scale, MT LTR transcripts alone in fully-grown oocytes are more abundant than the top 50 highest-expressed conventional genes combined. Ostensibly it appears oocytes and early embryos tolerate massive onslaught of TEs expression. While the ultimate factors causing this massive increase in TEs expression are not completely clear, there are a few candidate proximate mechanisms that may account for the increase in TE transcriptional activity.
Figure 2.
Differential expression of various TEs during the mouse egg-to-embryo transition and preimplantation development. Only the most abundant types of TEs are represented. Mouse Transcript (MT) transposons and Origin-region repeat (ORR) transposons belong to MaLR group of LTR retrotransposons; MuERV-L are endogenous retroviruses; Early Transposons (ETn) are non-autonomous LTR retrotransposons; IAP are endogenous retroviruses, also known as RLTR2 TEs; LINE are Long Interspersed Nuclear Elements; Other include all TEs outside the previous six categories.
One parsimonious proximate mechanism possibly underlying the expression of TEs in the female germline of mice is genome-wide DNA demethylation during germ cell development. Loss of cytosine methylation during mammalian gametogenesis is particularly profound in mouse oocytes [48,49], setting the stage for TE reactivation due to de-repression of epigenetic silencing. However, stage-restricted expression of TEs during the transition from oogenesis to early development (Figure 2) does not neatly fit this model because under these conditions, all existing TEs, not a select few, would be expected to be reactivated. An alternative model has been recently proposed tying regulation of mammalian TEs expression, specifically LTR retrotransposons, to a family of rapidly evolving mammalian DNA binding repressors, Krüppel-associated-box zinc finger (KRAB-ZF) proteins [50,51]. While conceptually attractive, at the moment the model has some caveats, such as relative paucity of KRAB-ZF loci in non-eutherian genomes, and notably, KRAB-ZFs do not drive suppression of all LTRs in the female germline and early development.
The potential resolution to the conundrum of high TEs expression in oocytes and early embryos is species exaptation of TEs’ regulatory sequences for their own needs, a view proposed by Roy Britten and Eric Davidson [52,53]. Indeed, setting aside the presumption of intrinsic parasitic nature of TEs [6,7], as well as their dependence on germline quality, a case can be argued that germline-TE interactions is either a mutually beneficial relationship, or TE suppression reduces germline fitness. In concordance with this idea, in conjunction with the massive upregulation of TEs themselves, there is a concomitant increase in TE-driven expression of conventional genes in mouse oocytes and preimplantation embryos, with TEs acting as robust, often cell-specific, promoters for transcription [2,3]. In fact, our current estimate is about five percent of conventional genes are driven by LTRs, or other TEs, acting as alternative promoters during oogenesis and early development in mammals (Lockhart and Evsikov, unpublished observations). For example, abundant expression of Spin1 in mouse oocytes is driven by an MT LTR retrotransposon found in its third intron, whose insertion is specific to mice [2]. Interestingly, there is an overall, albeit not perfect, concordance between the type of TEs expressed at specific stages of oogenesis and early development, and TEs that act as alternative promoters at these stages. For instance, MT LTRs drive expression of genes in oocytes, but MuERV-L LTRs drive expression of another subset of genes in the 2-cell-stage embryos [2]. Thus, differential expression of TEs may co-evolve to fulfill the need for spatio-temporal regulation of gene expression, an undisputable constraint and paradigm of developmental genetics and developmental biology [54]. Among notable exceptions to the rule of stage-specific TEs driving stage-specific gene expression is an MT2B LTR. MT2B promoter drives maternal expression of Zbed3 gene in oocytes, whereas another MT2B LTR drives expression of Rpl41 after embryonic genome activation at the 2-cell stage [2]. However, despite being classified as the same repetitive element, there is definitely enough sequence difference in the sequences upstream of the first exons encoded by these two LTRs to imply potential differences in their transcriptional regulation (Figure 3). These observations suggest that in mice, TEs and particularly LTRs are driving exaptation of new promoters for the needs of the female germline and early development. This neofunctionalization of TEs as alterative promoters may potentially serve as a deterrent for selection against silencing all LTRs in the host mammalian genomes. Robust support for this model of TE-host genome interaction would be bolstered by an observation that TE-driven expression of a host gene is essential for oogenesis and early development. To date, there is at least one confirmation of functional significance for specific TE-driven gene expression in oogenesis. Dicer1, a central component of the classic epigenetic RNAi pathway [55], is expressed from an alternative, MT LTR retrotransposon promoter in mouse oocytes. Deletion of this MT LTR element by homologous recombination results in compromised fitness due to defective oocytes that fail to properly assemble a meiotic spindle [56]. Consequently, deletion of a TE promoter delivers a perfect phenocopy of oocyte-specific Dicer1 mutation generated using traditional conditional knockout technology [57]. Hence, this observation strengthens critical regulatory epigenetic function of retrotransposons within the developmental context of biological processes underlying the egg-to-embryo transition.
