Abstract
In animals, PIWI-interacting RNAs (piRNAs) play a crucial role in genome defense. Moreover, because piRNAs can be maternally transmitted, they contribute to the epigenetic profile of inheritance. Multiple studies, especially in Drosophila, have demonstrated that the machinery of piRNA biogenesis is often the target of positive selection. Because transposable elements (TEs) are a form of genetic parasite, positive selection in the piRNA machinery is often explained by analogy to the signatures of positive selection commonly observed in genes that play a role in host-parasite dynamics. However, the precise mechanisms that drive positive selection in the piRNA machinery are not known. In this review, we outline several mechanistic models that might explain pervasive positive selection in the piRNA machinery of Drosophila species. We propose that recurrent positive selection in the piRNA machinery can be partly explained by an ongoing tension between selection for sensitivity required by genome defense and selection for specificity to avoid the off-target effects of maladaptive genic silencing by piRNA.
Keywords: piRNA, transposable element, selfish elements, positive selection, Red Queen, host-parasite
Introduction
Transposable elements (TEs) are selfish elements that encode the capacity to copy themselves across genomes and they pose a significant threat in the form of mutation. Moreover, because of their repetitive nature, they can mediate harmful chromosome rearrangements. In response to these threats many eukaryotes utilize mechanisms of RNA silencing to limit TE proliferation. In the animal gonad, a specialized form of RNA silencing mediated by PIWI-interacting RNAs (piRNAs) plays a critical role in maintaining TE repression across generations. Strikingly, multiple studies, especially in Drosophila, have demonstrated that the machinery of piRNA biogenesis is often the target of recurrent positive selection. This signature is often explained by analogy to the signatures of positive selection commonly observed in genes that play a role in host-parasite dynamics. Since TEs can be considered genomic parasites, a Red Queen evolutionary arms race has been proposed to explain this signature of adaptive evolution. Under this scenario, recurrent evolution in the piRNA machinery would be driven by ongoing evolution on the part of TEs to evade piRNA silencing. In turn, the piRNA machinery would show a strong signature of adaptation driven by recurrent selection to maintain TE silencing.
While plausible, no specific mechanism supports a Red Queen model for the evolution of the piRNA machinery. In this review, we outline several mechanistic models that might explain pervasive positive selection in the piRNA machinery of Drosophila species. Finally, we discuss the evolution of the piRNA machinery in light of off-target effects. While most piRNAs are dedicated to TE silencing, recent studies have shown that there are significant off-target effects of gene silencing. Under varying circumstances, piRNAs can target and silence genes as well as TEs. This indicates that there is a cost to TE control by piRNAs. We designate this form of off-target gene silencing as "genomic autoimmunity." Genomic autoimmunity is analogous to classic forms of autoimmunity, which are caused by an immune response that incorrectly targets self. In the case of genomic autoimmunity, genes rather than TEs become the target of repression. We propose that the strong evolutionary tension driven by genomic autoimmunity contributes to the signature of adaptive evolution observed in the piRNA machinery. We argue that recurrent positive selection in the piRNA machinery can be partly explained by ongoing tension that leads to cycles of selection for sensitivity required by genome defense and selection for specificity to avoid the off-target effects of maladaptive genic silencing by piRNA.
Transposable Elements: Endogenous Selfish Mutagens
Sexual reproduction plays an important role in producing variation that enables hosts to adapt to rapidly evolving parasites [1]. Therefore, it is an evolutionary irony that sexual reproduction also establishes a condition ripe for exploitation by genetic parasites known as TEs [2]. TEs come in two major types. Retrotransposons (known as Class I elements) transpose via RNA that is reverse transcribed into DNA and inserted elsewhere into the genome. Class I retrotransposons are classified as long terminal repeat (LTR) elements, which are similar to retroviruses; LINE-like retrotransposons, which lack LTRs; and SINE-like elements, which do not code for reverse transcriptase (RT) and instead hijack the RT from other families. DNA transposons (known as Class II elements) move via DNA intermediates. The most well understood DNA transposons encode transposases that "cut-and-paste" insertions from one location to the next. If DNA transposons move during DNA replication, copy number may increase if elements jump ahead of the replication fork. A second class of DNA transposons, Helitrons, replicate via rolling circle amplification.
TEs are major determinants of variation in genome architecture [3] and there is a positive relationship between bulk TE content and genome size [4]. For example, salamander genomes are notoriously large and much of this can be explained by a great proportion of LTR TEs [5,6]. It has also been estimated that nearly 70 percent of the human genome is comprised of repetitive sequences, many of which are derived from TEs [7]. TEs can sometimes be exapted for functions beneficial to the host. For example, TE sequences play critical roles in telomere function in Drosophila [8] and have been recruited to mediate V(D)J recombination in jawed vertebrates [9].
Nonetheless, TEs are mostly harmful. In humans, TEs can cause a variety of diseases [10]. For example, a case of haemophilia has been caused by an insertion of an L1 retroelement [11]. In addition, several cases of colorectal cancer have been found to be caused by insertion of L1 elements into the APC tumor suppressor [12]. TEs can also harm the host by mechanisms independent of insertional mutation. For example, overexpression of Alu RNA can cause macular degeneration by activating the NLRP3 inflammasome [13]. Finally, due to their repetitive nature, TEs pose a challenge to genome stability [14,15]. Ectopic recombination among dispersed repeat sequences can lead to significant chromosomal damage [16]. In fact, natural selection against chromosomal damage caused by ectopic recombination is proposed to be a major force limiting TEs from completely overwhelming genomes [14].
Genome Defense by PIWI Proteins
In the face of this threat, different mechanisms of genome defense have evolved to protect the genome from TEs. The primary modes of genome defense are chromatin modification, DNA methylation and RNA silencing, though there are others [17,18]. These modes of genome defense are mechanistically connected and also function as epigenetic regulators of gene expression. In fact, many of the epigenetic mechanisms discussed in this issue were likely derived from ancestral mechanisms of genome defense.
Genome defense against TEs by RNA silencing is seen widely across eukaryotes. In plants, fungi, and metazoans, diverse pathways of genome defense by RNA silencing are centered on the Argonaute proteins [19]. Argonaute proteins are characterized by their capacity to carry a small 20 to 30 nt guide RNA that mediates recognition of reverse complement RNA. Upon target recognition, Argonaute proteins have the capacity to slice the recognized RNA. In this way, RNA silencing by Argonaute proteins can function as an adaptive immune system. The slicing function of Argonaute proteins can target a wide array of harmful RNA species, both viruses and TEs, via diverse guide RNAs.
Despite the core similarity of small RNA-based genome defense shared across eukaryotes, these pathways have significantly diversified. Some of this diversification can be explained by the fact that multicellular animals, unlike many other eukaryotes, commonly have a germline specified early in development. This (with some exceptions) results in a soma that does not contribute to the next generation. This has important consequences for the evolution of genome defense in animals. Because TEs are transmitted vertically through genomes passed to offspring, transposition in the soma does not confer an advantage to a TE lineage. Instead, somatic harm caused by TE insertions can only reduce the chance that genomic copies will be represented in the next generation. For this reason, in animals with an early specified germline, natural selection will act on TE lineages to be active only within the germline compartment. In turn, when TEs are primarily active within the germline, selection will favor genome defense mechanisms that are also specified to the germline. Consistent with this expectation, a unique mode of genome defense has specialized within the reproductive tissues of animals. This mode of genome defense is mediated by a specialized class of Argonaute proteins known as the PIWI proteins.
PIWI proteins are a specialized clade of Argonaute proteins that play a critical role in genome defense in animals and, for the most part, are only found in reproductive tissues [20,21]. Similar to other Argonautes, PIWI proteins carry small guide RNAs and mediate a slicing reaction of the target RNA. Guide RNAs loaded into PIWI proteins are commonly derived from TEs and, when derived from an anti-sense TE transcript, can target sense TE mRNAs for destruction. PIWI proteins are also unique in how they receive their guide RNA. While other Argonaute proteins receive either siRNAs or miRNAs via Dicer proteins, PIWI proteins receive their guide RNAs (known as PIWI-interacting RNAs, or piRNAs) from a complex machinery of piRNA biogenesis [21]. piRNAs are significantly longer than siRNAs and miRNAs, ranging from 23 to 30 or more nucleotides.
