Abstract
Many thousands of Circular RNAs (circRNAs) have recently been identified in metazoan genomes by transcriptome-wide sequencing. Most circRNAs are generated by back-splicing events from exons of protein-coding genes. A great deal of progress has recently been made in understanding the genome-wide expression patterns, biogenesis, and regulation of circRNAs. To date, however, few functions of circRNAs have been identified. CircRNAs are preferentially expressed in neural tissues and some are found at synapses, suggesting possible functions in the nervous system. Several circRNAs have been shown to function as microRNA “sponges” to counteract microRNA mediated repression of mRNA. New functions for circRNAs are arising, including protein sequestration, transcriptional regulation, and potential functions in cancer. Here, we highlight the recent progress made in understanding the biogenesis and regulation of circRNAs, discuss newly uncovered circRNA functions, and explain the methodological approaches that could reveal more exciting and unexpected roles for these RNAs.
Keywords: circRNAs, RNA-seq, transcriptome, non-coding RNA, alternative splicing, aging, EIciRNAs, ciRNAs, microRNA, ceRNA
Introduction
Individual examples of circular RNAs (circRNAs) have been sparsely described in the literature starting in the early 1990s [1-3]. Long written off as low abundance splicing errors with no function, circRNAs have recently been thrust into the spotlight as a newly appreciated class of non-coding RNAs. This reemergence sprung from new RNA-seq technology and analysis methods that have revealed pervasive, and in some cases abundant expression of circRNAs in various organisms, including plants, Caenorhabditis elegans, Drosophila melanogaster, mice and humans [4-7].
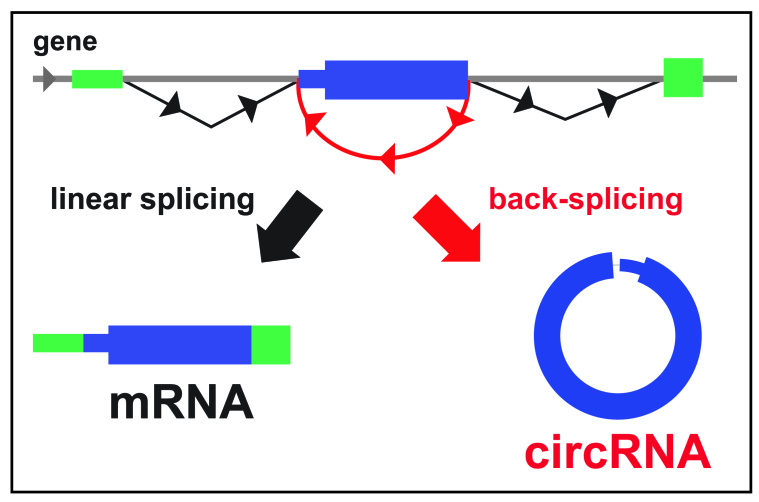
Today, the term “circRNA” is commonly applied to describe exonic circRNAs that arise from direct back-splicing events that covalently link the 3´ end of an exon with the 5´ end of either the same exon, or a further upstream exon (Figure 1). This generates a single or multi-exon RNA that usually has intronic sequence spliced out. These exonic circRNAs are distinct from intron lariat precursor RNAs (intronic circRNAs) that are also circular in form [8]. Other species of RNA in circular form that have long been appreciated, include single stranded circular RNA viruses [9] and plant viroids [10]. Due to the lack of free ends, circRNAs are not capped, and thus are not predicted to be translated by cap-dependent mechanisms. Thus, they are classified as non-coding RNAs. Another consequence of the lack of free ends is that circRNAs avoid degradation by exonucleases and thus have enhanced stability compared to linear RNAs [5,11].
Figure 1.
CircRNA biogenesis. In-order splicing occurs to produce linear mRNAs with the introns removed. CircRNAs are most commonly produced from back-splicing events, usually from exons of protein-coding genes. Shown is an example of a single exon circRNA. Note that circRNAs can contain multiple exons and the intervening intronic sequence in multi-exon circRNAs is usually removed.
As is the case in general with non-coding RNAs, the big question to answer is, “what do they do?” The first examples of functional circRNAs were found to act as microRNA sponges in 2013 [5,12]. Now, only a few years later, additional functions of circRNAs are being uncovered. Information on the mechanism of circRNA biogenesis, regulation and expression patterns are implicating them in multiple aspects of biology and disease, including potential roles in cancer, heart disease, synaptic transmission, and aging.
Properties of CircRNAs
Methods to Detect CircRNAs
RNA-seq, a high-throughput sequencing technology for sequencing RNA, is a widely used technology to annotate new RNA species and quantify RNA abundance [13]. Continually advancing progress in high-throughput sequencing and quantification methods has led to the discovery of a plethora of diverse coding and non-coding RNA species [14,15]. Until recently, RNA-seq methods to detect long RNAs were confined to the polyadenylated fraction of the transcriptome, which is dominated by messenger RNAs (mRNAs). This approach, which uses an oligodT primer for reverse transcription, is used to enrich for mRNAs, but most importantly, to avoid ribosomal RNA, which is highly abundant and constitutes > 90 percent of total RNA in eukaryotic cells. More recent library cloning methods have been established that employ effective ribosomal RNA depletion and random priming for cDNA synthesis (ribo-depleted total RNA-seq) [16]. The resulting library thus includes large RNAs that are not polyadenylated. It is through analysis of such libraries using custom mapping tools that the compendium of circRNAs in the genomes of diverse species has been revealed [17].
Back-splicing is much less favorable than linear splicing [11], causing most circRNAs to be of low abundance in cells. Thus, the detection and quantification of circRNAs by ribo-depleted total RNA-seq requires a much greater depth of sequencing than what would be required to quantify the protein-coding transcriptome. The simplest method for quantification counts split back-spliced reads, and does not include reads that align directly to exons as these could emanate from circRNA or mRNA (Figure 2) [5,7,18,19]. Multiple algorithms have been devised to annotate and quantify circRNAs from RNA-seq data. These can yield quite different results with regard to detection and quantification, and thus many groups are working toward improved bioinformatics pipelines [17,19,20]. Split, back-spliced reads tend to constitute less than 1 percent of the reads generated from a total RNA-seq experiment, whereas the majority of reads align to expressed mRNAs [20,21]. Because most published and publically available RNA-seq datasets are derived from polyA+ selected RNA or lack sufficient depth, they are unsuitable for circRNA profiling.