Figure 3.
Divergence of nucleotide sequence during transposon exaptation. Multiple sequence alignment of MT2B LTR-derived alternative first exons, and flanking genomic sequences, of mouse zinc finger, BED type containing 3 (Zbed3) and ribosomal protein L41 (Rpl41) genes [2], and the full MT2B consensus sequence from RepBase [9]. Note the rapid diversification of upstream regulatory sequences, and conservation of putative TATA box. Surprisingly, despite dissimilarities in the first exons, transcription start sites and splice site boundaries are conserved between these genes. In mouse egg-to-embryo transition, these two genes have differential expression patterns, with Zbed3 transcribed in oocytes, while Rpl41 transcribed from 2-cell stage to blastocyst stage [2]. MSA was performed using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/). First exons are highlighted with tan background; TATA box is boxed; conserved nucleotides are highlighted in yellow; transcription start sites are in green.
Abundant expression of TEs is not restricted to mice; our observations on embryonic genome activation in cow, Bos taurus, morula-stage embryos revealed EGA-specific, abundant upregulation of the bovine-specific retrotransposons, ERV1-1_BT, ERV1-2_BT, and a yet unannotated bovine ERV similar to human HERVL66 [17,58]. Furthermore, low expression of these LTRs in ‘cloned’ bovine embryos created using somatic cell nuclear transfer correlate with poor embryo quality and developmental competency, suggesting a link between TE activity during embryonic genome activation, epigenetic remodeling of the genome, and embryo fitness in mammals [17]. Incomplete epigenetic “reprogramming” of persistent repressive epigenetic marks in the somatic cell nuclei chromatin microsurgically transferred to the oocytes is considered as one of the major reasons for developmental deficiencies in cloned embryos [17,59]. Indeed, studies on gene expression in mice reveal deficiency of expression during embryonic genome activation in cloned mouse embryos [59,60], which is caused by persistent repressive epigenetic marks on the MuERV-L loci in somatic cell nuclei [59]. Thus, aptitude to correctly activate EGA-specific transposons correlates with viability in cloned embryos, and may explain substantially lower viability seen in cloned mouse compared to bovine embryos.
More recent findings in mice confirm correlation between MuERV-L expression and the epigenetically undifferentiated, totipotent state of MuERV-L-expressing embryonic stem (ES) cells [8]. Recent progress in availability and affordability of next-generation sequencing (NGS) technologies opened a door to broaden the studies of oocyte and preimplantation embryo transcriptomes in unprecedented detail (e.g., [61,62]). Undoubtedly, TE expression data from such studies, while considerable to retrieve, will reveal the repertoire of TEs active in oogenesis and early development across many mammals in the near future.
Epigenetic Regulation of Transposons
As noted earlier, direct relationship between DNA methylation and TE activity was described decades ago for IAP LTR retrotransposons [42]. Indeed, DNA cytosine methylation is touted to be the major barrier sheltering mammalian genomes from the destructive, damaging effects of endogenous TEs [7]. More recently, the discovery of the “histone code” [63] revealed that specific posttranslational modifications of histones, specifically a core nucleosome subunit histone 3 (H3) protein, affect expression of TEs as well. Thus, TEs expression is regulated at the levels of DNA cytosine methylation [64], and specific core histone modifications, such as trimethylation of lysine 9 of histone H3 (a.k.a. H3K9me3) [65,66]. The third level of anti-TE defense, post-reverse-transcriptional deamination of single-stranded DNA by APOBEC3G proteins, is discussed in an exhaustive review [67]. However, all three of major epigenetic suppressors of TEs expression necessitate recognition of TEs loci by the host genome.