How are anti-sense piRNAs generated from TEs, rather than genes? To provide defense against only TEs, natural selection has identified the Achilles heel of TE biology. Unlike other coding sequences in the genome, TEs increase in number by copying themselves to other locations. Thus, TEs can be distinctly recognized by their capacity to land in different regions of the genome. TEs that transpose into a piRNA "trap", known as a piRNA cluster, are thus recognized. piRNA clusters typically reside in the boundary between euchromatin and heterochromatin and have distinct chromatin signatures. They are also replete with diverse TE fragments, representing the history of TE invasion within a species. TE sequences, residing in these piRNA clusters, then become the source of anti-sense TE piRNAs. Loaded into PIWI-proteins, they can mediate repression of sense TE mRNAs.
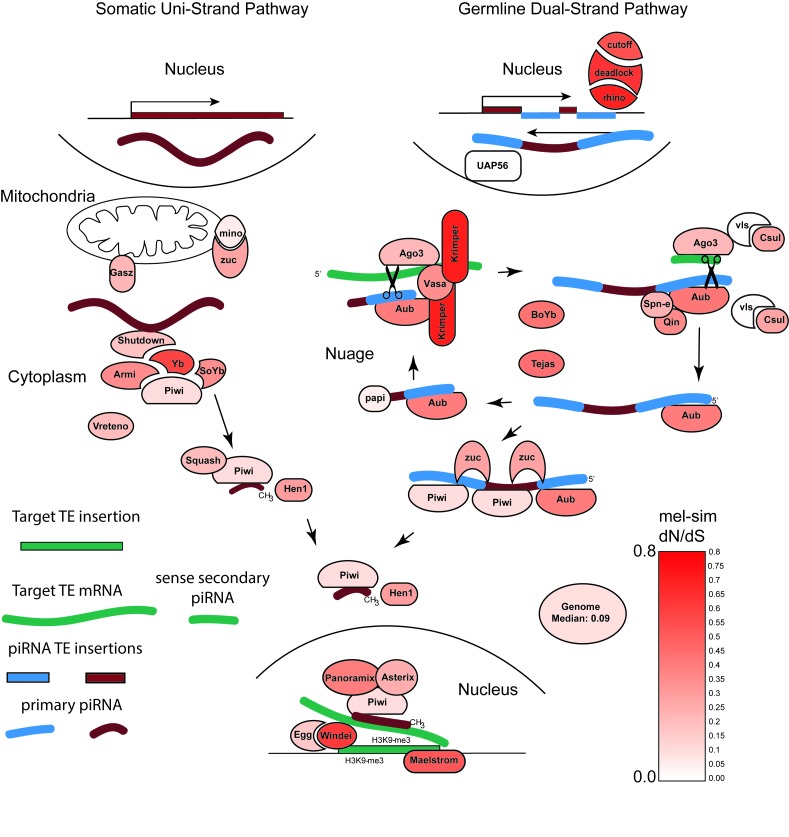
The mechanisms underlying piRNA cluster designation, transcript processing and piRNA biogenesis have been extensively reviewed elsewhere [20-24]. An overview of the somatic and germline pathways, as understood in female Drosophila, is shown in Figure 1. Many components of the piRNA biogenesis pathway evolve rapidly and this rapid evolution is also demonstrated in the figure. Several salient features of piRNA biogenesis should be noted. First, genome defense by piRNA is specialized across different parts of the female gonad. Within the somatic follicle cells that surround the ovary, piRNAs are derived from loci that produce single-stranded anti-sense transcripts. These yield piRNAs that are found exclusively in Piwi, the sole Drosophila PIWI protein expressed in these cells. The most well understood locus that is the source of these somatic piRNAs is the flamenco locus. Examination of the TE fragments that comprise the flamenco locus explains the existence of piRNA mediated genome defense in somatic cells on the ovary exterior; the anti-sense piRNAs derived from the flamenco locus target gypsy, idefix and ZAM, as well as other elements [25-28]. A unique property of these elements is that they can all be considered endogenous retroviruses with the capacity to move between cells. For example, gypsy elements are known to form virus-like particles that can cross cell membranes [29,30]. Thus, control of somatic elements by piRNAs derived from the flamenco locus can protect the germline against invasion of endogenous retroviruses derived from flanking follicle cells.
Figure 1.
The piRNA biogenesis and silencing pathway. dN/dS estimates, indicated with varying intensity of red, between D. melanogaster and D. simulans piRNA pathway components were obtained from the flyDIVaS web server [81], with the exception of estimates for ago3, deadlock, Yb, maelstrom, rhino, squash, vasa, and valois. dN/dS estimates for these missing genes were obtained by through our analysis.
Within the germline proper, a more complex pathway of piRNA biogenesis maintains genome defense. Here, piRNAs are derived from dual-strand, rather than single strand clusters. This leads to the production of both sense and anti-sense piRNAs that target sense TE mRNAs in a biogenesis loop denoted as ping-pong biogenesis. Anti-sense piRNAs loaded into the Drosophila Aubergine protein destroy sense TE mRNAs by post-transcriptional gene silencing (PTGS). And while anti-sense piRNAs loaded into Drosophila Piwi protein can also mediate PTGS, they also enter the nucleus and target TE insertions for transcriptional gene silencing (TGS). Since Drosophila Piwi is a nuclear protein and found in both the somatic follicle cells and the germline, this form of transcriptional silencing is shared across these two compartments of the ovary. In the germline, this form of transcriptional silencing is also transmitted to the next generation via maternally provisioned of Piwi-piRNA complexes.
piRNA Silencing as an Epigenetic Phenomenon
Before the discovery of piRNA silencing, syndromes of hybrid dysgenesis demonstrated an important connection between genome defense and epigenetic control of gene expression. This is because TEs inherited only via the paternal germline were known to become activated in progeny and cause sterility. Genetically identical reciprocal progeny, with maternally inherited TEs, were completely fertile. These observations indicated that while sperm failed to maintain TE control, the maternal lineage maintained TE repression across generations. This maternal effect was designated a 'cytotype' [31,32] and it is now known that the cytotype is the presence of piRNAs targeting strain-specific TE populations [33]. Since females exclusively transmit the cytoplasm in the egg, piRNAs that target TEs in the germline are transmitted alongside genomic TE insertions in females. In contrast, piRNAs are not transmitted through male sperm and TE families only residing in the paternal genome find themselves in an egg from a mother lacking piRNAs that target the TE family. In this way, the female germline maintains TE repression across generations in an epigenetic fashion.
The way TEs are silenced at the transcriptional level also has connections to epigenetic control of gene expression. Nuclear piRNAs, in complex with Piwi, mediate transcriptional silencing by directing histone methylation at TE insertion sites. This transcriptional silencing is facilitated by asterix/DmGTSF-1 [34-36] and panoramix/Silencio [37,38]. While these mechanisms of transcriptional silencing can efficiently maintain TE repression, there are also significant off-target effects of piRNA mediated transcriptional silencing on genes. Off-target gene silencing by piRNA can be considered a form of "genomic autoimmunity" because the piRNA machinery targets self rather than non-self. For example, in some strains of D. virilis, the center divider gene has become silenced by being converted into a piRNA cluster, presumably caused by flanking TE sequences located within the telomere [39-41]. This conversion is associated with local H3K9 tri-methylation, a mark of heterochromatin. It has been further shown that TE insertions exert an epigenetic effect on local genic chromatin states, with genes closer to TEs more likely to yield genic piRNAs and enrichment for silencing by H3K9 tri-methylation [42,43]. For these reasons, piRNA mediated genome defense can be considered an important epigenetic regulator of gene expression. In this way, piRNA-mediated genome defense is costly, since genes important for host fitness can be caught in the crossfire by this form of "genomic autoimmunity". We propose that the cost of genomic autoimmunity contributes to the pervasive signature of positive selection in the piRNA machinery.
