Abstract
Pancreatic cancer is the third leading cause of cancer mortality in the U.S. with close to 40,000 deaths per year. Pancreatic ductal adenocarcinoma (PDAC) represents approximately 90 percent of all pancreatic cancer cases and is the most lethal form of the disease. Current therapies for PDAC are ineffective and most patients cannot be treated by surgical resection. Most research efforts have primarily focused on how genetic alterations cause, alter progression, contribute to diagnosis, and influence PDAC management. Over the past two decades, a model has been advanced of PDAC initiation and progression as a multi-step process driven by the acquisition of mutations leading to loss of tumor suppressors and activation of oncogenes. The recognition of the essential roles of these genetic alterations in the development of PDAC has revolutionized our knowledge of this disease. However, none of these findings have turned into effective treatment for this dismal malignancy. In recent years, studies in the areas of chromatin modifications, and non-coding RNAs have uncovered mechanisms for regulating gene expression which occur independently of genetic alterations. Chromatin-based mechanisms are interwoven with microRNA-driven regulation of protein translation to create an integrated epigenetic language, which is grossly dysregulated in PDAC. Thus in PDAC, key tumor suppressors that are well established to play a role in PDAC may be repressed, and oncogenes can be upregulated secondary to epigenetic alterations. Unlike mutations, epigenetic changes are potentially reversible. Given this feature of epigenetic mechanisms, it is conceivable that targeting epigenetic-based events promoting and maintaining PDAC could serve as foundation for the development of new therapeutic and diagnostic approaches for this disease.
Keywords: pancreatic cancer, epigenomics, gene expression, chromatin remodeling, Non-coding RNAs
Introduction
Pancreatic cancer is currently the third leading cause of cancer-related death in the United States [1] and is predicted to be the second leading cause by 2030 [2]. The vast majority of pancreatic cancer cases (~90 percent) are diagnosed as pancreatic ductal adenocarcinoma (PDAC), while other forms of pancreatic cancer (e.g., neuroendocrine tumors, squamous, pseudopapillary, and acinar cell carcinomas) are much less frequent [3-6]. PDAC remains one of the most intractable and devastating of all malignancies, with a median survival time of six months after diagnosis and a five-year survival rate of 5 to 7 percent, PDAC kills close to 40,000 people in just the United States each year [1,4]. The bleak prognosis of PDAC is due to lack of early detection, and an aggressive biology with extensive stromal involvement and almost inevitable dissemination, rapidly leading to an incurable stage, for which therapeutic intervention is a challenge [7-9]. Notably, these aggressive neoplasms are highly resistant to conventional chemotherapy and radiation [10,11]. Surgical resection is the only curative modality; however, less than 20 percent of PDAC patients are eligible for surgery, and even with adjuvant chemotherapy, five-year survival in this group is only 20 to 25 percent [12,13].
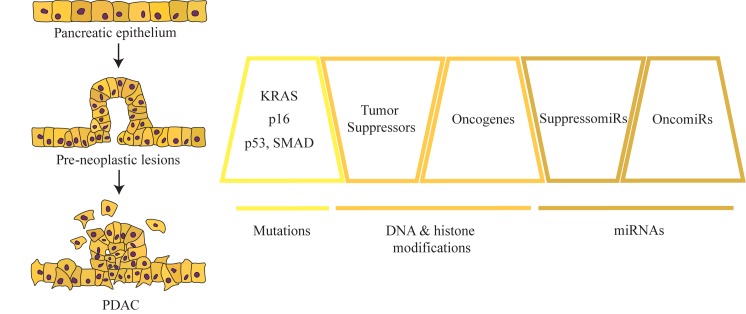
Certain genetic alterations (e.g., KRAS G12D) in pancreatic epithelial cells leading to the expression of mutant forms of the encoded protein are key drivers in PDAC development (See Figure 1). These alterations initiate in noninvasive precursor lesions. The most well characterized of these are microscopic lesions consisting of proliferated epithelial cells called pancreatic intraepithelial neoplasia (PanIN), which are classified into PanIN 1-3 based on the severity of their histopathological dysplasia [14,15]. Less commonly, PDAC may also derive from macroscopic intraductal papillary mucinous neoplasms (IPMNs) or mucinous cystic neoplasms (MCNs) [16]. At the PanIN-1 stage, some mutations in KRAS are detectable, whereas alterations in CDKN2A, SMAD4, and TP53 are acquired in PanIN-2 or 3 grades [17,18]. The past several decades of PDAC research have yielded substantial knowledge about the genetics of tumor cells. Genome-wide DNA sequencing efforts have demonstrated that each PDAC patient tumor has a unique mutational landscape, with an average of 26-119 mutations per tumor reported in several studies [19-22]. In addition, PDAC patient samples have been shown to contain many chromosomal rearrangements, including gene deletions and amplifications [20,23]. This knowledge of mutational drivers has led to new understanding of the basis of oncogenesis in PDAC and has suggested new potential therapeutic approaches targeting the molecular pathways disrupted by mutation [19,22,24,25]. However, these findings have not yet not translated into effective strategies for PDAC treatment. For example, there has been a lack of success in the development of clinically applicable direct inhibitors of KRAS, and attempts to disrupt the KRAS pathway through the use of inhibitors of kinases downstream of KRAS [e.g., RAF, mitogen kinase kinase 1 [MEK], phosphoinositide 3-kinase [PI3K]) alone or in conjunction with cytotoxic agents such as gemcitabine have led to disappointing results in clinical trials thus far [7,26-32].
Figure 1.
Diagram of major genetic and epigenetics mechanisms mediating PanINs and PDAC development. This review focuses on three specific epigenetic mechanisms to alter gene expression: DNA methylation, histone-based epigenetics, and ncRNAs.
Therefore, additional approaches beyond genomic characterization are needed to elucidate the dysfunctional biology of PDAC and identify novel targets for therapeutic intervention. PDAC tumors exhibit significant changes in gene expression compared to normal pancreatic exocrine cells [25,33], even at loci that are not genetically altered. Thus, genomic alterations do not directly account for all of the phenotypic and molecular aberrations demonstrated by PDAC cells. In recent years, PDAC research has turned to the field of epigenetics to attempt to further understand these alterations in gene expression observed in this disease. Epigenetics was initially defined as the inheritance of cell phenotype unrelated to the DNA sequence [34]. A revised version of the definition of epigenetics refers to the mechanisms by which cells stably maintain or alter their gene expression (i.e., phenotype) without changes in DNA sequence [35]. The processes included under the epigenetic mechanism “umbrella” are DNA methylation, histone post-translational modification, nucleosome remodeling and regulation by non-coding RNAs (ncRNAs) [35,36]. In this review we will discuss recent findings in the field of PDAC epigenetics and their impact on the biology and translational significance for this devastating malignancy.
Overview of Epigenetic Mechanisms
Epigenetic processes can be divided mechanistically into the regulation of DNA methylation, histone-based mechanisms which include post-translational modifications and nucleosome remodeling, and regulation of transcription or translation by ncRNAs.