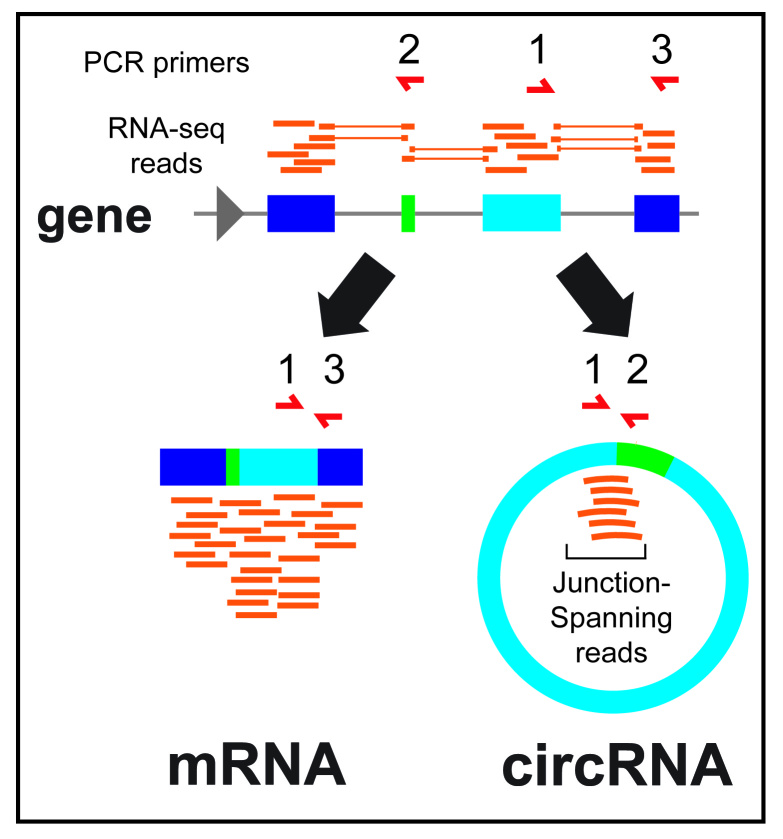
Figure 2.
Quantification of circRNA expression. RNA-seq reads (orange) can align directly to the genome or be split, indicating a spliced junction. Back-spliced split reads that do not align in linear fashion are consistent with detection of a circRNA. PCR primers (red) can be used to detect linear RNAs (inward facing orientation), or circRNAs (outward facing orientation) from cDNA preparations.
CircRNAs can also be detected using microarrays that employ probes spanning back-spliced junctions. Several studies have employed this approach, although it is not clear to what degree these probes also might erroneously detect linear species [22-25]. Individual circRNAs can be distinguished from their linear counterparts by RT-PCR using “outward facing” primer sets (Figure 2) and Northern blotting [7,26,27]. RNase R, a 3´ to 5´ exoribonuclease that preferentially degrades linear over circular RNAs, has been used to validate whether a detected RNA is in fact circular, and to enrich for circRNAs from a total RNA pool [21-28]. However, there is debate as to the efficacy of RNase R as an enrichment method for circRNAs, since some linear RNAs are resistant to RNase R [29], different circRNAs can show drastically different levels of RNase R resistance [30], and RNase R can lead to an artifactual enrichment of circRNA levels, potentially due to interference with reverse transcription reactions [21]. One of the best characterized circRNAs, CDR1-as, is sensitive to RNaseR treatment [28].
Genomic Properties of CircRNAs
CircRNAs have been annotated in a diverse set of species, tissues and cell types. Several general features have arisen from these studies. Perhaps the most surprising is the breadth and diversity of circRNA molecules. CircRNAs can contain one to many exons, and it is common that multiple circRNAs can be produced from a single gene [5,28]. For instance, 18 circRNAs are generated from the pangolin gene in Drosophila [7]. For multiexon circRNAs, the vast majority lack retained introns [5,7]. There is, however, a subclass of nuclear retained circRNAs called Exon-Intron circRNAs (EIciRNAs) that retain introns [31]. In addition, intronic lariats that are normally a degraded byproduct of splicing generate intronic circular RNAs [8].
What genomic features determine circRNA biogenesis? Early work noted that back spliced exons are followed by unusually large introns [2], and in Drosophila, flanking intron length is highly predictive of whether the contained exons will circularize [7]. It is possible that overall intron length influences back-splicing dynamics, and also that the increased intronic landscape provides more binding sites for splicing factors. Complementary Alu elements in introns flanking back-spliced exons have been uncovered as critical factors for RNA circularization in mouse and human [28,32]. These elements on either side of the circularizing exon(s) base pair together to bring splice acceptor and donor sites in close proximity. It is estimated that ~80 percent of human circRNA loci are flanked by Alu elements [32]. Several studies have shown that the entire sequence of the Alu element is not necessary but only a highly complementary region along with the canonical splicing elements [27]. This idea has been successfully tested for some human circles such as the ZKSCAN1 circRNA using mini-gene constructs with small flanking introns [33]. Flanking, intronic short repeats, including Alu elements, were also found to play a role in Drosophila circRNA biogenesis [33]. Alu repeats are only found in a subset of vertebrates, thus they do not explain back-splicing in all organisms. However, in C. elegans, which lack Alu repeats, it was found that reverse complementary matches (RCMs) promote circularization. These RCMs were found to be more predictive of circularization than intron length [32].
Abundance of CircRNAs
In most organisms, the number of individual circRNAs is greater than the count of protein coding genes. Recent work on circRNA annotation from various brain regions and neural cells detected 15,849 distinct circRNAs in mouse, and 65,731 in human [26]. Note that the differences in the number of circRNAs in human versus mouse might be attributed to different read depth among experiments. CircRNAs are expressed in all eukaryotes tested [34] and, in addition to mice and human, detailed annotations are available for Drosophila (2,513 circRNAs), and C. elegans (1,111 circRNAs) [5,7]. These model organisms are advantageous systems for studying the regulation and function of circRNAs.