Positive Selection in the piRNA Machinery
Despite the essential function of piRNA mediated genome defense, a number of studies, especially in Drosophila, have demonstrated that the machinery of piRNA biogenesis and function is rapidly evolving [44-52]. Moreover, this rapid evolution can largely be explained by the action of positive selection. The first of these studies identified a strong signature of positive selection in the rhino protein, even before piRNAs were known [45]. Since this study, multiple studies have used diverse approaches to characterize natural selection on the piRNA machinery at different time scales. These range from strict population-based studies of patterns of linked polymorphism to McDonald-Kreitman based tests of polymorphism and divergence to molecular evolutionary based tests of synonymous and non-synonymous substitution. Table 1 indicates that this signature of adaptation is pervasive across the machinery of piRNA biogenesis. In some cases, such as for spn-E, diverse and independent tests identify positive selection across different time scales.
Table 1. Previous studies that have identified signatures of positive selection in the Drosophila piRNA machinery.
| Study | Species | Test of Neutrality | Genes w/significant tests | Genes w/non-significant tests |
| Lee and Langley [44] | mel and/or sim | MK test | armi, aub, krimper, spn-E, vasa | ago3, hen1, mael, piwi, rhino, squash, zuc |
| Vermaak et al. [45] | mel, sim | Sliding window dN/dS, MK test | rhino | N/A |
| mel to pseudo | PAML: M1 vs M2, M7 vs. M8 | rhino | N/A | |
| Lewis et al. [46] | Nematocera, Brachycera | PAML: M8a vs M8 | N/A | ago3, piwi/aub |
| Heger and Ponting [47] | mel, sim, yak, ere, ana | SLR | spn-E | N/A |
| Obbard et al. [49] | mel, sim | MK test | aub, armi | N/A |
| Kolaczkowski et al. [50] | mel | SweepFinder | armi, aub, hen1, piwi, rhino, spn-E, squash, zuc | ago3, krimper, mael |
| 12 species | PAML: Branch-sites | aub, krimper, spn-E, squash, zuc | ago3, armi, mael, hen1, piwi, rhino | |
| Obbard et al. [48] | mel, sim | MK test | armi, aub, piwi, mael, spn-E | ago3, zuc, squash, hen1, krimper |
| Simkin et al. [52] | mel | Polymorphism | aub, armi, rhino, spn-E, vasa, zuc | ago3, krimper, piwi, squash |
| mel,sim | MK test | aub, armi, krimper, spn-E, vasa | ago3, piwi, rhino, squash, zuc | |
| mel, sim, sech, yak, ere, ana | PAML: M7 vs. M8 | ago3, aub, piwi, rhino | armi, krimper, spn-E, squash, vasa, zuc |
What explains this pervasive signal of positive selection in the piRNA machinery? Since TEs can be considered genomic parasites, an analogy is frequently made to the adaptation often observed in immune systems. This model proposes that hosts and TEs are in an evolutionary arms race and positive selection is driven by Red Queen dynamics of adaptation and counter-adaptation. Most descriptions of the Red Queen process describe it as a co-evolutionary process. In the case of host-parasite relationships, positive selection on host immune systems will occur when parasites evolve strategies to evade the host immune system. In turn, selection favors immune strategies that maintain control of parasites. While this analogy to other host-parasite dynamics seems a plausible explanation for recurrent positive selection in the piRNA machinery, it lacks a clear mechanistic explanation.
Mechanisms to Explain Positive Selection in the piRNA Machinery
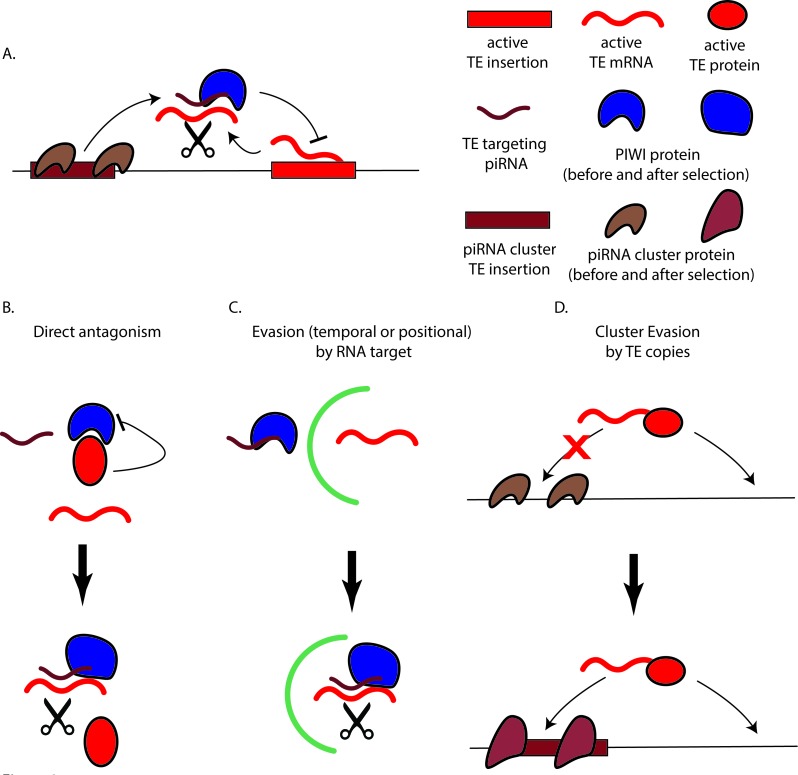
If positive selection is driven by a canonical host-parasite Red Queen process, then TEs and the host piRNA machinery must be co-evolving. Specifically, adaptation in TE lineages must enable TE evasion of piRNA silencing, followed by corresponding adaptation in the piRNA machinery to the new TE variant. Several mechanisms of TE evasion are plausible and described in Figure 2. Importantly, since we are mostly concerned with evolution of the piRNA machinery itself, we restrict our discussion to adaptation in protein sequence.
Figure 2.
Models for TE evasion of the piRNA machiney. A. General pathway for piRNA silencing with cluster insert that becomes source of anti-sense piRNA, followed by transcriptional and post-transcriptional gene silencing. B. Direct antagonism. A TE encoded antagonist of a piRNA effector. Here, a TE encoded protein (red) interferes with a PIWI protein (blue) directed slicing reaction. An evolutionary change in the PIWI protein (blue) allows it to silence the mRNA of the TE. C. Evasion. Specialized localization of the TE mRNA (either temporal or physical) allows the TE mRNA to avoid being a target. Separation in space or time is indicated with the green line. Adaptation in the PIWI protein (blue) allows it re-localize and silence the TE mRNA. D. Cluster evasion. A newly arriving TE proliferates within the genome, but avoids inserting into a cluster. A change in protein sequence (perhaps in rhino, as has been proposed [52]) facilitates TE insertion into the cluster, leading to subsequent piRNA silencing of the TE family.
One possibility is that TEs directly antagonize the piRNA machinery. This is demonstrated in Figure 2B. Here, a TE-encoded protein may directly antagonize a PIWI protein and disrupt the association of the PIWI protein with a guide piRNA. This would lead to global loss of TE piRNA silencing by the target protein. We have proposed that TE-encoded suppressors of piRNA silencing may explain the general release of diverse elements during hybrid dysgenesis [53]. Viruses in plants and animals are known to antagonize RNA silencing pathways in this fashion. In plants, virally encoded suppressors of RNA silencing (VSRs) can act through diverse mechanisms that include sequestering siRNAs and directly antagonizing Argonaute proteins [54-56]. Different modes of VSR action, including the antagonism of Argonaute proteins and their corresponding slicer activity, are also observed in insect viruses [57-59]. Natural selection to escape VSRs has been proposed to explain a strong signature of positive selection in the RNAi machinery that is devoted to defense against viruses [48,60]. Thus, if TEs encode suppressors of piRNA silencing, natural selection on the piRNA machinery to avoid TE antagonism may drive the pervasive signature of positive selection [52].