In order to be epigenetically silenced, a specific locus, or loci, in the genome need to be either recognized or demarcated. Years of mechanistic studies of epigenetic regulation of germ cell development, oogenesis, and early development, while mostly focusing on imprinted genes, revealed a number of players involved in suppression of TEs. These players can be sorted, accordingly, into three levels of epigenetic control mechanisms: sequence recognition, initial silencing, and establishment of constitutive epigenetic repression. In mammals, it appears that TE sequence recognition in germline, and by extension, early development, occurs at the RNA level, specifically via MILI-interacting RNA [68] piRNA pathway [69-71]. Recent articles suggest that tRNA fragments introduced by sperm may also be directly involved in epigenetic TE repression in the nascent zygote, and notably MuERV-L, which is embryonically expressed post-fertilization at the 2-cell stage [72,73]. While the exact mechanism and machinery underlying targeted TE silencing in mammalian genomes is still under investigation, it appears that possible epigenetic resolutions are either fast-evolving, TE-sequence-specific KRAB-ZFPs mentioned earlier [50], or the RNA-guided identification of cognate TEs in the genomes. Subsequent silencing of TEs in female germline involves recruitment of chromatin-modifying players, a multitude of which have been identified. Most of these putative chromatin-remodeling players are also active at different stages of oogenesis, development, and have disparate functions. Among the first identified regulators of TE silencing in female germline are DNA methyltransferase 1 (Dnmt1) and lymphoid-specific helicase LSH (Hells), both required for de novo DNA methylation of specific TEs in addition to some conventional genes. Targeted inactivation of either Dnmt1 or Hells disrupts the egg-to-embryo developmental transition by causing oogenesis defects and embryonic lethality [74-77]. The PRMT5/Blimp1 pathway is another critical regulator of TEs in female germline epigenome, acting via arginine methylation of histones H2A and H4, as well as DNA methylation [78-80]. One of the important pleiotropic regulators of TE activity is KAP1 (Trim28), whose interactions with various KRAB-ZF proteins regulate TEs, imprinted loci, and possibly other targets [81-85].
Applications of Epigenetics: Transposons as Environment Biosensor
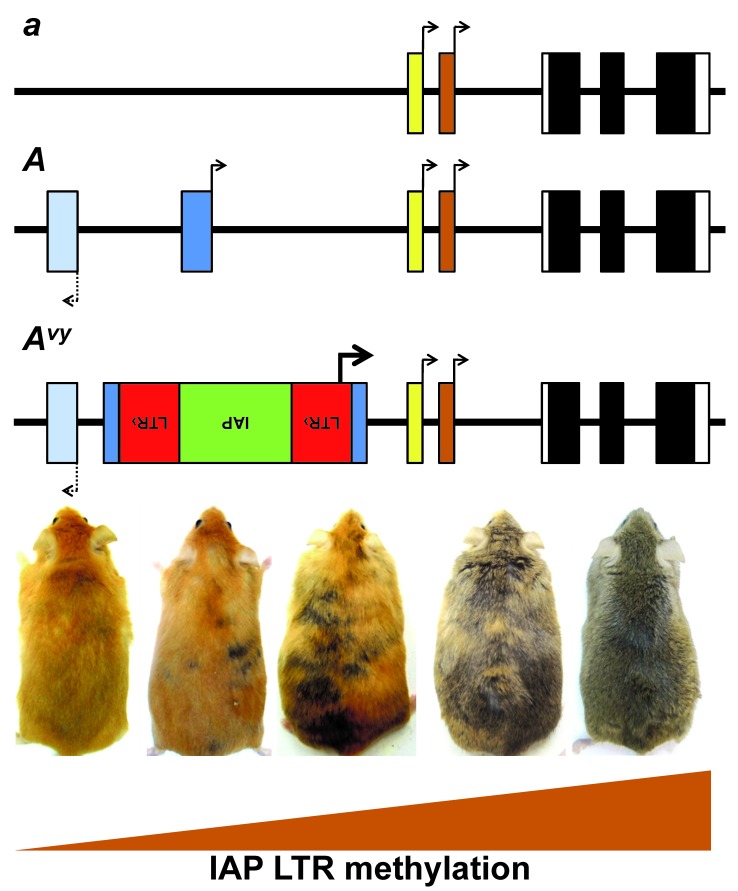
As mentioned above, TEs act as bridges connecting genetics to epigenetics to determine variation in phenotypic traits. Originally discovered by Barbara McClintock in maize studies [5], TEs activity can be influenced by developmental or environmental factors. In mammals, there are naturally occurring mutations whose phenotypic expression depends upon the epigenetic status of retrotransposons, which in turn can be controlled by the environment [86]. Mutations in the genome, either naturally occurring or engineered, may reflect the status of specific epigenetic marks at individual genetic loci [87]. In mammals, among first discovered and most studied models of this type is the coat color mutation, Agouti viable yellow (Avy) allele in mice. The mutation arose spontaneously in a colony of C3H/HeJ mouse strain at the Jackson Laboratory in 1960, with color coat ranging from a pseudoagouti (brown) to yellow, with variegation in brown and yellow fur patches, despite all individual mice being isogenic at the agouti locus (Figure 4) [88,89]. The Avy allele resulted from an IAP retrotransposition event, which inserted IAP copy upstream of the Agouti gene. It is the first reported mammalian model whose phenotype reflects DNA methylation status of this IAP LTR retrotransposon insertion at the Agouti locus [87]. Phenotypic manifestation of the Avy allele is driven by the methylation status of the inserted IAP LTR, whose expression results in a yellow coat color (Figure 4).