As sequencing methods improve with greater depth, accuracy, and read length [35], additional circRNAs will undoubtedly be uncovered. These annotations of circRNAs includes extremely low abundance species, which one might assume to be by-products of splicing as opposed to functional RNAs. Annotation of as little as two unique back-splice junction spanning reads have been used as a minimum cutoff for annotation [5,18], whereas other groups have been more conservative, requiring a 10 read cutoff [7], or only annotating a circRNA if it constitutes > 10 percent of the fraction of transcript isoforms from a given gene [20].
Not all circRNAs are of low abundance. For some genes the major product is not a protein coding mRNA, but instead is a circRNA. This is the case for hundreds of genes found in human cell lines [6]. In Drosophila, a single circRNA is the major product of the mbl gene [7]. In another study of cell line derived circRNAs, 57 circRNAs were found to constitute more than half of transcript isoforms from their parent gene, including CDR1-as [20]. In brain tissues, circRNAs were found to be the major isoform of multiple genes, including Rims2, Tulp4 and Elf2 [26]. Interrogation of functions is likely to focus on the most highly expressed circRNAs and ones that are regulated under particular cell conditions.
Regulation of CircRNA Abundance
Neural Determinants of CircRNA Abundance
Profiling of circRNAs among diverse tissues and developmental timepoints in Drosophila revealed that the nervous system is enriched for circRNAs compared to other tissues. Of the ~2500 circRNAs annotated in Drosophila, > 90 percent were detected in the head [7]. Moreover, the categories of genes from which circRNAs were expressed were enriched for nervous system functions as determined by gene ontology analysis [7,18]. This suggests that circRNAs might have neural functions in Drosophila.
Importantly, this neural enrichment trend also applies to mammals. Profiling of multiple tissues in mouse has revealed that brain tissue expresses the most circRNAs compared to other tissues. Of five tissues profiled in mice (brain, heart, liver, lung, testis) for circRNA expression, occurrence of back-splicing, and number of tissue specific circRNAs, brain ranked first [36]. Interestingly, testis was the tissue that ranked second. In human, RNA-seq profiling of cortex, muscle, thyroid and liver revealed that cortex contains the most back-spliced reads [26]. Other studies have found similar trends of a bias for brain expressed circRNAs over other tissues in mice and human [11,19]. Thus, this enrichment pattern appears to be a phenomenon conserved in evolution.
Is the enrichment of circRNAs detected in brain tissue due to neural-specific expression? RNA profiling of cells during neural differentiation revealed strong patterns for increased circRNAs in neural tissue, as assayed in P19 and SH-SY5Y cells undergoing neural differentiation, and in primary cortical neurons in early versus late stages of differentiation [26]. This suggests that factors expressed in neurons might be directly enhancing circRNA biogenesis and stability. Slow cell-turnover and limited neurogenesis are thought to contribute to the stability of circRNAs in the brain, and may explain why circRNAs are less abundant in other tissues like the liver and lung, which have a higher capacity for regeneration.
Some circRNAs in neurons appear to be enriched at synapses. RNA sequencing of synaptosomes and microdissected synaptic neuropil have revealed enhanced expression of circRNAs [26,36]. Interestingly, some genes were found to express circRNAs preferentially at synapses, whereas mRNAs from the same genes were found to be primarily cytoplasmic [26]. This suggests that circRNAs might bind to proteins and molecular motors that direct them to synapses. CircRNAs also appear to be regulated by neuronal activity. Cultured primary hippocampal neurons treated with bicuculline (an antagonist to inhibitory GABAA receptors) to induce homeostatic plasticity caused upregulation of 37 circRNAs whereas only five were downregulated [36]. What could be the function of circRNAs at synapses is an open question. Roles as scaffolds for RNA-protein complex assembly have been suggested [26].
In addition to the synaptic localization of circRNAs, several circRNAs have been detected by in situ hybridization in dendrites [36]. Global RNA profiling of neuronal compartments, including soma, axons, and dendrites [37,38] could reveal whether this is a general property of neuronal circRNAs. Localized translation can occur in axons and dendrites [39], thus circRNAs might be acting as scaffolds to assemble translational machinery and RNA-binding proteins.
Conditions/stresses Regulating CircRNA Abundance
Multiple recent studies have uncovered cellular conditions and stresses that can modulate circRNA expression levels. Specific regulation of circRNAs was first suggested by discordant changes in circRNA versus mRNA abundance among multiple cell lines on a genome-wide level [21]. These patterns provide clues that can help identify specific factors involved in circRNA biogenesis and regulation.
A key factor contributing to the levels of circRNAs in cells and tissues is circRNA stability and the proliferative status of the cells in which they are expressed. If one assumes a steady transcription rate for a gene, where mRNA products are continually degraded over time and circRNA products persist, then the ratio of circRNA to mRNA is expected to increase as time passes. In proliferative cells, this increased circRNA/mRNA ratio is diluted when cell division takes place and when cells die. In contrast, terminally differentiated cells that rarely turnover would be expected to have progressively increased levels of circRNAs.
In line with this hypothesis, proliferating cells, including cancer cells, express lower levels of circRNAs compared to terminally differentiated cells on a genome-wide level [30]. Further supporting this assertion, circRNAs are less abundant in human gliomas compared to healthy brain sample controls [40]. This expression trend has been studied in detail for a circRNA from the Foxo3 locus. Cancer cells show reduced expression of circ-Foxo3 compared to non-transformed cell lines, and the inverse pattern is found for Foxo3 mRNA [41]. In addition, Epidermal Growth Factor (EGF) treatment decreases circ-Foxo3, whereas treatment with an EGF inhibitor increases circ-Foxo3. CircRNAs were found to be regulated by Epithelial-mesenchymal transition (EMT) [42]. EMT is critical for morphogenesis, and understanding the mechanisms behind EMT has important relevance to metastasis of epithelial tumors [43]. In a model of EMT involving TGF-β treatment of human mammary epithelial cells, a strong trend for circRNA upregulation independent of mRNA expression from the same genes was identified [42]. This suggests that circRNAs might have functions related to the mesenchymal phenotype.