To date, however, there are few examples of TEs that encode anti-silencing function. One explanation for this contrast with viruses is that TE encoded suppressors of the piRNA machinery may have a net negative effect on TE fitness. Virally encoded suppressors of RNA silencing may reduce host fitness by increasing susceptibility to other viruses, but this cost may not significantly burden the virus lineage encoding the VSR. A weakened host with a higher viral titer may be tolerable to the viral lineage since viruses have the capacity to move from dying hosts to healthy ones. TEs do not have this luxury. A TE encoded suppressor of piRNA silencing that releases global TE control may only reduce host fertility, thereby reducing the chance of transmission to the next generation. Therefore, if TE-encoded suppressors of piRNA silencing exist, it seems most likely that they would act on specialized, rather than general components of the piRNA machinery.
The absence of known TE-encoded suppressors of piRNA silencing may also be explained by lower adaptive capacity of TEs, as compared to viruses. Viruses can achieve very large population sizes. However, at least in Drosophila where the signatures of positive selection in the piRNA pathway are pervasive, there may only be dozens of TE copies of each TE family per genome. Moreover, since each copy likely jumps about every 100 to 10,000 generations [61,62], the functional effective population size of the TE lineage (which measures the adaptive capacity) may be smaller than the host itself. Thus, compared to viruses, TEs may have less capacity to evolve suppressors of piRNA silencing. Nonetheless, the piRNA machinery might experience positive selection not through its function in genome defense, but through pleiotropic function to protect against viruses. In mosquitoes, the piRNA machinery appears to play an important role in somatic defense against viruses [63,64]. However, in D. melanogaster, recent studies have shown that the piRNA machinery provides little protection against viral challenge [65]. Alternately, positive selection in the piRNA machinery may be driven by the indirect effects of viral suppressors of RNA silencing that serendipitously target the piRNA machinery [50]. Overall, future studies are needed to formally test whether TEs or viruses can antagonize the piRNA machinery directly.
While positive selection in response to TE mediated suppression of genome defense may be an unlikely explanation for adaptive evolution of the piRNA machinery, several other mechanisms are plausible. Figure 2C demonstrates that TE evasion of the piRNA machinery may be achieved through temporal or positional regulation within the germline. TEs are known to carry regulatory sequences that can drive exquisite localization within the germline. For example, the I, G2 and jockey elements all carry gurken-like mRNA localization signals that allow them to localize to regions just flanking the oocyte nucleus within the developing egg chamber [66]. This shows that natural selection may act on TE lineages to ensure patterns of localization that enable proliferation. Similar mechanisms may also enable evasion of piRNA silencing. Such physical evasion of piRNA silencing by TEs may lead to positive selection on piRNA proteins to target new compartments within the germline. This form of selection is borne out by the fact that Aubergine and Piwi proteins are paralogs that have evolved specialized localization within the germline and the soma of the ovary, as well as the cytoplasm and the nucleus [25].
Lastly, evasion by TEs may simply happen by avoiding entry into the piRNA pathway altogether (Figure 2D). TEs become targets for piRNA biogenesis by inserting into designated piRNA clusters. Dual strand clusters are designated by the Rhino-Deadlock-Cutoff (RDC) complex [67-69]. Therefore, if TEs are capable of moving freely within the genome without landing in any of these dual strand clusters, they may avoid detection by the piRNA machinery altogether. This may place strong selection on the RDC complex to entice TE insertions into these dual strand clusters [52,67]. In fact, even before piRNAs were known, a model of TE entrapment was proposed to explain positive selection on rhino [45].
Considerations of Modes of Adaptation in the piRNA Machinery
A standard model of Red Queen driven adaptation in an immune system proposes that both host and parasite co-evolve with each other. As mentioned earlier, and in contrast to viruses, the capacity of a TE lineage to evolve to evade the piRNA machinery may be limited. Therefore, a co-evolutionary dynamic may not directly influence the evolution of the piRNA machinery. However, it is well known in Drosophila that genomes are constant targets to horizontal invasion by TEs [70-72]. For example, the P element likely became established in D. melanogaster through horizontal transfer from D. willistoni [73]. Therefore, adaptation in the piRNA machinery may not be driven by newly evolved strategies of resident TEs. Rather, adaptation in the piRNA machinery may be driven by novel strategies of transposition carried by horizontally transferred TEs. TE diversity itself has been proposed to be a driver of adaptation in the piRNA machinery [44,51]. This seems especially plausible given our recent simulation study demonstrating that horizontal transfer may be a more important determinant of TE abundance and diversity than drift [74].
We have performed a preliminary test of this hypothesis [75]. If TE diversity was a key determinant of adaptation in the piRNA machinery, one might expect that the piRNA machinery might evolve more quickly in species of Drosophila with a greater TE load. However, in a molecular evolutionary analysis of rates of evolution in the piRNA machinery across different species of Drosophila, we found the opposite to be true. In species with a higher TE burden, and also greater TE diversity, we found that the piRNA machinery is evolving more slowly. However, we found that there is a striking relationship between TE abundance and codon bias in the piRNA machinery [75]. It is widely known that highly expressed genes frequently show a greater degree of codon bias [76,77]. In species with a greater TE burden, codon bias in the piRNA machinery (but not the rest of the genome) is increased. This suggests that the primary response to increased TE abundance is not an increased rate of adaptive evolution. Rather, an increased TE burden appears to select for increased function of the piRNA machinery. This result suggests an alternate model to explain the rapid and adaptive evolution of the piRNA machinery.
The Genomic Autoimmunity Hypothesis for Positive Selection in the piRNA Machinery
Genomic autoimmunity is likely to be a significant cost of genome defense [42,43,78]. And because piRNA silencing can be transmitted as a maternal effect, this collateral silencing can be transmitted across generations as a "paramutation" in the absence of the original piRNA source allele [39-41,79,80]. This may lead to a population-level "paramutation load". Further insight into this tension is provided by studies of hybrid dysgenesis caused by TEs. As in the case of D. melanogaster [31], investigations of sterility induction in D. virilis have shown a strong relationship between the dose of paternal chromosomes carrying TEs and the induction of sterility [41]. Correspondingly, there is also a strong dose effect of maternal chromosomes carrying TE insertions that produce piRNAs [53]. These studies of hybrid dysgenesis suggest that the paternal dose of inherited TEs must be appropriately matched with a maternal piRNA dose that maintains TE silencing. These dosage effects can explain our observation that the piRNA machinery evolves increased codon bias in species of Drosophila with higher TE burden [75]. Species with a higher TE burden presumably experience increased selection for an increased dose of piRNA defense and this may be facilitated by increased codon bias in the piRNA machinery.
It seems likely that an increased dose of the piRNA machinery comes at a cost of increased off-target gene silencing. We propose that ongoing fluctuation in the TE load leads to cycles of oscillating selection favoring either specificity or sensitivity in piRNA-mediated genome defense. Rather than being driven by co-evolution and adaptation in TE lineages, we propose this oscillating mode of selection is driven by fluctuating TE exposure by horizontal transfer. In this way, natural selection drives the machinery continuously, as both TE load increases and decreases. We further propose that evolution in factors that determine specificity and sensitivity lead to natural selection that propagates through the entire network of piRNA biogenesis.
In Figure 1, we identify the fastest evolving components of the piRNA machinery in a comparison between D. melanogaster and D. simulans using results provided from the flyDIVAS database (http://igem.temple.edu/flydivas/index.html) [81]. This analysis shows that the fastest components are not the effectors of piRNA silencing (aubergine and piwi). Rather, they are krimper, rhino, deadlock, cutoff and Yb. The fast evolution of rhino, deadlock and cutoff is striking since these code for the three proteins that comprise the Rhino-Deadlock-Cutoff (RDC) complex [68,82]. The RDC complex plays an essential role in determining the identity of dual-strand clusters that are the source of piRNAs in the germline. We propose that fast evolution of proteins in the RDC complex is driven by an especially strong tension over the costs of transcriptional genic silencing that can spread into flanking genes; in contrast to PTGS, TGS is extremely dangerous. While off-target effects of cytoplasmic gene silencing can lead to the destruction of a single mRNA molecule, TGS can lead to the silencing of the sole genomic copies that the cell relies on. Natural selection against the effects of TE mediated TGS on neighboring genes has been seen in both corn [78] and Drosophila [43]. A dual-strand cluster may be especially potent in this respect. Thus, if TEs become more common within a species, natural selection may act on components of the RDC complex to become more discriminating in their capacity to define dual-strand clusters.