Figure 4.
Genomic locus of Agouti gene. The top line represents the recessive allele nonagouti, a, which encodes black coat color in mice. The second line represents the dominant allele, Agouti, A, which encodes brown coat color. The third line represents the dominant allele, Agouti viable yellow, vy, allele containing the antisense insertion of IAP-LTR retrotransposon, whose methylation levels at the LTR regulate ectopic expression producing coat colors that can range from pseudoagouti, yellow, or admixture of pseudoagouti and yellow patches despite identical DNA sequence.
Importantly, the window of environmental effects upon Avy allele appears to range from pre-conceptual to peri-implantation [86], which corresponds to the waves of global changes in DNA methylation, including genome de-methylation in germline, followed by its re-methylation during the egg-to-embryo transition [48,49]. The coat color phenotype of the offspring was demonstrated to be affected by the parental phenotype [89] and environment, and specifically by the amount of available methyl supplements in the maternal diet [91]. Subsequently, other similar phenotype and allelic forms to Avy, such as Aiapy and Aiy, have been reported in mouse colonies originating from the Jackson Laboratory [90,92]. Depending on coat color status, a subtle parent-of-origin effect was reported [89], although transgenerational inheritance of either Avy coat color phenotype, or its methylation status, by transmission through the germline is controversial [91,93,94]. Indeed, it was experimentally demonstrated that methylation levels at Avy are subject to erasure and re-establishment in patterns similar to non-imprinted genetic loci [95]. However, studies of this model clearly demonstrate that DNA methylation at specific TEs loci may act as environmental sensor tying genetics and phenotype manifestations via the epigenetic state.
Caveats & Conjecture
Among the most important challenges with both functional and bioinformatics analysis of TEs, is a general apparent lack of restricted evolutionary conservation, and more specifically, a lack of correlation between the phylogenetic history of individual species and specific TEs populating their genomes. In particular, this phenomenon applies to phylogenetically young and thus still active transposons, which unexpectedly are the largest contributor to the plasticity of gene expression signatures observed in oogenesis and early development in mammals [3,4,17].
While not dismissing the deleterious nature of de novo transposition, we postulate a new conceptual framework for the study of the TE-host genome interactions in mammals: mutually beneficial cooperativity on the grounds of increased genetic variability, and thus, consequently, increased phenotypic plasticity and adaptability [96], traits critical for fitness and survival – within a host species. This idea is concordant with a recent model of TE-driven speciation proposed by Jerzy Jurka et al. [97,98]. It seems that in mammals, this cooperative compromise is actively negotiated in the female germline as evidenced by massive upregulation of TEs during oogenesis. The TE contribution to the compromise is providing genetic novelty, which increases genetic diversity and variability essential for population and species stability over time. The species’ cost is allowing TEs to spread in their genomes. TEs provide a benefit to the species in the form of increased genetic diversity. To receive this benefit of increased genetic diversity, the species has to pay a fee to the TEs, which is incorporation of the TEs into germline and spreading in the species’ gene pool. While the host has developed molecular machinery to suppress TEs activity, under certain developmental and cellular circumstances, namely during the egg-to-embryo transition, the host genomes use TEs to increase their genetic diversity at the right place, the oocyte, and the right time, transition to embryonic activation.
This situation resembles a symbiotic relationship rather than parasitism. Indeed, there is increasing evidence that TEs expression is not just tolerated, but in some cases, is essential for oocyte quality and embryo viability [2,56], and thus is important in regulation and maintenance of germline and early development. Therefore, we propose that TE-host interactions should be studied with a symbiosis paradigm in mind. Finally, the strong evidence for rapid diversification of species-specific TEs suggests that novel regulatory functions arising from TEs as promoters in oocytes are shaped by the pressures of gametic and compatibility selection, adjusting and fine-tuning the processes unique to a given mammalian species. This prompts speculation that TEs may potentially be involved in the processes of molecular diversification leading to reproductive isolation, and consequently, the processes of speciation.