CircRNAs are dynamically expressed during development. Many circRNAs from maternal genes were found to be expressed prior to fertilization and were detected throughout human pre-implantation development [44]. This persistent expression of maternally derived circRNAs could be attributed to evasion of maternal degradation pathways due to enhanced stability. Multiple studies have found a general trend for increased circRNA levels during development. Ribo-depleted RNA-seq data from human fetal tissues ranging from 10 to 21 weeks of age revealed an upregulation trend for circRNAs in heart and lung tissues [19]. A global trend for upregulation of circRNAs was also uncovered during development of mouse brain between embryonic day 18 and postnatal day 30 [36]. Confirmation of individual circRNAs by RT-qPCR revealed that the upregulation is unique to the circRNAs and not reflective of increased transcription of the host genes. Upregulation of circRNAs was also evident during embryonic development in Drosophila [7]. It remains to be determined what specific factors underlie the increased circRNA levels during development in multiple tissues and species. Various interpretations have been proposed, including shifts in cell populations during these developmental timepoints, development of synapses, and increases in the population of post-mitotic cells.
CircRNAs appear to be regulated in response to aging. In a study that profiled circRNAs from comprehensive ribo-depleted total RNA datasets of various Drosophila tissues, cell types, and developmental timepoints, thousands of circRNAs were identified [7]. These samples included adult samples of various ages. Unexpectedly, a strong trend for upregulation of circRNAs during aging was uncovered. In Drosophila head samples, over 250 circRNAs were significantly upregulated greater than 2-fold during aging (comparing 1 day versus 20 day old flies). It will be interesting to determine if this trend holds in other organisms and whether it occurs in other tissues as well. It is possible that the increase of circRNAs concomitant with aging is due to intrinsic circRNA stability and the post-mitotic status of neurons, but this remains to be directly demonstrated. It is also possible that age-regulated trans-factors influencing circRNA stability or biogenesis might also play a role in this phenomena.
Additional stress events such as high versus low temperature, and oxidative stress can regulate circRNAs [18,45]. In summary, these emerging studies show that circRNA expression levels are highly regulated in different cellular contexts, in many cases independently of linear transcripts produced from their host gene. Undoubtedly, we are only beginning to uncover cellular conditions, stresses, physiological and pathological conditions that influence circRNA abundance.
Factors Regulating CircRNA Biogenesis
Multiple factors have been identified to regulate circRNA biogenesis. Here, we focus discussion on factors and mechanisms regulating the biogenesis of exonic circRNAs via direct back-splicing. There is an alternative to the direct back-splicing model to generate exonic circRNAs. The “exon-skipping” or “lariat-precursor” model has been found to regulate production of some exonic circRNAs [46,47], although from genome-wide studies this appears to not constitute a major mechanism of circRNA formation [6,28]. Stable intronic lariats are another class of circular RNAs identified genome-wide [8] that have unique mechanisms regulating their abundance [48,49].
Back-splicing appears to utilize the same canonical splicesomal machinery employed in linear splicing [18,27]. Thus, it is likely that many factors and mechanisms already known to regulate alternative splicing [50] might also influence circRNA biogenesis. Along these lines, it was found that multiple SR proteins (SR2, SRp54, B52) and an hnRNP protein (Hrb27c) suppress biogenesis of several Drosophila circRNAs in a combinatorial manner [51].
The regulation of circRNA abundance among cell types, tissues, and in response to stress suggests that there are tissue-specific trans-factors that can regulate circRNA biogenesis or stability. RNA binding proteins (RBPs) are the most likely candidates. In one case, the observation of circRNA regulation in response to a cellular stress led to the discovery of a novel circRNA biogenesis suppressing factor [42]. In the previously discussed study that found circRNA abundance increased during EMT, the authors also examined how RBPs were regulated by EMT. Each RBP that increased or decreased more than 2-fold was examined in a targeted screen for regulation of circRNA biogenesis using a dual-fluorescence reporter system for a circRNA from the SMARCA5 gene. Strikingly, out of 20 RBPs screened, knockdown of only one decreased circRNA biogenesis- the RBP Quaking (QKI). QKI is a member of the STAR family of KH domain RNA binding proteins. QKI has described roles in multiple steps of RNA processing, including splicing and translation [52,53]. Knockdown of QKI reduced many circRNAs, and those circRNAs reduced showed a bias for containing QKI target sites in their flanking introns. Mutation of QKI binding sites reduced circRNA biogenesis, and most convincingly, the insertion of QKI binding sites in introns flanking exons that do not normally circularize caused back-splicing to occur [42].
Although targeted screens or full genome-wide screens provide an unbiased approach to identifying new circRNA regulating factors, to date most have been identified through hypothesis-based tests. As previously mentioned, evidence for base-pairing between flanking introns is a strong indication of exon circularization [32,33]. Double-stranded RNA regions are targeted by the RNA editing enzyme ADAR1, and significant adenosine to inosine editing was found in introns flanking circularized exons [32]. It was thus a reasonable hypothesis to test if ADAR1 influences circularization. Knockdown of ADAR1 and ADAR2 by RNAi in HEK293 cells led to 84 circRNAs upregulated more than 2-fold. A model was devised in which ADAR1 binds to stem structures of these reverse complementary intron pairs, and “melts” or disrupts the pairing to suppress the formation of circRNAs [32].
Muscleblind (MBL) is another recently uncovered regulator of circRNA biogenesis [18]. The mbl gene expresses the most abundantly expressed circRNA in Drosophila heads, emanating from the second exon and flanked by large introns [7]. The mammalian homologues of MBL are well studied RBPs with roles in various steps of RNA processing, including RNA localization, alternative polyadenylation, and alternative splicing [6,54,55]. Target sites for MBL binding were identified in the introns flanking exon 2 of mbl. This presented the intriguing possibility that MBL can regulate circularization from its own gene. An elegant feedback system was uncovered in which MBL binds to the introns flanking exon 2 of mbl to enhance circRNA production. This back-splicing competes with linear splicing; thus, MBL downregulates its own expression by producing circRNA in expense of mRNA from the mbl gene. This is accomplished largely by MBL protein binding to introns, but some additional evidence suggested that the circRNA could also modulate MBL protein by directly binding it [18]. It remains to be determined if MBL regulation of circularization occurs on a genome-wide scale (several Drosophila circRNAs were found in another study to not be regulated by MBL [51]). Perhaps additional RBPs autoregulate their expression levels via enhanced back-splicing to produce circRNAs from their host genes.