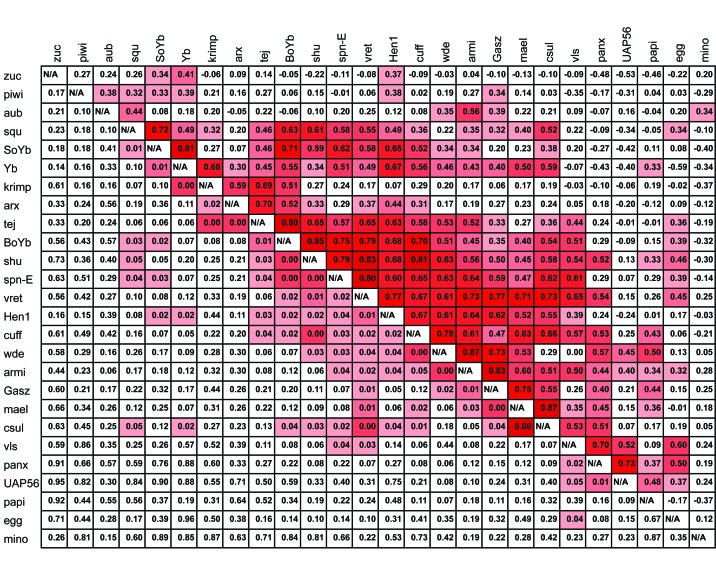
How might fast evolution in some components of the piRNA machinery drive pervasive adaptation across the whole pathway? We further propose that the key evolutionary innovations in proteins that modulate the balance between specificity and sensitivity also select for compensatory changes throughout the machinery of piRNA biogenesis. A propagating consequence of adaptive divergence in the piRNA machinery has also been proposed by others [50,83]. If adaptive changes lead to selection for compensatory changes in other components of the piRNA machinery, we would expect that components of the piRNA machinery show a correlated pattern of amino acid divergence across species. In particular, we would expect that this correlated divergence would be greater than that expected to arise simply due to the effects of changing demography and population size. We can test this hypothesis by performing an Evolutionary Rate Covariation analysis of piRNA biogenesis factors [84-86] (Figure 3). In this analysis, we utilized the Evolutionary Covariation Rate webserver (http://csb.pitt.edu/erc_analysis/) to determine the level of evolutionary covariation of the piRNA machinery within the 12 well sequenced species of the Drosophila genus. This method determines branch-specific levels of amino acid divergence and tests whether gene-by-gene pairwise divergence covaries. Statistical significance is determined empirically against the rest of the genome-wide pairwise correlations. If positively selected amino acid substitutions in some components of the piRNA machinery lead to selection for compensatory changes in other components, we would expect to see a significant pattern of evolutionary covariation in the piRNA machinery. While some genes, such as rhino and deadlock, are unfortunately excluded from this analysis due to their extreme divergence, there is strong evidence for co-evolution among piRNA biogenesis factors (Contrast to global pairwise ERC values, P << .001). Perhaps most interesting and also perplexing, the effector proteins Piwi and Aubergine show only a weak signal of co-evolution with other factors.
Figure 3.
Evolutionary Rate Covariation analysis of piRNA proteins across the Drosophila genus. Results were obtained from [84]. Proteins in the middle of the table share the greatest signature of co-evolution. Evolutionary Rate Covariation (ERC) analysis measures the degree to which changes in one protein (relative to background) are correlated with changes in another (also relative to background). The analysis is performed by estimating branch specific amino acid divergence across a phylogeny. Here, branch specific amino acid divergence was estimated for 12 sequenced species of the Drosophila genus. Significance is measured relative to the genomic background of all pairwise covariation estimates. Some proteins are not available due to lack of clear orthologs in divergent species. Above the diagonal: ERC values. Values range from -1 to 1, with values closer to 1 indicate higher levels of covariation. Red intensity above diagonal scales with strength of correlation. Below the diagonal: P values. P values were determined empirically relative to background. Red intensity below diagonal scales with degree of significance. Significant co-variation across the piRNA machinery is demonstrated by a Z-score of observed P values being equal to -15.4 (P << .001).
Is there a way to explicitly test the hypothesis that fluctuating TE loads can lead to oscillating natural selection between sensitivity and specificity? One possible way of testing this is to examine the evolution of piRNA lengths. Little is known about the specificity of piRNA targeting and how many mismatches may be tolerated between piRNA and target [87]. However, it is reasonable to propose that piRNAs of increasing length have increased specificity. In an analysis of oxidized small RNA sequencing data obtained from wild type Drosophila melanogaster [88], we determined the number of transcripts targeted by ~4,000,000 26 bp piRNAs and compared this to the number of transcripts targeted by the same pool of piRNA with one base removed from the 3' end (anti-sense mappers with no mismatches). We found that the 25 bp small RNAs could potentially target ~8 percent more transcripts (2284 transcripts vs. 2477 transcripts).
The inference that longer piRNAs have increased specificity is also supported by several aspects of RNA silencing biology. As previously mentioned, TGS is significantly more dangerous for the host than PTGS. In accordance with this hypothesis, piRNAs, which can mediate TGS, are longer than miRNAs and siRNAs. Moreover, in Drosophila, the class of piRNAs that directly mediate TGS (those bound to Piwi protein in the nucleus) are longer than those with exclusive roles in the cytoplasm (those bound to Aubergine) [25]. A similar difference is observed in mice among pre-pachytene piRNAs. Nuclear piRNAs that are bound to MIWI2 and likely directly methylate TE insertions are longer than piRNAs that are cytoplasmic and bound to MILI [89]. Strikingly, this pattern of silencing RNAs that target chromatin being longer than cytoplasmic small RNAs is also observed in plants. In plants, there are two classes of small RNAs (plants lack piRNAs). 21/22 nt small RNAs mediate PTGS whereas 24 nt small RNAs have a primary role in RNA-directed DNA methylation and TGS [90,91] (but see [92]). Again, it appears that small RNAs with primary roles in nuclear silencing are longer. This is consistent with the hypothesis that costs associated with off-target TGS require additional specificity.
Based on these observations, one might predict that natural selection would favor increasing specificity of piRNA silencing when the genomic TE burden is large. A true phylogenetic comparative analysis of piRNA size and TE abundance has yet to be performed, but some interesting trends arise when comparing available piRNA profiles across species. In Drosophila, the most common piRNAs are 25 to 26 nts. However, in mammals ranging from macaques and marmosets to mice and pigs, piRNAs are considerably longer, with the most common piRNAs being 28 to 30 nts [93]. This coincides with the fact mammalian genomes carry a significantly greater TE burden. This trend of greater TE burden being associated with longer piRNAs is also observed more narrowly within Dipterans. For example, mosquito genomes carry a significantly greater burden of TEs compared to Drosophila, and piRNAs in mosquitoes are longer than those in Drosophila [94,95]. Even more taxonomically narrow, within the genus Drosophila, this pattern holds. D. virilis has a significantly greater TE burden compared to D. melanogaster and D. virilis has a correspondingly longer piRNA profile (Table 2). Together, this overall pattern suggests that increased TE burden may select for increased specificity of the piRNA machinery. This is likely mediated by changes in the proteins that comprise the piRNA biogenesis pathway and define piRNA length distributions.