DNA Methylation
The methylation of cytosine to 5-methylcytosine (5mC) in cytosine-phosphate-guanine (CpG) regions of DNA sequences is a crucial epigenetic modification regulating gene activity. DNA methylation often occurs in extended stretches of CpG, called CpG islands [37]. DNA hypermethylation in gene promoters is often linked to the repression of transcription, whereas DNA demethylation of normally methylated promoters is frequently associated with increased gene expression [38]. Although the functions of gene body methylation are not fully elucidated, global demethylation is considered to increase genomic instability and elevate genetic mutation rates, leading to carcinogenesis [39]. DNA methylation is accomplished by DNA methyltransferases (DNMT1, DNMT3a, and DNMT3b). De novo DNA methylation is a mainly a product of the activity of DNMT3a and DNMT3b, whereas the maintenance of DNA methylation over the course of DNA replication is a result of the activity of DNMT1 [38]. Physiological DNA methylation is essential in embryonic development, genomic imprinting, X-chromosome inactivation for monoallelic expression, and preventing chromosomal instability by suppressing the expression of transposable elements [38,40]. In differentiated somatic cells, most CpG islands at gene promoters remain at low levels of methylation and these genes can be transcriptionally activated depending on the state of histone modifications, as discussed later in this review. However, a small percentage of CpG islands become methylated resulting in a long-term repressed state of the gene transcription. Most human tumors, including PDAC have been shown to exhibit aberrant DNA methylation, including both a global loss of DNA methylation, particularly at repetitive elements and at some gene promoters, and hypermethylation at CpG islands within specific gene promoters [41]. Demethylation of repetitive elements may increase genomic instability, leading to chromosomal deletions, translocations and amplifications typical of many cancers [38]. When present at gene promoters, demethylation may lead to the expression of normally silent genes, such as cancer drivers. In contrast, promoter DNA hypermethylation has been shown to be responsible for the repression of tumor suppressors in cancer.
Many genes are reported to be silenced by CpG methylation in PDAC. For example, the CDKN2A (p16) gene, a tumor suppressor functioning as a cell cycle regulator by inhibiting a cyclin-dependent kinase, is known to be inactivated in 98 percent of PDAC, either by genetic or epigenetic mechanisms [42]. While the genetic mechanisms consist of intragenic mutation and homozygous deletion, p16 is epigenetically silenced in cases without mutation by CpG methylation of its promoter in 14 to 21 percent of PDAC cases [42]. An example of DNA methylation alterations leading to expression of a pro-oncogenic protein is the expression of guanine nucleotide exchange factor, VAV1, in PDAC [43]. VAV1 in normally expressed only in hematopoietic cells but is expressed in human PDAC samples as a result of promoter DNA hypomethylation and gene body DNA hypermethylation [43-45]. Further, VAV1 expression and promoter hypomethylation/gene body hypermethylation is negatively correlated with PDAC patient survival [43,44]. Finally, the promoter hypomethylation of VAV1 leading to its increased expression was recently demonstrated to be a result of TGFβ signaling in PDAC, wherein TGFβ leads to a dissociation of DNMT1 from the VAV1 promoter [44].
A number of studies have used genome-wide approaches to identify genes that are differentially methylated in PDAC samples vs. normal pancreas. For example, Vincent and colleagues [46] performed methylated CpG island amplification followed by CpG island microarrays in conjunction with gene expression studies in PDAC cells vs. control pancreatic tissue samples. This study detected a number of genes silenced by DNA methylation, including genes involved in stem cell pluripotency (BMI1, BMP3, FOXD3), WNT signaling (WNT5A, APC2, SOX1) and cell adhesion (CDH2, CDH4, PCDH1, and PCDH10) [46]. Several genes were found to be hypomethylated and overexpressed in PDAC relative to normal cells including genes for chromatin enzymes (e.g., histone methyltransferase SETD8, histone deacetylase KDM6A and the histone acetylase EP400) and oncogenes (JUNB, MYB, and FOS) [46]. A recent study compared the methylation profiles of 167 resect PDAC samples from previously untreated patients with 29 samples from untransformed regions of pancreas and identified ~3500 aberrantly methylated genes in PDAC [47]. Pathway analysis of affected genes indicated the axonal guidance signaling pathway as one of the most effected processes with hypermethylation of the ROBO1, ROBO3, SLIT3, and SLIT2 genes. The axon guidance pathway, including SLIT/ROBO ligand/receptor signaling, is best known for its role in neuronal pathfinding and angiogenesis but has also been shown to be deregulated in a number of cancers [48,49]. Exome sequencing of human PDAC tumors have recently revealed frequent mutations in SLIT and ROBO genes [19]. Although the functions of SLIT and ROBO proteins in PDAC are still under investigation [19,50], some studies suggest that these proteins can play a tumor suppressive role [49,51]. Thus, silencing or mutation of these proteins in PDAC may support oncogenesis. Other pathways identified included TGFβ, integrin and WNT signaling [47]. Dutreal et al. compared gene promoter methylation patterns between a small number of samples of PDAC, chronic pancreatitis (CP), a condition predisposing to PDAC, and normal pancreas [52]. This approach identified some genes that were hypermethylated in PDAC only and some that were methylated in both PDAC and CP. One of the genes hypermethylated in both PDAC and CP was WNK2, a cytoplasmic serine/threonine kinase. Further study showed that WNK2 mRNA and WNK2 protein expression decreased progressively from normal pancreas, through PanIN to PDAC. Finally, WNK2 in tumor tissue was negatively correlated with extracellular signal-regulated kinase (ERK) phosphorylation, and WNK2 overexpression in PDAC cells in vitro inhibited growth. These finding suggest a role for WNK2 silencing by DNA hypermethylation that supports pancreatic oncogenesis.
Thus, genome-wide studies discovered many genes that are differentially methylated in PDAC. Each publication identified different, although sometimes overlapping, lists of genes that are aberrantly methylated, and subsets of genes that are regulated by methylation status. Differences between studies in individual samples (e.g., PDAC cell lines vs. patient samples), molecular methods (e.g., particular methylation arrays) and mode of data analysis (e.g., threshold values for significant differences in methylation or expression) may explain some variance between the studies. Nonetheless, these investigations have provided further insight into genes and pathways that are epigenetically regulated by DNA methylation. While some genes identified by global methylation profiles were known to be involved in PDAC (e.g.TGFβ, WNT) [24], other novel genes may provide important targets for future investigation. It should be emphasized that genome-wide scale studies of gene methylation in pancreatic cancer report that the expression of only some genes with altered methylation patterns in cancer vs. normal tissue are regulated simply by inverse correlation with methylation, indicating that other epigenetic mechanisms play roles in the net expression of these genes [46,47,53].
A number of investigations have evaluated whether changes in DNA methylation observed in PDAC are also detectable in precancerous stages (e.g., CP and PanIN) in the pancreas. For example, Peng et al. [54] compared the promoter methylation of a series of genes (e.g., CDKN2A, APC, BRCA1, GSTP1, TIMP3, CDH1, and DAPK1) in microdissected human samples of normal pancreatic duct epithelium, ducts of inflamed pancreas, PanIN and PDAC. Although individual samples of each tissue subtype exhibited some heterogeneity of methylation patterns, on average a progressive increase in methylation of these genes was found when comparing normal tissue to inflamed ducts, through PanIN to PDAC. Among the PDAC samples, higher numbers of methylated genes was associated with poorer tumor differentiation and also with higher expression of DNMT1 [54].