Conclusion & Outlook
In summary, TEs may initiate synchronous, developmentally regulated expression of multiple conventional genes in mammals. Apart from introducing genomic variation by self-propagation, retrotransposons appear to have an important role in transcriptional regulation during the egg-to-embryo transition, reflected in their differential expression. Expression of specific TEs in mammalian oocytes and early embryos is quite robust, despite activity of well-established TE silencing pathways via DNA methylation, histone modification, and activity of KRAB-ZF suppressors. Nevertheless, differential regulation of TE transcription is a puzzle yet to be solved. It appears that TE activity may reflect the host interaction with the environment, and at least in some cases, certain TE loci may act as “environmental biosensors”. It is still unclear whether the TEs are the “drivers” or “passengers” of epigenetic genome restructuring occurring during oogenesis and early development in mammals. Indeed, in mice, for example, MuERV-L emerges as a “driver of pluripotency”, albeit the molecular mechanism is not understood. However, if we put aside the mechanistic details of precise molecular interactions, it seems that epigenetic regulation of TEs may act as a proximate mechanism by which evolutionary forces, and selection pressure, utilize TEs to increase the species’ “hidden reserve of variability” [96,99]. Thus, TEs, beyond being mutagens that reshuffle the genomes to increase genome size and variation, also act as fine tuners able to adjust gene expression levels and timing in early development, thereby allowing individual genomes, and by proxy whole species, to adapt to their environment.
Glossary
- EGA
embryonic genome activation
- ERV
endogenous retrovirus
- ETn
early transposon
- IAP
intracisternal A-particle
- KRAB-ZFP
Krüppel-associated-box zinc finger protein
- LINE
Long Interspersed Nuclear Element
- LTR
long terminal repeat
- MaLR
mammalian apparent long terminal repeat retrotransposon
- MuERV-L
murine endogenous retrovirus L
- ORF
open reading frame
- SINE
Short Interspersed Nuclear Element
- TEs
transposable elements
- TIR
terminal inverted repeat
Author Contributions
Both authors equally drafted the manuscript: AVE drafted the half of the manuscript on transposons, its corresponding figures, and conclusion whereas CMdE drafted the remaining half on epigenetics, its corresponding figures, abstract, and introduction. Both authors revised the manuscript and approved the final version. Both authors were supported by intramural funds provided by the Morsani College of Medicine, University of South Florida, Tampa.
References
- De Vries WN, Evsikov AV, Brogan LJ. et al. Reprogramming and Differentiation in Mammals: Motifs and Mechanisms. Cold Spring Harb Symp Quant Biol. 2008;73:33–38. doi: 10.1101/sqb.2008.73.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH. et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7(4):597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Evsikov AV, de Vries WN, Peaston AE. et al. Systems biology of the 2-cell mouse embryo. Cytogenet Genome Res. 2004;105(2-4):240–250. doi: 10.1159/000078195. [DOI] [PubMed] [Google Scholar]
- Evsikov AV, Graber JH, Brockman JM. et al. Cracking the egg: molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo. Genes Dev. 2006;20(19):2713–2727. doi: 10.1101/gad.1471006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federoff N, Botstein D, editors. The Dynamic Genome: Barbara McClintock's Ideas in the Century of Genetics. Cold Spring Harbor: CSHL Press; 1992. [Google Scholar]
- Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284(5757):601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13(8):335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S. et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487(7405):57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A. et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110(1-4):462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Newman JC, Bailey AD, Fan HY. et al. An abundant evolutionarily conserved CSB-PiggyBac fusion protein expressed in Cockayne syndrome. PLoS Genet. 2008;4(3):e1000031. doi: 10.1371/journal.pgen.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan HJ, Smith JD, Hubley RM. et al. PiggyBac-ing on a primate genome: novel elements, recent activity and horizontal transfer. Genome Biol Evol. 2010;2:293–303. doi: 10.1093/gbe/evq021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, Feschotte C, Pagan HJ. et al. Multiple waves of recent DNA transposon activity in the bat, Myotis lucifugus. Genome Res. 2008;18(5):717–728. doi: 10.1101/gr.071886.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin E, Lithwick G, Levy AA. Structure and evolution of the hAT transposon superfamily. Genetics. 2001;158(3):949–957. doi: 10.1093/genetics/158.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF, Riggs AD. Tiggers and DNA transposon fossils in the human genome. Proc Natl Acad Sci U S A. 1996;93(4):1443–1448. doi: 10.1073/pnas.93.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B. et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E. et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Bui LC, Evsikov AV, Khan DR. et al. Retrotransposon expression as a defining event of genome reprograming in fertilized and cloned bovine embryos. Reproduction. 2009;138(2):289–299. doi: 10.1530/REP-09-0042. [DOI] [PubMed] [Google Scholar]