Table 2. Comparison of piRNA length between D. virilis and D. melanogaster. Mode was the most common length class among small RNAs 23 to 30 nt, post adapter trimming and filtering against ncRNA, miRNA, miscRNA and tRNA. Only including one of each replicate library for the comparison (indicated with * for each library included), the difference in small RNA mode is significant (p < .01, t test among modes). § indicates a library generated by oxidation by sodium periodate.
| Species | Sample | Tissue | Source | Small RNA mode |
| Drosophila virilis | Strain 160 (rep 1) | 0-2 hr embryos | Erwin et al. [41] | 26* |
| Drosophila virilis | Strain 160 (rep 2) | 0-2 hr embryos | Erwin et al. [41] | 26 |
| Drosophila virilis | Strain 9 (rep 1) | 0-2 hr embryos | Erwin et al. [41] | 26* |
| Drosophila virilis | Strain 9 (rep 2) | 0-2 hr embryos | Erwin et al. [41] | 26 |
| Drosophila virilis | wt | follicle cells | Chirn et al. [93] | 27* |
| Drosophila virilis | wt (rep 1) | 0-4 hr unfertilized embryos | Ninova et al.[96] | 27* |
| Drosophila virilis | wt (rep 2) | 0-4 hr unfertilized embryos | Ninova et al.[96] | 27 |
| Drosophila virilis | Strain 9 | 0-2 hr embryos | Rozhkov et al. [97] | 26* |
| Drosophila melanogaster | Oregon-R | follicle cells | Chirn et al. [93] | 26* |
| Drosophila melanogaster | wt (rep 1) | 0-4 hr unfertilized embryos | Ninova et al.[96] | 26* |
| Drosophila melanogaster | wt (rep 2) | 0-4 hr unfertilized embryos | Ninova et al.[96] | 26 |
| Drosophila melanogaster | Ral 437 | ovaries§ | Song et al.[98] | 26* |
| Drosophila melanogaster | Ral 375 | ovaries§ | Song et al.[98] | 26* |
| Drosophila melanogaster | Ral 714 | ovaries§ | Song et al.[98] | 25* |
| Drosophila melanogaster | Ral 707 | ovaries§ | Song et al.[98] | 25* |
| Drosophila melanogaster | Ral 427 | ovaries§ | Song et al.[98] | 25* |
| Drosophila melanogaster | Ral 313 | ovaries§ | Song et al.[98] | 25* |
| Drosophila melanogaster | Ral 732 | ovaries§ | Song et al.[98] | 25* |
| Drosophila melanogaster | Ral 391 | ovaries§ | Song et al.[98] | 25* |
This signature supports a model of piRNA machinery evolution driven by an ongoing tension between sufficient genome defense and the costs of off-target gene silencing. As TE burden fluctuates within a species, we propose the piRNA machinery must be constantly fine-tuned. In this case, natural selection will follow a mode of positive selection when the TE burden increases but also when the TE burden decreases, driving protein sequence evolution in specific components of the piRNA machinery. In turn, this would lead to cascading positive selection on other parts of the machinery to compensate for these changes. This correlated response across diverse components of the piRNA machinery is supported by the strong signature of correlated evolution.
A weakness of the argument outlined here is that evidence for the tension between specificity and sensitivity lacks a phylogenetic perspective. To test this general hypothesis, it will be critical to use more phylogenetically informed approaches to the analysis of evolution of the piRNA machinery. For example, future studies must test whether a reduced TE load coincides with changes in sensitivity in the form of smaller piRNAs. It will also be critical to perform functional analysis of the amino acid changes that appear to have been driven by selection for increased or decreased specificity. The acid test of any proposed explanation for recurrent positive selection in the piRNA machinery will be to identify the functional consequences of amino acid substitutions that have been fixed by natural selection.
Conclusions and Outlook
We propose that the pervasive signature of positive selection in the piRNA machinery is in part driven by selection that fluctuates alternately for sensitivity and specificity in genome defense. An ever changing TE assemblage, dynamically driven by horizontal transfer, may lead to ongoing positive selection. This would not represent a formal Red Queen process, because TE lineages would not co-evolve with the piRNA machinery. Rather, the piRNA machinery would evolve in response to a fluctuating TE burden. This hypothesis is supported by the current observation that species with a greater TE burden have piRNA length distributions that favor specificity over sensitivity.
It is important to consider that evolution can happen at both the protein sequence level and at the expression level. Our examination of codon bias suggests that increasing expression of the piRNA machinery may be another adaptive response to increasing TE burden. This hypothesis has yet to be tested experimentally. In fact, changing size distributions of piRNAs could be driven either by altered mechanisms of piRNA biogenesis, or changing expression levels of different Piwi proteins. For example, an increased size distribution of piRNAs in D. virilis could be explained either by protein evolution leading to preference for longer piRNAs in Piwi or increased expression of Piwi, which carries longer piRNAs compared to Aubergine and Ago3 [25]. A test of the genomic autoimmunity hypothesis for positive selection in proteins of the piRNA machinery will require teasing apart these two modes of adaptive evolution.
Acknowledgments
This work was funded by NSF MCB-1413532 and the University of Kansas
Glossary
- TE
Transposable element
- piRNA
PIWI-interacting RNA
- siRNA
small interfering RNA
- miRNA
microRNA
- RNAi
RNA interference
Author Contributions
J.P.B wrote the paper. AAE and LWH contributed to analysis.
References
- Morran LT, Schmidt OG, Gelarden IA. et al. Running with the Red Queen: host-parasite coevolution selects for biparental sex. Science. 2011;333(6039):216–218. doi: 10.1126/science.1206360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey DA. Selfish DNA: A sexually-transmitted nuclear parasite. Genetics. 1982;101(3-4):519–531. doi: 10.1093/genetics/101.3-4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Ross-Ibarra J. Perspective - Selection on major components of angiosperm genomes. Science. 2008;320(5875):484–486. doi: 10.1126/science.1153586. [DOI] [PubMed] [Google Scholar]
- Munoz-Diez C, Vitte C, et al. Using nextgen sequencing to investigate genome size variation and transposable element content. In: Grandbastien M-A, Casacuberta JM, editors. Plant transposable elements. Berlin Heidelberg: Springer; 2012. pp. 41–58. [Google Scholar]
- Metcalfe CJ, Casane D. Accommodating the load: The transposable element content of very large genomes. Mob Genet Elements. 2013;3(2):e24775. doi: 10.4161/mge.24775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Shepard DB, Chong RA. et al. LTR retrotransposons contribute to genomic gigantism in plethodontid salamanders. Genome Biol Evol. 2012;4(2):168–183. doi: 10.1093/gbe/evr139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA. et al. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7(12):e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E, Pardue ML. Transposon telomeres are widely distributed in the Drosophila genus: TART elements in the virilis group. Proc Natl Acad Sci U S A. 2003;100(6):3363–3368. doi: 10.1073/pnas.0230353100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Tao X, Yuan S. et al. Discovery of an Active RAG Transposon Illuminates the Origins of V(D)J Recombination. Cell. 2016;166(1):102–114. doi: 10.1016/j.cell.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH. Roles for retrotransposon insertions in human disease. Mobile DNA. 2016;7(1):1–28. doi: 10.1186/s13100-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH Jr., Wong C, Youssoufian H. et al. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332(6160):164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- Miki Y, Nishisho I, Horii A. et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52(3):643–645. [PubMed] [Google Scholar]
- Tarallo V, Hirano Y, Gelfand BD. et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149(4):847–859. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Langley CH. The population genetics of Drosophila transposable elements. Annu Rev Genet. 1989;23:251–287. doi: 10.1146/annurev.ge.23.120189.001343. [DOI] [PubMed] [Google Scholar]
- Langley CH, Montgomery E, Hudson R. et al. On the role of unequal exchange in the containment of transposable element copy number. Genet Res. 1988;52(3):223–235. doi: 10.1017/s0016672300027695. [DOI] [PubMed] [Google Scholar]