The mechanisms by which DNA methylation is altered in PDAC are not fully understood. One possible proximal cause of hypermethylation of certain genes is that DNMTs are overexpressed in PDAC, as they are in some other cancer types [55]. Several investigators have reported that DNMT1, DNMT3a and/or DNMT3b are elevated in PDAC [56-59], although one report suggests that DNMT3b is decreased [60]. Thus, it is possible that DNMT overexpression may drive hypermethylation at gene loci. One mechanism for DNMT overexpression in PDAC is that DNMTs may be elevated by oncogene signaling [61]. In addition, nuclear protein 1 (Nupr1), a chromatin protein that is overexpressed in pancreatitis and PDAC, binds to the DNMT1 promoter, enhancing the expression of this gene [62]. The DNMT3b promoter is report to be hypomethylated with coordinate overexpression of DNMT3b in some PDAC samples [53], suggesting the importance of DNA methylation mechanisms in regulating this gene. In addition, DNMT3b is reported to be amplified in some PDAC samples, leading to enhanced activity [59]. Finally, DNMT activity could be altered without changes in mRNA or protein expression by the interaction between DNMTs and other proteins.
The alteration of methylation status of the cellular epigenome may begin early in the development of PDAC. Peng et al. 2005 [58] found that DNMT1 expression increased from normal pancreatic ducts through inflamed pancreatic ducts and PanIN to PDAC. The methylation of genes studied by Peng et al. 2006 [54] was also found to increase progressively from normal duct through PanIN to PDAC. Increasing DNA methylation of specific genes was also reported when comparing low to high grade PanINs [63]. The progressive DNA methylation and silencing of individual genes (e.g., WNK2) from normal tissue through PanIN to PDAC has also been demonstrated, as has the systematic hypomethylation and enhanced expression of genes (e.g., MUC4) in the transition to PDAC [64]. Thus, the epigenetic dysregulation of the PDAC methylome appears to be an additive process that begins at preneoplastic stages of the disease. The mechanisms by which DNMT1 expression may be increased during PDAC progression are not fully known, but are likely to arise secondarily to oncogene signaling and/or interactions between stromal and tumor cells [57,61,65].
Histone Posttranslational Modifications and Nucleosomal Dynamics
Chromatin consists of the genomic DNA systematically wound and condensed by association with histones and other proteins [66,67]. Nucleosomes are considered to be the basic unit of chromatin. Each nucleosome is an octamer of histones (two molecules each of histones H2A, H2B, H3 and H4) upon which ~146 base pairs of genomic DNA are wound [35,68]. Chromatin remodeling, i.e., reconfiguring chromatin structure into a more open or closed state, is an important mechanism that contributes to the regulation of gene transcription. Two important components of chromatin remodeling are post-translational histone modification, and nucleosome remodeling, a process in which particular regions of DNA are spooled onto or off of nucleosomes [66,69,70].
Post-translational chemical modification on histone tails is an evolutionally conserved mechanism involved in transcriptional regulation [71-73]. Several distinct types of histone modification are known, such as acetylation, methylation, phosphorylation, ubiquitination and SUMOylation, all of which are generally referred to as “histone marks” [35]. The concept of the “histone code” has been extensively reviewed [36,74-77]. Briefly, this hypothesis proposes that each type of histone mark present at specific locations relative to genes (i.e., promoters, enhancers, gene bodies) conveys specific information to the transcriptional machinery about whether a gene should be expressed or repressed. The concepts of “writers” (enzymes that place the marks), “readers” (proteins that recognize the mark, and likely recruit other proteins to the site), and “erasers” (enzymes that remove the mark) have been important for the development of mechanistic models of histone-based regulation of gene transcription. Although there is debate over whether histone marks directly influence gene transcription or are simply reflective of the present transcriptional state of genes (e.g., see [78]), clear evidence exists that particular histone marks recruit specific protein complexes (e.g., methylated histone 3, lysine 9 recruits heterochromatin 1 proteins and acetylated lysines bind bromodomain-containing proteins) that can repress or activate transcription [79,80].
The most widely studied histone modification is lysine alteration, including lysine methylation, acetylation and phosphorylation. Specific lysine methylation marks on chromatin associated with promoter regions have been shown to be correlated with transcriptional activation [e.g., histone 3, lysine 4 trimethylation and histone 3, lysine 36 trimethylation (H3K4me3 and H3K36me3)] or silencing (e.g., H3K9me3 or H3K27me3), usually through recruiting co-effectors, such as heterochromatin protein 1 (HP1) binding to H3K9me3 and Polycomb-group proteins to H3K27me3 [81]. One, two, or three methyl groups can be added to the lysine residues on a histone tail, and each of these modifications can have different significance. In addition, a regulatory region may be associated with multiple marks. For example, genes whose promoters are marked with H3K4me3 and H3K27me3 are called bivalent genes. Such genes express negligible or low transcript but are said to be poised to be activated upon a stimulating signal. Bivalent genes are a frequent feature in stem cells and pluripotent cells, where these genes can be activated by loss of H3K27me3, or repressed by loss of H3K4me3, upon signals for differentiation or development [35,77]. Similarly, H3K4me1 and H3K27me3 have been demonstrated to be indicative of poised enhancers.
In PDAC there are numerous examples of code writers and erasers that are aberrantly expressed and/or activated. For example, EZH2, a well-studied trimethylation writer of H3K27, is a component of Polycomb Repressive Complex 2 (PRC2), which is known to regulate cell-cycle checkpoints and DNA damage repair pathways [82]. EZH2 overexpression is associated with poor prognosis of cancer, including PDAC [83], as a result of silencing tumor suppressor genes, such as E-cadherin [84], CDKN1C [85] and BRCA1 [86]. In PDAC, poorly differentiated PDAC is known to exhibit nuclear accumulation of EZH2 more frequently [87], in which context tumor suppressor p27Kip1 is implicated to be a target gene of EZH2. EZH2 is frequently overexpressed in PDAC compared to normal pancreas [87,88]. RAS oncogenic signaling increases EZH2 expression in PDAC [89], suggesting a possible mechanism for EZH2 upregulation. High EZH2 expression in PDAC is significantly associated with decreased E-cadherin expression and more aggressive disease [90]. In contrast, depletion of EZH2 by RNAi in PDAC cells inhibits cell growth, increases chemosensitivity, and decreases the growth of PDAC cancer stem cells [87,91]. The abovementioned studies indicate that EZH2 can play a pro-oncogenic role in PDAC. However, a study using genetic mouse models of pancreatic neoplasia showed that mice with pancreas-specific expression of KRASG12D plus knockout of EZH2 accelerated the incidence and stage of neoplasia compared to mice with KRASG12D alone [92]. Results from that study also suggested that EZH2 may be involved in the repair of pancreatic tissue after injury, and proposed that loss of EZH2 may contribute to pancreatic carcinogenesis by inhibiting tissue regeneration. In addition, investigations have shown that H3K27me3 levels are decreased PDAC vs. normal pancreas and that lower levels of H3K27me3 are associated with a poorer prognosis among PDAC patients [88,93]. Thus, although EZH2 is generally elevated in PDAC, its enzymatic product, H3K27me3, is decreased. The reasons for these seemingly anomalous findings are not known, but might involve imbalances in other components of the PRC2 complex (e.g., EED, SUZ12) that could affect the activity of EZH2. In summary, EZH2 levels are altered in pancreatic cancer; however, reports of its role in oncogenesis are so far conflicting. Further studies are needed to determine if other PRC2 proteins are altered in PDAC. The possible role of EZH1, a homolog of EZH2 that can also catalyze the trimethylation of H3K27 [94,95], in PDAC should also be investigated.