- Piriyapongsa J, Jordan IK. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS One. 2007;2(2):e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan GT. Identification in the human genome of mobile elements spread by DNA-mediated transposition. J Mol Biol. 1995;254(1):1–5. doi: 10.1006/jmbi.1995.0593. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Rolling-circle transposons in eukaryotes. Proc Natl Acad Sci U S A. 2001;98(15):8714–8719. doi: 10.1073/pnas.151269298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Phillips CD, Baker RJ. et al. Rolling-circle transposons catalyze genomic innovation in a mammalian lineage. Genome Biol Evol. 2014;6(10):2595–2610. doi: 10.1093/gbe/evu204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 2008;9(5):411–412. doi: 10.1038/nrg2165-c1. [DOI] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH Jr.. Active human retrotransposons: variation and disease. Curr Opin Genet Dev. 2012;22(3):191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH Jr.. Roles for retrotransposon insertions in human disease. Mob DNA. 2016;7:9. doi: 10.1186/s13100-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taskesen M, Collin GB, Evsikov AV. et al. Novel Alu retrotransposon insertion leading to Alstrom syndrome. Hum Genet. 2012;131(3):407–413. doi: 10.1007/s00439-011-1083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA. Alu repeats and human disease. Mol Genet Metab. 1999;67(3):183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- Wagstaff BJ, Hedges DJ, Derbes RS. et al. Rescuing Alu: recovery of new inserts shows LINE-1 preserves Alu activity through A-tail expansion. PLoS Genet. 2012;8(8):e1002842. doi: 10.1371/journal.pgen.1002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Engel AM. A tale of an A-tail: The lifeline of a SINE. Mob Genet Elements. 2012;2(6):282–286. doi: 10.4161/mge.23204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benit L, Lallemand JB, Casella JF. et al. ERV-L elements: a family of endogenous retrovirus-like elements active throughout the evolution of mammals. J Virol. 1999;73(4):3301–3308. doi: 10.1128/jvi.73.4.3301-3308.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 1993;21(8):1863–1872. doi: 10.1093/nar/21.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brulet P, Condamine H, Jacob F. Spatial distribution of transcripts of the long repeated ETn sequence during early mouse embryogenesis. Proc Natl Acad Sci U S A. 1985;82(7):2054–2058. doi: 10.1073/pnas.82.7.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brulet P, Kaghad M, Xu YS. et al. Early differential tissue expression of transposon-like repetitive DNA sequences of the mouse. Proc Natl Acad Sci U S A. 1983;80(18):5641–5645. doi: 10.1073/pnas.80.18.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D, Dewannieux M, Heidmann T. An active murine transposon family pair: retrotransposition of "master" MusD copies and ETn trans-mobilization. Genome Res. 2004;14(11):2261–2267. doi: 10.1101/gr.2924904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager DL, Freeman JD. Novel mouse type D endogenous proviruses and ETn elements share long terminal repeat and internal sequences. J Virol. 2000;74(16):7221–7229. doi: 10.1128/jvi.74.16.7221-7229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, Romanish MT, Gagnier L. et al. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2(1):e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J. The origin of human cancers. Nature. 1981;289(5796):353–357. doi: 10.1038/289353a0. [DOI] [PubMed] [Google Scholar]
- Nevers P, Saedler H. Transposable genetic elements as agents of gene instability and chromosomal rearrangements. Nature. 1977;268(5616):109–115. doi: 10.1038/268109a0. [DOI] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang L. et al. Landscape of somatic retrotransposition in human cancers. Science. 2012;337(6097):967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco PG, Brown EH. An ultrastructural and cytological study of preimplantation development of the mouse. J Exp Zool. 1969;171(3):253–283. doi: 10.1002/jez.1401710303. [DOI] [PubMed] [Google Scholar]
- Miller GG, Makarova IV, Iazykov AA. [Detection of endogenous intracisternal type A particles in early mouse zygotes.] Biull Eksp Biol Med. 1983;96(7):94–96. [PubMed] [Google Scholar]