Alternative splicing is known to be regulated by the kinetic rate of RNA pol II, which is influenced by events such as phosphorylation targeting the pol II carboxy-terminal domain [56]. It was found that Drosophila bearing the pol II C4 mutation, previously shown to slow elongation rate [57], expressed less circRNA overall than control flies [18]. Thus, pol II elongation rate is positively correlated with circRNA production. Studies in cultured mouse cells support these findings: transcription elongation rate was found to be faster for circRNA producing genes, and expression profiling of nascent circRNAs from cells expressing either fast or slow pol II mutants strongly revealed a positive correlation between transcription elongation rate and circRNA biogenesis [11]. Although transcription elongation influenced circRNA biogenesis suggesting co-transcriptional biogenesis of circRNAs, the majority of back-splicing was found to occur post-transcriptionally [11]. Post-transcriptional biogenesis of circRNAs is also supported by a study that found 3´ end processing signals on pre-mRNA were required to generate the ZKSCAN1 circRNA [33].
The knowledge gained in understanding the regulation of circRNA expression points the way toward what functions circRNAs might have in cells. Next, we discuss functions of circRNAs that have recently been uncovered.
Arising Functions of CircRNAs
Methods to Interrogate CircRNA Functions
New approaches to overexpress and knock-down circRNAs have been recently developed (Figure 3). One of the challenges for circRNA overexpression has been that vectors that overexpress circRNA also overexpress linear RNA, making it difficult to determine if a circular species, per se, is responsible for a given phenotype. Newly developed expression vectors that drive almost exclusively circular and not linear exons [51] by employing flanking introns with base-pairing repeats will be useful for identifying new circRNA functions (Figure 3A).
Figure 3.
Methods to manipulate circRNA expression. A) Expression constructs are available that include intronic elements flanking the laccase2 or ZKSCAN1 circRNAs [51]. These express circularized over linear exons almost exclusively. Overexpression and knockdown approaches to study circRNA function must be designed to specifically alter circRNAs, and not linear RNAs from the same gene. B) siRNA strategies to knockdown circRNAs must have the siRNA designed to specifically target the back-spliced junction. C) Deletion of one pair of a flanking intronic reverse complementary matches can disrupt circularization without impeding linear mRNA [11].
Experimentally suppressing circRNA function has particular technical hurdles to overcome. The challenge is to disrupt circRNA expression without affecting linear protein coding RNAs generated from the same locus. One method that has been commonly applied is siRNA knockdown of circRNAs (Figure 3B). In this case the siRNA is designed to specifically span the back-spliced junction so that the protein coding mRNA is unaffected. However, this severely hampers design of siRNA, and makes targeting in some cases impossible. Off-targeting of siRNAs is a common problem which is usually accounted for by performing experiments with multiple unique siRNAs. Since it is usually not possible to design multiple siRNAs to a circRNA junction, one must be cautious in interpreting the results of circRNA knockdown studies.
The pioneering work that uncovered the key importance of complementary pairing of intronic elements to circRNA generation [28,32] has formed the basis for new methods to disrupt or ectopically express circRNAs. It was recently demonstrated that disrupting one intronic complementary sequence using CRISPR abolished the expression of circGCN1L1 from its endogenous locus [11]. Although this was demonstrated in cultured cells, the strategy should work in vivo as CRISPR is now routinely used to create deletion mutants in model organisms. The strategy could thus be widely employed to create circRNA knockout animals (Figure 3C).
Competition with Linear Splicing
Although most researchers are focused on identifying circRNA trans-functions, circRNA biogenesis itself might have functions in the cell. Back-splicing of exons generally occurs in expense of linear splicing to produce mRNAs. Thus, the production of back-spliced circRNAs has a cis function--the downregulation of protein coding mRNAs. Given the sizeable fraction of genes that generate circRNAs, this has been suggested to constitute a major gene regulatory mechanism. Exons that generate circRNAs are seldom removed by alternative splicing mechanisms [6,28], thus the decision to back-splice or splice linearly serves as a switch point that typically cannot be bypassed.
As previously discussed, back-splicing of mbl exon 2 is directed by the MBL protein, forming an autoregulatory loop [18]. Previous studies have established that alternative splicing can be used to generate mRNA isoforms that do not encode proteins as a way to regulate gene expression. This non-productive splicing can result in RNAs that are targeted for degradation by the nonsense-mediated decay (NMD) pathway. This has previously been termed Regulated Unproductive Splicing and Translation (RUST), and occurs for thousands of genes [58,59]. In some cases, RUST can be autoregulated. For instance, alternative splicing to include exon2 in the tra2β gene leads to a non-translatable protein, which is regulated by TRA2β protein, thus forming an autoregulatory loop [60].
Future work will reveal whether the mbl circRNA autoregulatory loop is particularly selected for to generate circRNA with trans function, or whether the major consequence is tailoring MBL protein levels in the cell. Target sites for MBL are found in the circularized mbl exon. It is enticing to speculate that in addition to being produced in expense of linear protein coding mbl transcripts, the mbl circRNA might itself sequester, or “sponge” MBL protein [18].
MicroRNA Sponges
MicroRNAs are a class of small RNAs that negatively regulate gene expression of mRNAs. This occurs most commonly through interaction with target sites in mRNA 3´ UTRs that leads to deadenylation, decreased mRNA stability and translation suppression [61]. Multiple lines of evidence have shown that other RNAs with microRNA target sites can compete with mRNAs for microRNA binding [62]. These competitive endogenous RNAs (ceRNAs), however, are themselves targets of destabilization and degradation by microRNAs. In contrast, circRNAs have no free ends, and thus would be predicted to evade microRNA-mediated deadenylation (however, see [63]).
The strongest evidence for a functional circRNA sponge comes from studies on the CDR1-as circRNA (also known as cIRS-7) [5,12] (Figure 4F). In contrast to most circRNAs, CDR1-as is expressed antisense to a protein-coding gene in mammals. Remarkably, the CDR1-as locus harbors > 70 highly conserved target sites for the neural microRNA miR-7. Convincing evidence that CDR1-as acts as a sponge in vivo comes from studies where it was ectopically expressed in zebrafish (which do not express CDR1-as) [5]. This remarkably resulted in a decrease in midbrain size, that was shown to be partially rescued by overexpressing miR-7. This in vivo evidence for a circRNA function thrust circRNAs into the spotlight and has spurred a slew of studies searching for functional circRNA sponges.