Components of the Polycomb complex 1 (PRC1), such as Bmi1, also have been shown to be important for the PDAC development. Bmi1 is increased in PDAC, as well as in chronic pancreatitis and PanIN lesions [96], suggesting that the overexpression of Bmi1 is an early event in the ontogeny of PDAC. Using a conditional knockout of Bmi1 in combination with a Kras(G12D)-driven PDAC mouse mode, Bednar et al. [97] demonstrated that the requirement for Bmi1 in PDAC carcinogenesis is independent of the Ink4a/Arf locus and at least partially mediated by dysregulation of reactive oxygen species.
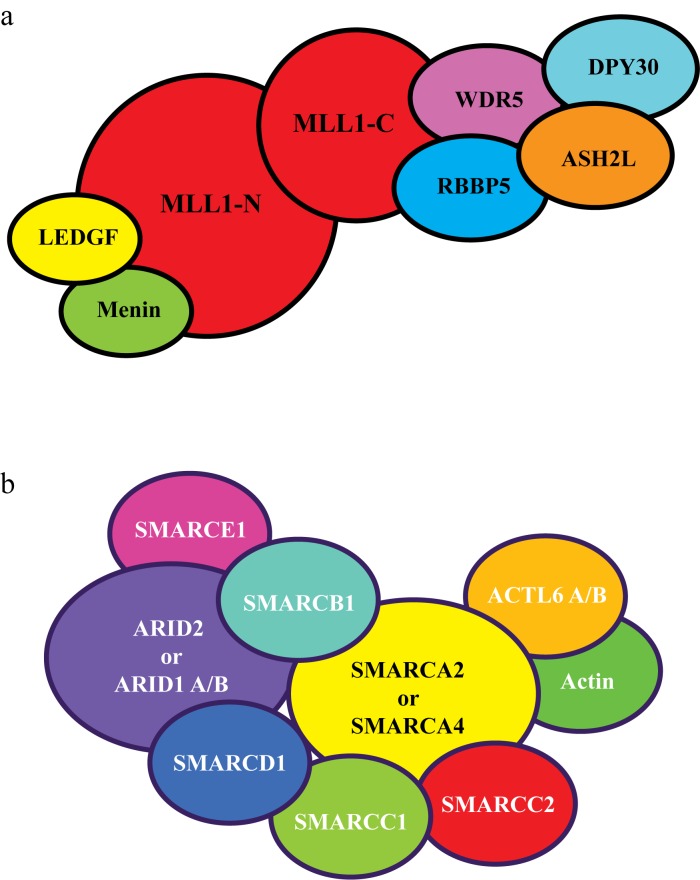
H3K4me activating marks are written by SET1 protein complexes, including the mixed lineage leukemia (MLL) protein family. While chromosomal translocation of MLL family genes have been well established as a cause of leukemia, there is increasing evidence that this family of proteins is frequently mutated in other cancers, including PDAC [98]. The MLL family consists of four members with histone methyltransferase activity, referred to as MLL1-4 [99]. Due to inconsistency in the literature between the nomenclature of MLL2 and MLL4, we clarify that we will refer to gene KMT2B on chromosome 19 as MLL2, and gene KMT2D on chromosome 12 as MLL4. Each MLL forms a large complex with additional protein subunits (e.g., WDR5, ASH2L, DPY30, and RBBP5) and with the ability to di- and tri-methylate H3K4 (see Figure 2) [99]. Current evidence demonstrates that each MLL complex has different functions but the roles of these proteins are not yet fully understood [100-102]. Of note, the MLL1 and MLL2 complexes bind to the transcriptional regulator, menin, a protein in which germline mutations lead to inherited multiple endocrine neoplasia type 1, a cause of pancreatic neuroendocrine tumors [98,103-105]. Genome-wide sequencing studies have demonstrated that some PDAC tumors contain mutations in MLL proteins [19,21,22,24,106]. One study reported that out of ~100 PDAC samples tested, mutations in MLL3, MLL4 and MLL1 occurred in 7, 5, and 2 percent of patients, respectively [106]. In addition to point mutations, truncations, frameshifts and amplification of MLL genes have been reported in PDAC [20,106,107]. Sausen et al. [106] reported that mutations in MLL1, MLL3, and MLL4 were correlated with improved overall survival in PDAC. In addition, Dawkins et al. [108] found that decreased MLL3 and MLL4 expression in tumors was correlated with increased survival time in PDAC patients. Studies by this same group showed that knockdown of MLL4 in PDAC cells in vitro led to reduced cell growth with an inhibition of the cell cycle and increased apoptosis. Thus, it appears that loss of expression or mutation of MLL proteins may have a negative impact on tumorigenesis or prognosis and that the presence of the wild type protein in some way supports oncogenesis. However, further studies are needed to refine these observations.
Figure 2.
MLL and SWI/SNF complexes. A) Representation of the MLL1 protein complex. MLL1 is expressed as a single protein but cleaved to an N- and C-terminal fragment by the enzyme, taspase. MLL2 has a similar subunit structure. MLL3 and MLL4 are not cleaved by taspase and are not associated with menin or LEDGF, but maintain association with WDR5, RBBP5. ASH2L and DPY30, and possess other unique subunits [99,105]. B) Generalized representation of a SWI/SNF complex. Several different complexes have been characterized with the alternatively exclusive presence of SMARCA2 or SMARCA4, and the presence of either ARID1A, ARID1B, or ARID2 plus PBRM [118].