- Ribet D, Louvet-Vallee S, Harper F. et al. Murine endogenous retrovirus MuERV-L is the progenitor of the "orphan" epsilon viruslike particles of the early mouse embryo. J Virol. 2008;82(3):1622–1625. doi: 10.1128/JVI.02097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojman-Montes de Oca F, Lasneret J, Dianoux L. et al. Regulation of intracisternal A particles in mouse teratocarcinoma cells: involvement of DNA methylation in transcriptional control. Biol Cell. 1984;52(3):199–204. doi: 10.1111/j.1768-322x.1985.tb00337.x. [DOI] [PubMed] [Google Scholar]
- Lasneret J, Canivet M, Hojman-Montes de Oca F. et al. Activation of intracisternal a particles by 5-azacytidine in mouse Ki-BALB cell line. Virology. 1983;128(2):485–489. doi: 10.1016/0042-6822(83)90275-1. [DOI] [PubMed] [Google Scholar]
- Sim GK, Kafatos FC, Jones CW. et al. Use of a cDNA library for studies on evolution and developmental expression of the chorion multigene families. Cell. 1979;18(4):1303–1316. doi: 10.1016/0092-8674(79)90241-1. [DOI] [PubMed] [Google Scholar]
- Taylor KD, Piko L. Patterns of mRNA prevalence and expression of B1 and B2 transcripts in early mouse embryos. Development. 1987;101(1):877–892. doi: 10.1242/dev.101.4.877. [DOI] [PubMed] [Google Scholar]
- Rothstein JL, Johnson D, Jessee J. et al. Construction of primary and subtracted cDNA libraries from early embryos. Methods Enzymol. 1993;225:587–610. doi: 10.1016/0076-6879(93)25038-4. [DOI] [PubMed] [Google Scholar]
- Evsikov AV, Marin de Evsikova C. Gene expression during the oocyte-to-embryo transition in mammals. Mol Reprod Dev. 2009;76(9):805–818. doi: 10.1002/mrd.21038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean W. DNA methylation and demethylation: a pathway to gametogenesis and development. Mol Reprod Dev. 2014;81(2):113–125. doi: 10.1002/mrd.22280. [DOI] [PubMed] [Google Scholar]
- Dean W, Santos F, Reik W. Epigenetic reprogramming in early mammalian development and following somatic nuclear transfer. Semin Cell Dev Biol. 2003;14(1):93–100. doi: 10.1016/s1084-9521(02)00141-6. [DOI] [PubMed] [Google Scholar]
- Thomas JH, Schneider S. Coevolution of retroelements and tandem zinc finger genes. Genome research. 2011;21(11):1800–1812. doi: 10.1101/gr.121749.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, Greenberg D, Macfarlan TS. Spotting the enemy within: Targeted silencing of foreign DNA in mammalian genomes by the Kruppel-associated box zinc finger protein family. Mob DNA. 2015;6:17. doi: 10.1186/s13100-015-0050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165(3891):349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 1971;46(2):111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- Gilbert SF. Developmental Biology. 6th ed. Sunderland, MA: Sinauer Associates; 2000. [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM. et al. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Flemr M, Malik R, Franke V. et al. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell. 2013;155(4):807–816. doi: 10.1016/j.cell.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z. et al. Critical roles for Dicer in the female germline. Genes Dev. 2007;21(6):682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. et al. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Liu Y, Lu F. et al. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell. 2014;159(4):884–895. doi: 10.1016/j.cell.2014.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Minami N, Kono T. et al. Zygotically activated genes are suppressed in mouse nuclear transferred embryos. Cloning Stem Cells. 2006;8(4):295–304. doi: 10.1089/clo.2006.8.295. [DOI] [PubMed] [Google Scholar]
- Guo H, Zhu P, Yan L. et al. The DNA methylation landscape of human early embryos. Nature. 2014;511(7511):606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Sun J, Dong H. et al. Transcriptional profiles of bovine in vivo pre-implantation development. BMC Genomics. 2014;15:756. doi: 10.1186/1471-2164-15-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Martens JH, O'Sullivan RJ, Braunschweig U. et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24(4):800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S. et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D. et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31(6):785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, van der Heijden GW, Castaneda J. et al. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 2009;5(12):e1000764. doi: 10.1371/journal.pgen.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaneda J, Genzor P, Bortvin A. piRNAs, transposon silencing, and germline genome integrity. Mutat Res. 2011;714(1-2):95–104. doi: 10.1016/j.mrfmmm.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM. et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351(6271):391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Yan M, Cao Z. et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351(6271):397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Geiman TM, Tessarollo L, Anver MR. et al. Lsh, a SNF2 family member, is required for normal murine development. Biochim Biophys Acta. 2001;1526(2):211–220. doi: 10.1016/s0304-4165(01)00129-5. [DOI] [PubMed] [Google Scholar]