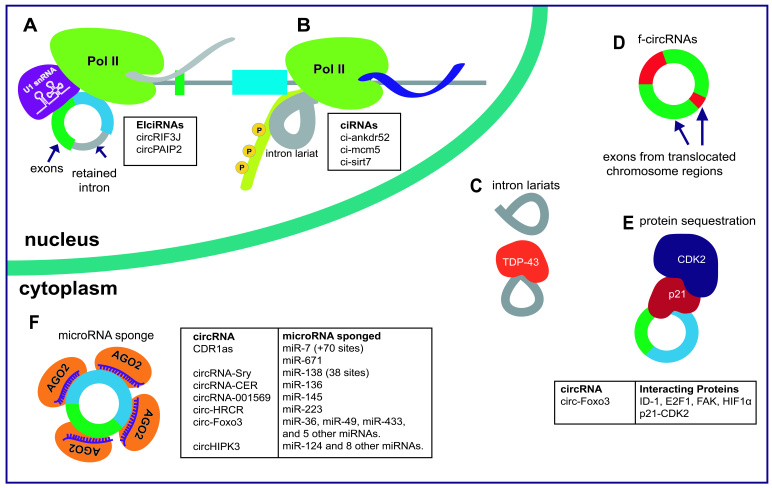
Figure 4.
CircRNA localization and molecular functions. A) EIciRNAs retain introns and seem to interact with U1 snRNA and Pol II, acting as regulators of their parental genes’ expression [31]. B) Circular intronic RNAs (ciRNAs) have a characteristic 7nt GU-rich element near the 5’ splice site and an 11nt C-rich element close to the branch point, and interact with phosphorylated Pol II regulating parental gene mRNA expression [8]. C) Not all the circRNAs generated from intronic lariats are in the nucleus. In Xenopus tropicalis oocytes circular lariats are abundant in the cytoplasm [71]. Lariat species can interact with proteins such as TDP-43 [48]. D) f-circRNAs arise from chromosomal translocated regions. They stimulate proliferation and contribute to cellular transformation and tumorigenesis [70]. E) Recent reports refer to the interaction of circ-Foxo3 with several proteins to regulate senescence [45] and cell cycle progression [41]. F) Multiple circRNAs have been shown to have activity as microRNA sponges, most notably CDR1-as [5,12].
Could microRNA sponge function be a general feature of circRNAs? Bioinformatics studies have led to mixed support for this idea. One study found conservation in the third nucleotide position codons of circularizing exons [5], suggesting these sequences might be conserved for reasons other than protein coding potential. Another found decreased single nucleotide polymorphisms in microRNA target sites of circularized exons, showing they are under similar selective pressure as microRNA target sites within 3´ UTRs [64]. However, another study failed to observe enhanced conservation of microRNA target sites in circRNA exons versus control protein coding exons [20]. Although preferred conservation of 7-mer sequences has been found for circRNAs in another study, enhanced conservation of specifically microRNA target sites was not [26]. If sponging is an important function for circRNAs, one would expect that enrichment of microRNA target sites would be found for circRNAs regulated by cellular stresses, cell differentiation or development. However, Conn et al. described that circRNAs enhanced during EMT do not show enrichment of circRNAs with reiterated microRNA target sites [42]. Another argument against microRNA sponging being a major function of circRNAs is that their low abundance makes them unsuitable to effectively sponge microRNAs or other proteins, with perhaps only a few exceptions [20].
Despite these perhaps disappointing genome-wide trends, evidence for functional circRNA sponges is emerging. In vivo evidence for a circRNA sponge with relevance to heart failure has recently been uncovered. This circRNA, named HRCR (mm9-circ-012559), was found to harbor six predicted target sites for miR-223. Mice expressing a miR-223 transgene exhibited severe cardiac hypertrophy and HRCR was found to be downregulated in failing mouse hearts [65]. The authors hypothesized that HRCR might act as a sponge for miR-223. To test this, a construct predicted to induce circularization of the HRCR exon was overexpressed in cardiomyocytes in vivo. Remarkably, this mitigated isoproterenol-induced cardiac hypertrophy. Overexpression of HRCR also increased levels of the ARC protein, which is strongly regulated in vivo by miR-223 through interaction with target sites in the Arc 3´ UTR. Overexpression of HRCR might prove to be an attractive therapeutic avenue for treating heart failure. Although perhaps not critically relevant to its ability as a potential therapeutic, from this study it is not clear what amount of linear versus circular RNA was produced from the overexpression constructs. Determination of the importance of HRCR as an endogenous microRNA sponge in the heart awaits additional circRNA-specific loss of function evidence for its protective role against heart failure.
Several circRNAs have been found to sponge microRNAs and enhance cell growth, implicating a role for circRNAs in cancer. Recently, a circRNA from the HIPK3 gene (circHIPK3) was found to sponge multiple circRNAs, including miR-124. Remarkably, siRNA knockdown of circ-HIPK3 reduced proliferation of cultured HEK-293T cells [66]. Thus, a mechanism was proposed in which circHIPK3 sponges miR-124 to enhance proliferation. Other studies have linked circRNA sponge activity to cell growth. CircRNAs that promote cell survival have purported roles in cancer including circRNA-CER, which sponges miR-136 [67], and circRNA_001569, which sponges miR-145 [68]. Another circRNA, circZNF292 was found to enhance proliferation, but seemingly not through microRNA sponging [69]. It is unclear whether fusion circRNAs (see Other Functional CircRNAs, below) might exert their tumor promoting properties via interaction with microRNAs [70].
Sequestering Proteins
Mounting evidence suggests most circRNAs are preferentially expressed in the cytoplasm versus the nucleus [28,70]. Several studies of circ-Foxo3 provide evidence that it can sequester proteins in the cytoplasm and prevent their nuclear entry (Figure 4E). Circ-Foxo3 was found to interact with stress-related proteins FAK and HIF-1α [45]. The authors demonstrated that knockdown of circ-Foxo3 enhanced nuclear expression of these proteins. It appears that circ-Foxo3 can bind these proteins in the cytoplasm and prevent their nuclear translocation to inhibit their anti-senescence and anti-stress functions [45]. Future work is required to uncover the cis-elements mediating the circRNA to protein interaction and whether this association truly depends on the noncoding RNA of interest being circular in form. It is especially encouraging that siRNA knockdown of circ-Foxo3 attenuated multiple markers of doxorubicin-induced cardiomyopathy in mice. These hallmarks of cardiomyopathy were exacerbated by circ-Foxo3 overexpression. Thus, circ-Foxo3, in contrast to HRCR (see above) appears to be detrimental to heart function.