Acetylation (Ac) of lysines (e.g., H3K9Ac and H3K27Ac) is usually associated with gene activation [109,110]. Lysine acetylation neutralizes the positive charge on histone tails, which is proposed to releases chromatin compaction, thus opening up chromatin for gene transcription [111]. However, histone acetylation has other more specific roles such as recruiting bromodomain proteins to chromatin [112]. A large number of histone acetylases and histone deacetylases (HDACs) are involved in the writing and erasing of histone acetylation marks [59,110,113]. Compared to histone methylation, the role of histone acetylation has been far less studied in PDAC. One report evaluated H4K12 and H3K18 acetylation in PDAC samples by immunohistochemistry and found that these acetylation marks were indicators of lower overall survival in patients [114]. This study suggests that significant alterations in the acetylation machinery occur in PDAC that impact upon tumor biology. Mutations in the EP300 histone acetylase have been reported in PDAC [115]. High expression of the NAD-dependent histone deacetylase, Sirtuin 1 (SIRT1), has been shown to be associated with poorly differentiated PDAC and is negatively correlated with PDAC patient survival [116]. SIRT1 expression in PDAC cells increased their viability, while small molecule inhibition of SIRT1 repressed growth in these cells [116]. Similarly, HDAC7 was reported to be overexpressed in PDAC and its expression was associated with poorer patient survival and disease recurrence [117]. Further investigations are required to determine what specific genes are affected by alterations in acetylation in PDAC that account for the impact of histone deacetylases on PDAC cell phenotype and patient prognosis.
Nucleosomal remodeling is accomplished by large protein complexes, such as SWI/SNF (switch-defective/sucrose non-fermentable) complexes containing BRG1 (SMARCA4) or BRM (SMARCA2) ATPases (see Figure 2) [118]. These protein complexes utilize the energy of splitting ATP to alter nucleosome position on the DNA strand, resulting in more open or closed chromatin structure [119]. SWI/SNF complexes are composed of ~10 protein subunits and several distinct complexes have been identified which are differentiated by the presence of specific alternate subunits (see Figure 2) [120]. For example, SWI/SNF complexes contain either BRG1 or BRM, and either ARID1A, ARID1B, or PBRM1 plus ARID2. In addition to their nucleosome remodeling abilities, SWI/SNF complexes have been shown to interact with epigenetic cofactors (e.g., HDACs, arginine methyltranserases) at gene promoters and thus are able to modulate gene expression by other means besides nucleosome positioning [121,122].
Multiple investigations have identified mutations in SWI/SNF components in PDAC [22,24,106]. For example, Bailey et al. [24] reported that 14 percent of PDAC samples had mutations in the SWI/SNF proteins, ARID1A, PBRM1 or SMARCA4. In addition, the ARID1B gene has been demonstrated to be silenced by hypermethylation in PDAC [123], while BRG1 is reported to be expressed in PDAC tumor tissue but not in neighboring benign pancreas [124]. These observations suggest that SWI/SNF proteins can be altered in PDAC by epigenetic mechanisms as well as by mutation. Several studies have attempted to address the role of SWI/SNF proteins in PDAC. Numata et al. [125] investigated SWI/SNF protein expression in PDAC samples vs. patient clinical characteristics. This study showed that high BRM and low PBRM1 were independent indicators of poorer survival, however BRG1 levels were not correlated with survival. In contrast, Dal Molin et al. found that surgically ~50 percent of resected IPMNs out of 60 samples exhibited reduced Brg1 immunostaining compared to normal pancreatic tissue. Using genetically engineered mouse models, Brg1 was selectively deleted in pancreatic exocrine cells in the context of KRAS mutant expression [126]. Loss of Brg1 reduced PanIN formation but increased IPMN development, leading to the suggestion that Brg1 promotes tumor formation in the case of PDAC derived from acinar cells, but plays a role in impeding the differentiation and hyperplasia of pancreatic duct epithelial cells [126,127]. In vitro, knockdown of Brg1 decreased the growth of PDAC cells in vitro and in xenografts, led to reduced AKT and p21cip/waf activation and increased chemosensitivity [124], further supporting the concept that Brg1 supports oncogenesis in PDAC. Together, these findings suggest that SWI/SNF proteins have cell context-dependent effects on pancreatic oncogenesis. Brg1 has been shown to regulate c-MYC [128,129], a protein frequently over-expressed in PDAC and known to regulate key oncogenic pathways [130,131]. Brg1 binds directly to the c-MYC promoter, and also interacts with c-MYC at MYC target gene promoters [122,128,129,132]. While BRG1 expression is usually negatively related to c-MYC expression, BRG1 increases c-MYC expression in hematopoietic cells by binding to a 3’ enhancer of the c-MYC gene [133]. Thus, BRG1 mutation or loss of expression in PDAC has to potential to affect c-MYC expression. However, to our knowledge, this possibility has not yet been investigated.
Non-coding RNAs (ncRNAs)
The coding mRNA and noncoding tRNA and rRNAs have been extensively characterized in regards to their protein translation functions. However, only a minority of RNA transcripts from the human genome are protein coding [134,135] and the functions of the remaining noncoding RNA transcripts are just beginning to be recognized. ncRNAs can be broadly categorized into short (sncRNAs; < 200 bases) and long forms (lncRNAs; > 200 bases) or pragmatically considered by their biological function. ncRNA transcripts are flexible and fold into 3D conformations that allow exact interactions with proteins, DNA and other RNA. These interactions regulate functional events of translation, splicing, replication, transcription and chromosome structure [135,136]. Thus, ncRNAs are important for many cellular processes including normal development and disease states.
lncRNAs are able to interact with DNA, RNA and proteins and have been demonstrated to have a number of cellular functions. Hundreds of lncRNA molecules have been functionally characterized to regulate gene expression and serve in diverse roles such as scaffolds, guides, tethers, decoys, and miRNA sponges [136]. Of major interest to PDAC are specific lncRNAs that interact with chromatin modifier complexes and thus affect gene regulation. For example, several lncRNAs have been demonstrated to interact with EZH2. MALAT-1 is an intergenic lncRNA shown to bind and recruit EZH2 to the E-cadherin promoter and suppress its expression in PDAC cells [137]. Loss of E-cadherin is associated with increased tumor metastasis [138]. Similarly, HOTAIR is an overexpressed lncRNA in many cancer types including PDAC and its increased expression correlates to tumor invasiveness and poor prognosis [139-141]. HOTAIR binds to the PRC2 complex causing an increase in H3K27me3 and a decrease in expression of genes such as SNAIL that repress EMT and invasion [140]. HOTTIP is another lncRNA overexpressed in PDAC but also detectable as stable fragments in plasma [142]. Since PDAC is characteristically asymptomatic until advanced stages, HOTTIP may provide a biomarker for early detection and monitoring. These lncRNAs may provide novel therapeutic targets for PDAC at some point, although the technologies for inhibiting ncRNAs in vivo is still under development [143-145].