- Montgomery EA, Huang SM, Langley CH. et al. Chromosome rearrangement by ectopic recombination in Drosophila melanogaster: genome structure and evolution. Genetics. 1991;129(4):1085–1098. doi: 10.1093/genetics/129.4.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U. et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281(31):22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- Jacobs FMJ, Greenberg D, Nguyen N. et al. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516(7530):242–245. doi: 10.1038/nature13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9(1):22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Ku H-Y, Lin H. PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. Nat Sci Rev. 2014;1(2):205–218. doi: 10.1093/nsr/nwu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. One Loop to Rule Them All: The Ping-Pong Cycle and piRNA-Guided Silencing. Trends Biochem Sci. 2016;41(4):324–337. doi: 10.1016/j.tibs.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki YW, Siomi MC, Siomi H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu Rev Biochem. 2015;84:405–433. doi: 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]
- Theron E, Dennis C, Brasset E. et al. Distinct features of the piRNA pathway in somatic and germ cells: from piRNA cluster transcription to piRNA processing and amplification. Mob DNA. 2014;5(1):28. doi: 10.1186/s13100-014-0028-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Toth KF, Aravin AA. To be or not to be a piRNA: genomic origin and processing of piRNAs. Genome Biol. 2014;15(1):204. doi: 10.1186/gb4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A. et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128(6):1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Zanni V, Eymery A, Coiffet M. et al. Distribution, evolution, and diversity of retrotransposons at the flamenco locus reflect the regulatory properties of piRNA clusters. Proc Natl Acad Sci U S A. 2013;110(49):19842–19847. doi: 10.1073/pnas.1313677110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mével-Ninio M, Pelisson A, Kinder J. et al. The flamenco Locus Controls the gypsy and ZAM Retroviruses and Is Required for Drosophila Oogenesis. Genetics. 2007;175(4):1615–1624. doi: 10.1534/genetics.106.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goriaux C, Théron E, Brasset E. et al. History of the discovery of a master locus producing piRNAs: the flamenco/COM locus in Drosophila melanogaster. Front Genet. 2014:5. doi: 10.3389/fgene.2014.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalvet F, Teysset L, Terzian C. et al. Proviral amplification of the Gypsy endogenous retrovirus of Drosophila melanogaster involves env-independent invasion of the female germline. EMBO J. 1999;18(9):2659–2669. doi: 10.1093/emboj/18.9.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syomin BV, Fedorova LI, Surkov SA. et al. The endogenous Drosophila melanogaster retrovirus gypsy can propagate in Drosophila hydei cells. Mol Gen Genet. 2001;264(5):588–594. doi: 10.1007/s004380000344. [DOI] [PubMed] [Google Scholar]
- Engels WR. Hybrid dysgenesis in Drosophila melanogaster - Rules of inheritance of female sterility. Gen Res. 1979;33(3):219–236. doi: 10.1017/S0016672308009592. [DOI] [PubMed] [Google Scholar]
- Kidwell MG. Hybrid dysgenesis in Drosophila melanogaster - The genetics of cytotype determination in a neutral strain. Genetics. 1981;98(2):275–290. doi: 10.1093/genetics/98.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA. et al. An Epigenetic Role for Maternally Inherited piRNAs in Transposon Silencing. Science. 2008;322(5906):1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donertas D, Sienski G, Brennecke J. Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev. 2013;27(15):1693–1705. doi: 10.1101/gad.221150.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani H, Iwasaki YW, Shibuya A. et al. DmGTSF1 is necessary for Piwi-piRISC-mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev. 2013;27(15):1656–1661. doi: 10.1101/gad.221515.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Guzzardo PM, Gillis J. et al. A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila. Mol Cell. 2013;50(5):736–748. doi: 10.1016/j.molcel.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Batki J, Senti KA. et al. Silencio/CG9754 connects the Piwi-piRNA complex to the cellular heterochromatin machinery. Genes Dev. 2015;29(21):2258–2271. doi: 10.1101/gad.271908.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Gu J, Jin Y. et al. Panoramix enforces piRNA-dependent cotranscriptional silencing. Science. 2015;350(6258):339–342. doi: 10.1126/science.aab0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov NV, Aravin AA, Sachidanandam R. et al. The RNA interference system differently responds to the same mobile element in distant Drosophila species. Dokl Biochem Biophys. 2010;431:79–81. doi: 10.1134/s1607672910020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Marinov GK, Aravin AA. A transgenerational process defines piRNA biogenesis in Drosophila virilis. Cell Rep. 2014;8(6):1617–1623. doi: 10.1016/j.celrep.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin AA, Galdos MA, Wickersheim ML. et al. piRNAs Are Associated with Diverse Transgenerational Effects on Gene and Transposon Expression in a Hybrid Dysgenic Syndrome of D. virilis. PLoS Genet. 2015;11(8):e1005332. doi: 10.1371/journal.pgen.1005332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S, Ryazansky S, Olovnikov I. et al. Euchromatic transposon insertions trigger production of novel Pi- and endo-siRNAs at the target sites in the Drosophila germline. PLoS Genet. 2014;10(2):e1004138. doi: 10.1371/journal.pgen.1004138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC. The Role of piRNA-Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in Drosophila melanogaster. PLoS Genet. 2015;11(6):e1005269. doi: 10.1371/journal.pgen.1005269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC, Langley CH. Long-term and short-term evolutionary impacts of transposable elements on Drosophila. Genetics. 2012;192(4):1411–1432. doi: 10.1534/genetics.112.145714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D, Henikoff S, Malik HS. Positive selection drives the evolution of rhino, a member of the Heterochromatin Protein 1 family in Drosophila. PLoS Genet. 2005;1(1):96–108. doi: 10.1371/journal.pgen.0010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SH, Salmela H, Obbard DJ. Duplication and Diversification of Dipteran Argonaute Genes, and the Evolutionary Divergence of Piwi and Aubergine. Genome Biol Evol. 2016;8(3):507–518. doi: 10.1093/gbe/evw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger A, Ponting CP. Evolutionary rate analyses of orthologs and paralogs from 12 Drosophila genomes. Gen Res. 2007;17(12):1837–1849. doi: 10.1101/gr.6249707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Gordon KHJ, Buck AH. et al. The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci. 2009;364(1513):99–115. doi: 10.1098/rstb.2008.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Welch JJ, Kim K-W. et al. Quantifying Adaptive Evolution in the Drosophila Immune System. PLoS Genet. 2009;5(10):e1000698. doi: 10.1371/journal.pgen.1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski B, Hupalo DN, Kern AD. Recurrent Adaptation in RNA Interference Genes Across the Drosophila Phylogeny. Mol Biol Evol. 2010;28(2):1033–1042. doi: 10.1093/molbev/msq284. [DOI] [PubMed] [Google Scholar]
- Yi M, Chen F, Luo M. et al. Rapid evolution of piRNA pathway in the teleost fish: implication for an adaptation to transposon diversity. Genome Biol Evol. 2014;6(6):1393–1407. doi: 10.1093/gbe/evu105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin A, Wong A, Poh YP. et al. Recurrent and recent selective sweeps in the piRNA pathway. Evolution. 2013;67(4):1081–1090. doi: 10.1111/evo.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstiel JP, Hartl DL. Evidence for maternally transmitted small interfering RNA in the repression of transposition in Drosophila virilis. Proc Natl Acad Sci U S A. 2005;102(44):15965–15970. doi: 10.1073/pnas.0508192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Xie ZX, Allen E. et al. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev Cell. 2003;4(2):205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- Burgyan J, Havelda Z. Viral suppressors of RNA silencing. Trends Plant Sci. 2011;16(5):265–272. doi: 10.1016/j.tplants.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Guo W, Liew JY, Yuan YA. Structural insights into the arms race between host and virus along RNA silencing pathways in Arabidopsis thaliana. Biol Rev Camb Philos Soc. 2014;89(2):337–355. doi: 10.1111/brv.12057. [DOI] [PubMed] [Google Scholar]