Circ-Foxo3 also appears to bind to cell-cycle proteins. Consistent with the general trend for reduced expression of circRNAs in proliferative cells [30], cancer cell lines showed reduced expression of circ-Foxo3 versus linear Foxo3 compared to non-cancer cell lines [41]. Circ-Foxo3 overexpression reduced cell proliferation. Circ-Foxo3 was found to form a ternary complex with the cell cycle associated proteins CDK2 and P21, offering a potential mechanism for the anti-proliferative effect [41]. The generation and study of specific circ-Foxo3 knockout animals will be essential to demonstrate that Foxo3 has endogenous functions in vivo.
Other Functional CircRNAs
Some of the circRNAs arising from lariat precursors have been described as possible interacting partners with RBPs (Figure 4C). Amyotrophic lateral sclerosis (ALS) involves an accumulation of the RBP TDP-43 in degenerating neurons. It has been shown that in yeast with suppressed activity of the debranching enzyme Dbr1, circular lariats accumulate and interact with TDP-43. This maintains circular lariats in the cytoplasm and mitigates cytotoxicity of the protein [48]. A similar cytoplasmic accumulation of circular intronic lariats has been reported for in the oocytes of Xenopus tropicalis and Xenopus laevis, but their function roles have not been investigated [71].
It appears that EIciRNAs can function as regulators of transcription. EIciRNAs are mainly confined to the nucleus, and retain intronic sequences between back-spliced exons [31]. EIciRNAs interact by RNA-RNA base paring with U1 snRNP and this complex has been shown to bind to Pol II and regulate the transcription of their parental genes [31] (Figure 4A). It is possible that the intronic retained sequences promote the interaction of this class of circRNAs with multiple RBPs.
A very recently described class of circRNAs have been dubbed fusion-circRNAs (f-circRNAs) (Figure 4D). These originate from cancer-associated chromosomal translocations. f-circRNAs were shown to be confer resistance to apoptosis-inducing drug therapy, and remarkably have tumor promoting effects in vivo [70].
Future Studies of CircRNA Function
It is notable that many circRNAs are expressed in the testis [7,36]. Could circRNAs mediate transgenerational epigenetic inheritance of acquired traits [72]? Although transgenerational epigenetic inheritance is a controversial topic, the inherent stability of circRNAs make them an enticing candidate to be transmitted across generations. Moreover, as we have discussed, circRNAs are regulated by stress events and aging. Increased levels of circRNAs resulting from experienced stress could be passed down to offspring and thus constitute an epigenetically transmitted memory. There is some early evidence that circRNAs can be transmitted across generations. Circular intronic lariats in Xenopus tropicalis oocytes are highly stable and are transmitted to the fertilized egg after the germinal vesicle breakdown, persisting until the blastulae stage of embryogenesis [71] (Figure 4C). Undoubtedly, whether circRNAs mediate the inheritance of experience-acquired traits, as has been described for some small tRNA derived fragments [73], is under intense investigation.
The consequence of the enhanced stability of circRNAs is that they accumulate as time passes in post-mitotic cells such as neurons [7,11]. Could this accumulation be deleterious for nervous system function? Could it have an impact on the cognitive decline and susceptibility to neurodegenerative disease that is associated with aging?
Finally, it might be that some circRNAs are in fact not “non-coding.” Several studies have shown that circular RNAs are capable of protein synthesis in artificial contexts when an Internal Ribosome Entry Site (IRES) is placed within the circularized RNA to allow for translation initiation [42,74,75]. The field awaits conclusive evidence that any endogenous circRNA can produce a protein, and whether such proteins have functions, perhaps detrimental ones. This could have important implications for disease as protein-generating circRNAs could theoretically undergo multiple rounds of translation due to their exceptional stability and perhaps lead to pathological levels of accumulated protein in old age.
Concluding Remarks
CircRNAs are gaining considerable interest and we have here highlighted very recent work providing evidence that individual circRNAs have biologically relevant roles. The next few years will be interesting times for the circRNA field. RNA-seq analysis has revealed the identity and expression patterns of circRNAs in major model organisms, diverse cellular conditions have been identified to regulate circRNAs, and new methodologies have been devised to overexpress and knockdown circRNAs. The future is thus ripe for uncovering the in vivo functions of circRNAs. As is so often the case for diverse classes of new RNAs being discovered, there will certainly be many more fascinating twists in the circRNA story.
Acknowledgments
Thanks to members of the Miura lab for helpful discussions and especially to Daphne Cooper and Hannah Gruner for suggestions and editing of the manuscript.
Glossary
- circRNAs
circular RNAs
- mRNAs
messenger RNAs
- EIciRNAs
Exon-Intron circRNAs
- ciRNAs
circular intronic RNAs
- EGF
Epidermal Growth Factor
- EMT
Epithelial-mesenchymal transition
- QKI
Quaking
- MBL
muscleblind
- RBP
RNA binding protein
- RUST
Regulated Unproductive Splicing and Translation
- IRES
Internal Ribosome Entry Site
- f-circRNAs
fusion-circRNAs
Author Contributions
M.C. and P.M wrote the manuscript. This work was supported by the National Institute of General Medical Sciences grant P20 GM103650 and National Institute on Aging grant R15AG052931.