sncRNAs include microRNAs (miRNAs), small nuclear RNAs, small nucleolar RNAs and piwi RNAs [146]. In recent years, miRNAs have been extensively studied in PDAC. miRNAs are evolutionally conserved, 18 to 25 nucleotide, non-protein coding RNAs found in parts of the genome that were once thought to be non-functional. miRNAs were first discovered in nematodes [147] but in 2000 the let-7 miRNA was identified and found to be evolutionarily conserved in diverse species including vertebrates, mollusks and arthropods [148]. Primary miRNAs are usually transcribed by PolII as transcripts of several kilobases; mature miRNA are formed from these transcripts through a multi-step, multi-enzyme process [149]. MiRNAs function by binding complementary regions of mRNAs, usually in the 3’ region of these mRNAs, inhibiting the process of translation or sometimes decreasing the stability of the associated mRNA species [150]. The number of individual miRNA species transcribed from the human genome has been estimated as ranging from 1000 to 2000, and each miRNA may have many mRNA targets [151-153]. Further, the miRNA transcriptome is complex, with a number of miRNA species having multiple copies of identical or near identical sequences in the genome, and frequent clustering of several miRNAs in polycistronic sequences that are coordinately expressed under the control of a single promoter [153]. It has been suggested that each cell type may have an individual characteristic miRNA transcriptome, analogous to cell-specific proteomes. Further, there is increasing evidence that the dysregulation of the miRNA profile of cells is linked to cancer [153-155]. Upon comparison of the miRNAs in a tumor vs. surrounding tissue, a typical observation is that some miRNAs are increased, whereas others are decreased in the tumor tissue. Some miRNAs that are increased in tumors have cancer-promoting effects when overexpressed in normal cells, and are thus referred to as oncomiRs. In contrast, some miRNA species that are diminished in tumor cells have growth or invasiveness-inhibiting properties when overexpressed in cancer cells, and are thus called tumor suppressor miRs or suppressomiRs [156].
Altered expression of both oncomiRs and suppressomiRs has been demonstrated with the progression of PDAC from normal tissue through PanIN lesions to PDAC. For example, miR-221 overexpression in PDAC promotes invasiveness by decreasing expression of TIMP2 through directly targeting its 3’UTR. TIMP2 is an inhibitor of MMP2 and MMP9 proteins involved in matrix degradation. Overexpression of miR-221 mimics in PDAC cells caused an increase in migration and an increase in MMP2 and MMP9 gene expression [157]. miR-221 was also identified in a meta-analysis of miRNAs in PDAC as upregulated in seven studies with an average fold change of 6.7 [158]. Another example illustrates that miRNAs increase in preneoplastic lesions as well as in PDAC. miR-155, was increased in PanIN2 and PanIN3 lesions, respectively, showing the progressive expression of miR-155 expression with the development of PDAC [159]. Additional analysis of PanIN tissues by locked nucleic acid in situ hybridization verified the increase of mir155 in PanIN2 and PanIN3 graded tumors, respectively [159]. Meta-analysis of miRNA expression identified miR-155 as upregulated an average of five-fold (eight studies) in PDAC compared to normal tissue [158]. miR-155 has been shown to enhance tumorigenesis by decreasing a number of targets including TP53INP1, SEL1L and MLH1 [160-162]. Extension of this work demonstrated that a large number of miRNA species are up or downregulated in PanIN compared to normal pancreas [163], suggesting that aberrant expression of miRNA begins early in the development of PDAC.
A number of studies compared the global expression profile of miRNA in PDAC vs. normal pancreas and identified many differentially expressed miRNA species in PDAC (e.g., [164-167]). Meta-analyses of these studies have identified a few miRNAs that are reported in multiple studies as consistently altered in PDAC. For example, miR-21 and miR-23a were identified as upregulated, and miR-148a and miR-375 were shown to downregulated in multiple profiling studies as recognized by two separate meta-analyses [158,168]. Nonetheless, these meta-analyses also demonstrate the enormous variability in findings between miRNA profile investigations in terms of consistency of which miRNAs are most highly altered and even the direction of alterations of individual miRNAs. These inconsistencies might be a result of differences in miRNA profiling techniques, miRNA nomenclature, individual sample heterogeneity including genetic differences between PDAC subtypes [24], or patient sample characteristics (e.g., microdissected tumor vs. fine needle aspirate, previous chemotherapy vs. naive, tumor stage). In addition, some investigations of the biological impact of individual miRNAs in PDAC have reported contradictory results (e.g., the same miRNA is reported to be pro- or anti-oncogenic) [150,169-173]. Whether these inconsistencies are a result of differences in techniques or cell models is so far unclear. In summary, miRNAs are clearly altered in PDAC and play a role in its pathology, but additional diligent studies are required to further understand the significance of these noncoding RNAs in PDAC and their possible value as therapeutic targets and biological markers.
Crosstalk Between Different Epigenetic Mechanisms
Thus far, we have described the epigenetic mechanisms of DNA methylation, histone modification and miRNAs as discrete processes, each contributing to the aberrant phenotype of PDAC cells. In living eukaryotic cells, however, these processes are intertwined as part the cellular machinery that dynamically defines net gene and protein expression. For example, epigenetic proteins may physically complex with lncRNAs as described above for the interaction between EZH2 and HOTAIR, HOTTIP, and MALAT-1. In addition, miRNAs can regulate the expression of epigenetic enzymes by targeting their transcripts, and miRNA expression can be modulated at the transcriptional level by epigenetic proteins. Here, we provide examples to illustrate some of these modes of epigenetic crosstalk that act to regulate final phenotypic outcomes in PDAC.
miRNAs Target Epigenetic Enzymes
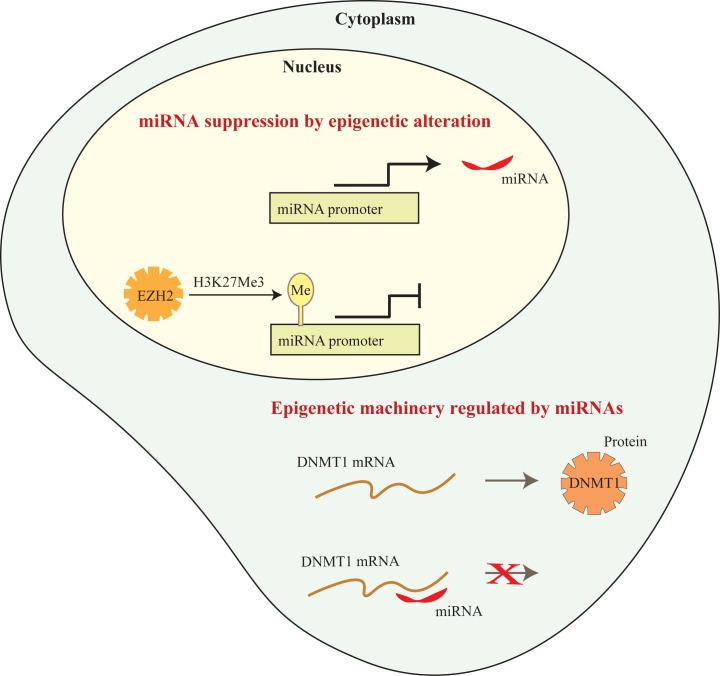
The first study to demonstrate that miRNAs could regulate the expression of epigenetic enzymes was that of Fabbri et al. [174] who showed that miR-29 family members (29a, 29b, and 29c) were decreased in lung cancer cells. This study further demonstrated that re-expression of miR-29 in this cell type decreased DNMT3a and DNMT3b levels by directly targeting their mRNAs, normalized gene methylation patterns and caused reexpression of methylation-silenced tumor suppressors. Several groups have identified miRNAs that directly regulate the expression of proteins involved in the epigenetic machinery in PDAC (see Figure 3). For example, miR-148b and miR-152 were identified as potential inhibitors of DNMT1 translation, and validated to target DNMT1 in PDAC cells [175]. A study showing that miR-148b is decreased in PDAC [176] supports the concept that decreasing these miRNAs could be a viable mechanism by which DNMT1 is frequently overexpressed in this malignancy [56,58]. EZH2 is frequently overexpressed in PDAC and is a well known target of miRNA regulation. For example miR-101, a regulator of EZH2, has been shown to be downregulated in PDAC [177]. Forced expression of miR-101 in PDAC cells decreases EZH2 expression and global H3K27me3, and increases expression of E-cadherin, a target of EZH2 regulation, as well as inhibiting cell growth [177]. Thus, decreases in miRNAs that target EZH2 in PDAC provide a potential mechanism for the overexpression of EZH2 in PDAC. Further, several studies show that miR-101 and other suppressomiRs can be induced in PDAC cells by certain drugs (e.g., metformin, difluorinated curcumin) [178,179], suggesting that it may be possible to decrease EZH2 expression via upregulation of miRNAs for therapeutic benefit. Finally, the PRC1 complex protein, Bmi1, is another pro-oncogenic chromatin regulator that is overexpressed in PDAC [97,180] and targeted by miRNAs. For example, both miR-15a and miR-135a are decreased in PDAC and directly target Bmi1 expression. Given the large numbers of miRNAs and the ability of each species of miRNA to inhibit multiple targets, it is likely that other chromatin enzymes and cofactors are subject to regulation by miRNAs in PDAC.