- van Mierlo JT, Overheul GJ, Obadia B. et al. Novel Drosophila Viruses Encode Host-Specific Suppressors of RNAi. PLoS Pathog. 2014;10(7):e1004256. doi: 10.1371/journal.ppat.1004256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li WX, WeiDing S. Induction and Suppression of RNA silencing by an Animal Virus. Science. 2002;296:1319–1320. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- Chao JA, Lee JH, Chapados BR. et al. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol. 2005;12(11):952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- Murray GG, Kosakovsky Pond SL, Obbard DJ. Suppressors of RNAi from plant viruses are subject to episodic positive selection. Proc Biol Sci. 2013;280(1765):20130965. doi: 10.1098/rspb.2013.0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin SV, Mackay TFC. The genomic rate of transposable element movement in Drosophila melanogaster. Mol Biol Evol. 1995;12(1):180–181. doi: 10.1093/oxfordjournals.molbev.a040188. [DOI] [PubMed] [Google Scholar]
- Nuzhdin SV. Sure facts, speculations, and open questions about the evolution of transposable element copy number. Genetica. 1999;107(1-3):129–137. [PubMed] [Google Scholar]
- Morazzani EM, Wiley MR, Murreddu MG. et al. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 2012;8(1):e1002470. doi: 10.1371/journal.ppat.1002470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnettler E, Donald CL, Human S. et al. Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. J Gen Virol. 2013;94(Pt 7):1680–1689. doi: 10.1099/vir.0.053850-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit M, Mongelli V, Frangeul L. et al. piRNA pathway is not required for antiviral defense in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2016;113(29):E4218–E4227. doi: 10.1073/pnas.1607952113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Bor V, Hartswood E, Jones C. et al. gurken and the I factor retrotransposon RNAs share common localization signals and machinery. Dev Cell. 2005;9(1):51–62. doi: 10.1016/j.devcel.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Xi HL, Li CJ. et al. The Drosophila HP1 Homolog Rhino Is Required for Transposon Silencing and piRNA Production by Dual-Strand Clusters. Cell. 2009;138(6):1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Sienski G, Handler D. et al. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell. 2014;157(6):1364–1379. doi: 10.1016/j.cell.2014.04.031. [DOI] [PubMed] [Google Scholar]
- Chen YC, Stuwe E, Luo Y. et al. Cutoff Suppresses RNA Polymerase II Termination to Ensure Expression of piRNA Precursors. Mol Cell. 2016;63(1):97–109. doi: 10.1016/j.molcel.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels SB, Peterson KR, Strausbaugh LD. et al. Evidence for Horizontal Transmission of the P-Transposable Element between Drosophila Species. Genetics. 1990;124(2):339–355. doi: 10.1093/genetics/124.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolome C, Bello X, Maside X. Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol. 2009;10(2):R22. doi: 10.1186/gb-2009-10-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaack S, Gilbert C, Feschotte C. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. 2010;25(9):537–546. doi: 10.1016/j.tree.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels SB, Peterson KR, Strausbaugh LD. et al. Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics. 1990;124(2):339–355. doi: 10.1093/genetics/124.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth SB, Blumenstiel JP. Horizontal transfer can drive a greater transposable element in large populations. J Hered. 2016 doi: 10.1093/jhered/esw050. [DOI] [PubMed] [Google Scholar]
- Castillo DM, Mell JC, Box KS. et al. Molecular evolution under increasing transposable element burden in Drosophila: a speed limit on the evolutionary arms race. BMC Evol Biol. 2011;11:258. doi: 10.1186/1471-2148-11-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quax TE, Claassens NJ, Soll D. et al. Codon Bias as a Means to Fine-Tune Gene Expression. Mol Cell. 2015;59(2):149–161. doi: 10.1016/j.molcel.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Gilchrist MA. Explaining complex codon usage patterns with selection for translational efficiency, mutation bias, and genetic drift. Proc Natl Acad Sci U S A. 2011;108(25):10231–10236. doi: 10.1073/pnas.1016719108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS. Epigenetic silencing of transposable elements: A trade-off between reduced transposition and deleterious effects on neighboring gene expression. Gen Res. 2009;19(8):1419–1428. doi: 10.1101/gr.091678.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vanssay A, Bouge AL, Boivin A. et al. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature. 2012;490(7418):112–115. doi: 10.1038/nature11416. [DOI] [PubMed] [Google Scholar]
- Ronsseray S, Lemaitre B, Coen D. Maternal inheritance of P-cytotype in Drosophila melanogaster - A pre-P cytotype is strictly extra-chromosomally transmitted. Mol Gen Genet. 1993;241(1-2):115–123. doi: 10.1007/BF00280208. [DOI] [PubMed] [Google Scholar]
- Stanley CE Jr., Kulathinal RJ. flyDIVaS: A Comparative Genomics Resource for Drosophila Divergence and Selection. G3 (Bethesda) 2016;6(8):2355–2363. doi: 10.1534/g3.116.031138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang J, Schultz N. et al. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell. 2014;157(6):1353–1363. doi: 10.1016/j.cell.2014.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Edelman NB, Barbash DA. Drosophila interspecific hybrids phenocopy piRNA-pathway mutants. PLoS Biol. 2012;10(11):e1001428. doi: 10.1371/journal.pbio.1001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe NW, Clark NL. ERC analysis: web-based inference of gene function via evolutionary rate covariation. Bioinformatics. 2015;31(23):3835–3837. doi: 10.1093/bioinformatics/btv454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NL, Alani E, Aquadro CF. Evolutionary rate covariation reveals shared functionality and coexpression of genes. Genome Res. 2012;22(4):714–720. doi: 10.1101/gr.132647.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GD, Sitnik JL, Wang W. et al. Evolutionary rate covariation identifies new members of a protein network required for Drosophila melanogaster female post-mating responses. PLoS Genet. 2014;10(1):e1004108. doi: 10.1371/journal.pgen.1004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post C, Clark JP, Sytnikova YA. et al. The capacity of target silencing by Drosophila PIWI and piRNAs. RNA. 2014;20(12):1977–1986. doi: 10.1261/rna.046300.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Liu J, Schnakenberg SL. et al. Variation in piRNA and transposable element content in strains of Drosophila melanogaster. Genome Biol Evol. 2014;6(10):2786–2798. doi: 10.1093/gbe/evu217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D. et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31(6):785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, He X, Wang X-J. et al. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443(7114):1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- Lewsey MG, Hardcastle TJ, Melnyk CW. et al. Mobile small RNAs regulate genome-wide DNA methylation. Proc Natl Acad Sci U S A. 2016;113(6):E801–E810. doi: 10.1073/pnas.1515072113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuthikattu S, McCue AD, Panda K. et al. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 2013;162(1):116–131. doi: 10.1104/pp.113.216481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirn G-W, Rahman R, Sytnikova YA. et al. Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals. PLoS Genet. 2015;11(11):e1005652. doi: 10.1371/journal.pgen.1005652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arensburger P, Hice RH, Wright JA. et al. The mosquito Aedes aegypti has a large genome size and high transposable element load but contains a low proportion of transposon-specific piRNAs. BMC Genomics. 2011;12(1):1–23. doi: 10.1186/1471-2164-12-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George P, Jensen S, Pogorelcnik R. et al. Increased production of piRNAs from euchromatic clusters and genes in Anopheles gambiae compared with Drosophila melanogaster. Epigenetics Chromatin. 2015;8(1):1–21. doi: 10.1186/s13072-015-0041-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninova M, Ronshaugen M, Griffiths-Jones S. MicroRNA evolution, expression, and function during short germband development in Tribolium castaneum. Genome Res. 2016;26(1):85–96. doi: 10.1101/gr.193367.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov NV, Aravin AA, Zelentsova ES. et al. Small RNA-based silencing strategies for transposons in the process of invading Drosophila species. RNA. 2010;16(8):1634–1645. doi: 10.1261/rna.2217810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Liu J, Schnakenberg SL. et al. Variation in piRNA and Transposable Element Content in Strains of Drosophila melanogaster. Genome Biol Evol. 2014;6(10):2786–2798. doi: 10.1093/gbe/evu217. [DOI] [PMC free article] [PubMed] [Google Scholar]