- De La Fuente R, Baumann C, Fan T. et al. Lsh is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells. Nat Cell Biol. 2006;8(12):1448–1454. doi: 10.1038/ncb1513. [DOI] [PubMed] [Google Scholar]
- Yu Y, McIntosh C, Lister R. et al. Genome-wide DNA methylation patterns in LSH mutant reveals de-repression of repeat elements and redundant epigenetic silencing pathways. Genome Res. 2014;24(10):1613–1623. doi: 10.1101/gr.172015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohinata Y, Payer B, O'Carroll D. et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436(7048):207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Ancelin K, Lange UC, Hajkova P. et al. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8(6):623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- Kim S, Gunesdogan U, Zylicz JJ. et al. PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Mol Cell. 2014;56(4):564–579. doi: 10.1016/j.molcel.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Hug K, Goff SP. TRIM28 mediates primer binding site-targeted silencing of Lys1,2 tRNA-utilizing retroviruses in embryonic cells. Proc Natl Acad Sci U S A. 2008;105(34):12521–12526. doi: 10.1073/pnas.0805540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D. et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463(7278):237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Messerschmidt DM, de Vries W, Ito M. et al. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335(6075):1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Kapopoulou A, Corsinotti A. et al. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 2013;23(3):452–461. doi: 10.1101/gr.147678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Friedli M, Offner S. et al. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development. 2013;140(3):519–529. doi: 10.1242/dev.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cellular Biol. 2003;23(15):5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewitt M, Whitelaw E. The use of mouse models to study epigenetics. Cold Spring Harb Perspect Biol. 2013;5(11):a017939. doi: 10.1101/cshperspect.a017939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickies MM. A new viable yellow mutation in the house mouse. J Hered. 1962;53:84–86. doi: 10.1093/oxfordjournals.jhered.a107129. [DOI] [PubMed] [Google Scholar]
- Wolff GL. Influence of maternal phenotype on metabolic differentiation of agouti locus mutants in the mouse. Genetics. 1978;88(3):529–539. doi: 10.1093/genetics/88.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry WL, Copeland NG, Jenkins NA. The molecular basis for dominant yellow agouti coat color mutations. Bioessays. 1994;16(10):705–707. doi: 10.1002/bies.950161002. [DOI] [PubMed] [Google Scholar]
- Wolff GL, Kodell RL, Moore SR. et al. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12(11):949–957. [PubMed] [Google Scholar]
- Michaud EJ, van Vugt MJ, Bultman SJ. et al. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev. 1994;8(12):1463–1472. doi: 10.1101/gad.8.12.1463. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HGE, Martin DIK. et al. Epigenetic inheritance at the agouti locus in the mouse. Nature Genetics. 1999;23(3):314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Travisano M, Tahiliani KG. Diet-induced hypermethylation at agouti viable yellow is not inherited transgenerationally through the female. FASEB J. 2007;21(12):3380–3385. doi: 10.1096/fj.07-8229com. [DOI] [PubMed] [Google Scholar]
- Blewitt ME, Vickaryous NK, Paldi A. et al. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet. 2006;2(4):e49. doi: 10.1371/journal.pgen.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Altenberg L. Perspective: complex adaptations and the evolution of evolvability. Evolution. 1996;50(3):967–976. doi: 10.1111/j.1558-5646.1996.tb02339.x. [DOI] [PubMed] [Google Scholar]
- Jurka J, Bao W, Kojima KK. Families of transposable elements, population structure and the origin of species. Biol Direct. 2011;6:44. doi: 10.1186/1745-6150-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Bao W, Kojima KK. et al. Distinct groups of repetitive families preserved in mammals correspond to different periods of regulatory innovations in vertebrates. Biol Direct. 2012;7:36. doi: 10.1186/1745-6150-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalhauzen II. Factors of Evolution: The Theory of Stabilizing Selection. Philadelphia: Blakiston Company; 1949. [Google Scholar]