Figure 3.
Crosstalk between epigenetic mechanisms. This diagram depicts the repression of miRNA transcription by the histone methyltransferase, EZH2, and the disruption of DNMT1 translation by an miRNA.
miRNAs are Regulated by DNA Methylation and Histone Modification
Increasing evidence has demonstrated that many miRNAs are transcribed by RNA polymerase II and regulated by similar transcriptional mechanisms to mRNAs [151]. Thus, miRNAs can be regulated by promoter DNA methylation and histone modification [181,182]. A number of studies have demonstrated that some suppressomiRs are silenced by DNA methylation. For example, miR-124, miR-148a, and miR-615-5p are each decreased in expression in PDAC, and their promoters have been shown to be highly DNA methylated in PDAC compared to normal tissue [183-185]. Further, expression of each of these miRNAs is increased upon treatment of PDAC cells with the DNMT inhibitor, 5-aza-2’-deoxcytidine. Finally, overexpression of these miRNAs were shown to inhibit PDAC cell growth [183,185-187], supporting the idea that methylation and subsequent silencing of these miRNAs plays a role in supporting oncogenesis.
Conversely, oncomiR expression may be induced by DNA hypomethylation during PDAC. For example, miR-200a and miR-200b are two miRNAs are processed from the same polycistronic transcript, and are well known for their roles in regulating epithelial-to mesenchymal transition by targeting ZEB1 and ZEB2. Li et al. [188] reported that miR200a and miR-200b are overexpressed in PDAC vs. normal pancreas. The promoter for miR-200 was found to be hypomethylated in PDAC cells vs. nonneoplastic cell lines, and in PDAC xenografts vs. normal pancreas [188]. Thus, the overexpression of miR-200a/b in PDAC is proposed to be a result of promoter demethylation. However, the mechanisms by which this miRNA promoter is demethylated in PDAC has not been reported. Nonetheless, promoter hypomethylation is likely contribute to the overexpression of additional miRNAs in PDAC.
A study by Zhu et al [189] suggests another possible mode for regulation of miRNA transcription. This group reported that miR-548an was downregulated in PDAC vs. normal tissue. In vitro studies showed that hypoxic conditions caused the increased binding of the transcription factor, HIF1α, and its cofactor, HDAC1, to the miR-548an promoter, resulting in reduced Pol II binding and decreased transcription. Treatment of hypoxia-treated cells with the HDAC inhibitor, trichostatin, increased the expression of miR-548an. Although changes in histone acetylation were not measured, this study suggests that the recruitment of HDAC1 to the miR-548an promoter instills a loss of acetylated histones in this region, thus decreasing expression of the miR.
miR-218 has been demonstrated to be reduced in PDAC samples vs. normal tissue and its reduction has been shown to be a predictor for poor prognosis [190]. Further, overexpression of miR-218 in PDAC cells reduces growth in vitro and tumor formation in mouse xenografts [191]. Li et al. [191], studied the mechanisms by which miR-218 is repressed in PDAC (see Figure 2). miR-218 was identified in a screen of miRNAs that were increased in PDAC cell lines treated with the methyltransferase inhibitor, DZnep, in a attempt to identify targets of EZH2. This study showed that miR-218 was decreased in expression in PDAC vs. normal pancreas tissue as well as in PDAC cell lines vs. the nontransformed pancreatic duct cell line, HPDE. Methylation-specific PCR showed that the promoter was methylated in the majority of PDAC cell lines and PDAC tissue, but not the HPDE cell line or normal pancreas. Expression of miR-218 was increased in PDAC cells by treatment with the DNMT inhibitor, 5-aza-cytidine, or by knockdown of DNMT1, DNMT3a or DNMT3b, confirming the regulation of miR-218 by DNA methylation. Repression of this miRNA was demonstrated to be pivotally regulated by EZH2. EZH2 and its product H3K27me3 were found to be bound to the miR-218 promoter in PDAC cells but not HPDE cells and knockdown of EZH2 in PDAC cells increased miR-218 expression. EZH2 depletion also reduced the occupancies of DNMT1, DNMT3a and DNMT3b at the miR-218 promoter. Finally, EZH2 loss also decreased the repressive histone mark, H3K9me2, and the associated repressive complex consisting of SUV39H1 methyltransferase, HP1-α and HP1-γ demonstrates in exquisite detail how an miRNA can be repressed in PDAC by a combination of DNA methylation and histone modifications. Surprisingly few studies have investigated the regulation of miRNAs at this level of detail, however, it is likely that the same mechanisms are involved in the suppression or silencing of many miRNAs in PDAC.