References
- Capel B, Swain A, Nicolis S. et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73(5):1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- Cocquerelle C, Daubersies P, Majerus MA. et al. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992;11(3):1095–1098. doi: 10.1002/j.1460-2075.1992.tb05148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro JM, Cho KR, Fearon ER. et al. Scrambled exons. Cell. 1991;64(3):607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- Lu T, Cui L, Zhou Y. et al. Transcriptome-wide investigation of circular RNAs in rice. RNA. 2015;21(12):2076–2087. doi: 10.1261/rna.052282.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL. et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm JO, Miura P, Olson S. et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9(5):1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang XO, Chen T. et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- Kos A, Dijkema R, Arnberg AC. et al. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323(6088):558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- Branch AD, Robertson HD. et al. A replication cycle for viroids and other small infectious RNA's. Science. 1984;223(4635):450–455. doi: 10.1126/science.6197756. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xue W, Li X. et al. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016;15(3):611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Rozowsky J, Yan KK. et al. Comparative analysis of the transcriptome across distant species. Nature. 2014;512(7515):445–448. doi: 10.1038/nature13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Gerhardt DJ, Dinger ME. et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2012;30(1):99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil D, Glowatz H, Schlumpberger M. Ribosomal RNA depletion for efficient use of RNA-seq capacity. Curr Protoc Mol Biol. 2013;4(4):19. doi: 10.1002/0471142727.mb0419s103. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Veno MT, Damgaard CK. et al. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016;44(6):e58. doi: 10.1093/nar/gkv1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti NR. et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Szabo L, Morey R, Palpant NJ. et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Agarwal V, Guo H. et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Chen RE, Olsen MN. et al. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SP, Ye S, Long Y. et al. Circular RNA expression alterations are involved in OGD/R-induced neuron injury. Biochem Biophys Res Commun. 2016;471(1):52–56. doi: 10.1016/j.bbrc.2016.01.183. [DOI] [PubMed] [Google Scholar]
- Qu S, Song W, Yang X. et al. Microarray expression profile of circular RNAs in human pancreatic ductal adenocarcinoma. Genom Data. 2015;5:385–387. doi: 10.1016/j.gdata.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sand M, Bechara FG, Sand D. et al. Circular RNA expression in basal cell carcinoma. Epigenomics. 2016;8(5):619–632. doi: 10.2217/epi-2015-0019. [DOI] [PubMed] [Google Scholar]
- Wang YH, Yu XH, Luo SS. et al. Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun Ageing. 2015;12:17. doi: 10.1186/s12979-015-0042-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, Stottmeister C, Glazar P. et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58(5):870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- Starke S, Jost I, Rossbach O. et al. Exon circularization requires canonical splice signals. Cell Rep. 2015;10(1):103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem. 2006;281(40):29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- Bachmayr-Heyda A, Reiner AT, Auer K. et al. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- Ivanov A, Memczak S, Wyler E. et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10(2):170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28(20):2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PL, Bao Y, Yee MC. et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9(6):e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S, McPherson JD, McCombie WR. et al. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17(6):333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X, Vlatkovic I, Babic A. et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18(4):603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy LF, Yeo GS, Tung YC. et al. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17(1):85–98. doi: 10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Dieterich DC, Ito HT. et al. Microfluidic local perfusion chambers for the visualization and manipulation of synapses. Neuron. 2010;66(1):57–68. doi: 10.1016/j.neuron.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Schuman EM. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron. 2013;80(3):648–657. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Zhang N, Han P. et al. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44(9):e87. doi: 10.1093/nar/gkw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du WW, Yang W, Liu E. et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn SJ, Pillman KA, Toubia J. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol. 2011;27:347–376. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- Dang Y, Yan L, Hu B. et al. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 2016;17(1):130. doi: 10.1186/s13059-016-0991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du WW, Yang W, Chen Y. et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2016 doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540. doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaphiropoulos PG. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol. 1997;17(6):2985–2993. doi: 10.1128/mcb.17.6.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armakola M, Higgins MJ, Figley MD. et al. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat Genet. 2012;44(12):1302–1309. doi: 10.1038/ng.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Ares M. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15(10):689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MC, Liang D, Tatomer DC. et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29(20):2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MP, Nagel RJ, Fagg WS. et al. Quaking and PTB control overlapping splicing regulatory networks during muscle cell differentiation. RNA. 2013;19(5):627–638. doi: 10.1261/rna.038422.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccomanno L, Loushin C, Jan E. et al. The STAR protein QKI-6 is a translational repressor. Proc Natl Acad Sci U S A. 1999;96(22):12605–12610. doi: 10.1073/pnas.96.22.12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra R, Charizanis K, Manchanda M. et al. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol Cell. 2014;56(2):311–322. doi: 10.1016/j.molcel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos MG, Batra R, Charizanis K. et al. Developments in RNA splicing and disease. Cold Spring Harb Perspect Biol. 2011;3(1):a000778. doi: 10.1101/cshperspect.a000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, Schor IE, Allo M. et al. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol. 2013;14(3):153–165. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- Coulter DE, Greenleaf AL. A mutation in the largest subunit of RNA polymerase II alters RNA chain elongation in vitro. J Biol Chem. 1985;260(24):13190–13198. [PubMed] [Google Scholar]
- Hillman RT, Green RE, Brenner SE. An unappreciated role for RNA surveillance. Genome Biol. 2004;5(2):R8. doi: 10.1186/gb-2004-5-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100(1):189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoilov P, Daoud R, Nayler O. et al. Human tra2-beta1 autoregulates its protein concentration by influencing alternative splicing of its pre-mRNA. Hum Mol Genet. 2004;13(5):509–524. doi: 10.1093/hmg/ddh051. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Rinn J, Pandolfi PP. et al. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Wiklund ED, Bramsen JB. et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30(21):4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LF, Saetrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics. 2014;30(16):2243–2246. doi: 10.1093/bioinformatics/btu257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Long B, Liu F. et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016 doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Bao C, Guo W. et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhang X, Hu X. et al. Circular RNA Related to the Chondrocyte ECM Regulates MMP13 Expression by Functioning as a MiR-136 'Sponge' in Human Cartilage Degradation. Sci Rep. 2016;6:22575. doi: 10.1038/srep22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Ren X, Xin S. et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckel JN, Jae N, Heumuller AW. et al. Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA. Circ Res. 2015;117(10):884–890. doi: 10.1161/CIRCRESAHA.115.306319. [DOI] [PubMed] [Google Scholar]
- Guarnerio J, Bezzi M, Jeong JC. et al. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell. 2016;165(2):289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- Talhouarne GJ, Gall JG. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. RNA. 2014;20(9):1476–1487. doi: 10.1261/rna.045781.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Otterdijk SD, Michels KB. Transgenerational epigenetic inheritance in mammals: how good is the evidence? FASEB J. 2016;30(7):2457–2465. doi: 10.1096/fj.201500083. [DOI] [PubMed] [Google Scholar]
- Chen Q, Yan M, Cao Z. et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351(6271):397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- Abe N, Matsumoto K, Nishihara M. et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman R, Ares M. Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA. 1998;4(9):1047–1054. doi: 10.1017/s135583829898061x. [DOI] [PMC free article] [PubMed] [Google Scholar]