Conclusions and Future Directions
Recent studies demonstrate that DNA methylation- and chromatin-based mechanisms are highly dysregulated in PDAC. Further, rather than operating independently, these epigenetic processes interact with each other, establishing a multilevel regulatory network altering the expression of genes and proteins beyond the changes imposed by genetic alterations alone. Unlike genetic changes, epigenetic alterations are potentially reversible. Thus, it has been suggested that small molecules that target epigenetic mechanisms could potentially used to reset the epigenetic state of PDAC cells [192,193]. Indeed, several clinical trials utilizing compounds to target DNMTs, and HDACs in PDAC are ongoing or completed [192,194]. New specific inhibitors of EZH2, such as GSK2816126 and tazemetostat have been developed and are beginning clinical trials for various cancers, although large-scale trials have yet to be performed in PDAC [192]. However, future studies will need to address many questions to optimize the use of epigenetic drugs. First, what is the optimal treatment regime? For, example, should epigenetic drugs be given simultaneously with cytotoxic drugs, or should patients be pretreated with epigenetic drugs and then receive conventional chemotherapies? Second, will epigenetic drugs have unanticipated negative effects on stromal fibroblasts or immune cells? In addition to the tumor cells, tumor associated fibroblasts, endothelial cells and immune cells have been demonstrated to undergo extensive epigenetic alterations in PDAC [195]. Extensive testing will be needed first in animal models and then in human subjects to evaluate the effects of epigenetic drugs on the tumor stroma in vivo. Finally, are there subsets of PDAC patients who would most benefit from certain epigenetic drug treatments? PDAC is an extremely heterogeneous disease. With the advent of individualized medicine, we often have extensive information on the individual mutations present in a PDAC patient that may suggest specific therapeutic approaches. For example, in some tumor cell types, inactivating mutations in SWI/SNF proteins (SMARCA4, ARID1A or SMARCB1) cause an increased sensitivity to EZH2 inhibitors, leading to the use of EZH2 inhibitors in patients with these alterations [196-199]. Similarly, mRNA sequencing or protein-specific methods (e.g., immunohistochemistry) might identify patients with high expression of targetable epigenetic proteins such as DNMTs or EZH2 who may benefit from treatments of inhibitors of those proteins. In conclusion, our increased understanding of the genetic and epigenetic aberrations that drive PDAC suggest novel avenues for treatment, but extensive basic and clinical studies are needed to translate these findings into improved patient outcome.
Acknowledgments
We would like to acknowledge the contributions of the authors of the excellent research studies that we have cited in this review. We apologize, due to space constrains, that we have omitted some of these studies. We thank Emily Porcher for the secretarial assistance. This work was supported by the Mayo Clinic Cancer Center, NCI CA136526, Mayo Clinic Pancreatic SPORE P50 CA102701 and Mayo Clinic Center for Cell Signaling in Gastroenterology P30 DK084567 to MEFZ.
Glossary
- CP
chronic pancreatitis
- CpG
cytosine-phosphate-guanine binucleotide
- DNMT
DNA methyltransferase
- EZH2
Enhancer of Zeste homolog 2
- HP1
heterochromatin protein 1
- HDAC
Histone deacetylase
- IPMN
Intraductal papillary mucinous neoplasm
- lncRNA
long non-coding RNA
- ncRNA
non-coding RNA
- miRNA
micro-RNA
- MLL
Mixed lineage leukemia
- MCN
Mucinous cystic neoplasm
- PDAC
Pancreatic ductal adenocarcinoma
- PanIN
Pancreatic intraepithelial neoplasia
- PRC
Polycomb repressive complex
- SWI/SNF
SwItch/sucrose non-fermentable
- TGFβ
Transforming growth factor β
References
- American_Cancer_Society. Cancer Facts & Figures. 2016
- Rahib L, Smith BD, Aizenberg R. et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- Fitzgerald TL, Hickner ZJ, Schmitz M. et al. Changing incidence of pancreatic neoplasms: a 16-year review of statewide tumor registry. Pancreas. 2008;37(2):134–138. doi: 10.1097/MPA.0b013e318163a329. [DOI] [PubMed] [Google Scholar]
- Hackeng WM, Hruban RH, Offerhaus GJ. et al. Surgical and molecular pathology of pancreatic neoplasms. Diagn Pathol. 2016;11(1):47. doi: 10.1186/s13000-016-0497-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishi A, Goggins M, Wood LD. et al. Pathological and molecular evaluation of pancreatic neoplasms. Semin Oncol. 2015;42(1):28–39. doi: 10.1053/j.seminoncol.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S, Sharma P, Zakalik D. Comparison of Demographics, Tumor Characteristics, and Survival Between Pancreatic Adenocarcinomas and Pancreatic Neuroendocrine Tumors: A Population-based Study. Am J Clin Oncol. 2016 doi: 10.1097/COC.0000000000000305. [DOI] [PubMed] [Google Scholar]
- Donahue TR, Dawson DW. Leveraging Mechanisms Governing Pancreatic Tumorigenesis To Reduce Pancreatic Cancer Mortality. Trends Endocrinol Metab. 2016;27(11):770–781. doi: 10.1016/j.tem.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharibi A, Adamian Y, Kelber JA. Cellular and molecular aspects of pancreatic cancer. Acta Histochem. 2016;118(3):305–316. doi: 10.1016/j.acthis.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Upadhyay G, Srivastava RK. et al. Recent advances in pancreatic cancer: biology, treatment, and prevention. Biochim Biophys Acta. 2015;1856(1):13–27. doi: 10.1016/j.bbcan.2015.04.003. [DOI] [PubMed] [Google Scholar]
- Garrido-Laguna I, Hidalgo M. Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat Rev Clin Oncol. 2015;12(6):319–334. doi: 10.1038/nrclinonc.2015.53. [DOI] [PubMed] [Google Scholar]
- Varghese AM, Lowery MA, Yu KH. et al. Current management and future directions in metastatic pancreatic adenocarcinoma. Cancer. 2016 doi: 10.1002/cncr.30342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettle H, Neuhaus P, Hochhaus A. et al. Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: the CONKO-001 randomized trial. JAMA. 2013;310(14):1473–1481. doi: 10.1001/jama.2013.279201. [DOI] [PubMed] [Google Scholar]
- Sener SF, Fremgen A, Menck HR. et al. Pancreatic cancer: a report of treatment and survival trends for 100,313 patients diagnosed from 1985-1995, using the National Cancer Database. J Am Coll Surg. 1999;189(1):1–7. doi: 10.1016/s1072-7515(99)00075-7. [DOI] [PubMed] [Google Scholar]
- Hruban RH, Maitra A, Goggins M. Update on pancreatic intraepithelial neoplasia. Int J Clin Exp Pathol. 2008;1(4):306–316. [PMC free article] [PubMed] [Google Scholar]
- Scarlett CJ, Salisbury EL, Biankin AV. et al. Precursor lesions in pancreatic cancer: morphological and molecular pathology. Pathology (Phila) 2011;43(3):183–200. doi: 10.1097/PAT.0b013e3283445e3a. [DOI] [PubMed] [Google Scholar]
- Kamisawa T, Wood LD, Itoi T. et al. Pancreatic cancer. Lancet. 2016;388(10039):73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- Kanda M, Matthaei H, Wu J. et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. 2012;142(4):730–733. doi: 10.1053/j.gastro.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda M, Sadakari Y, Borges M. et al. Mutant TP53 in duodenal samples of pancreatic juice from patients with pancreatic cancer or high-grade dysplasia. Clin Gastroenterol Hepatol. 2013;11(6):719–730. doi: 10.1016/j.cgh.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biankin AV, Waddell N, Kassahn KS. et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell N, Pajic M, Patch AM. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkiewicz AK, McMillan EA, Balaji U. et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre AJ, Brennan C, Bailey G. et al. High-resolution characterization of the pancreatic adenocarcinoma genome. Proc Natl Acad Sci U S A. 2004;101(24):9067–9072. doi: 10.1073/pnas.0402932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey P, Chang DK, Nones K. et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- He C, Jiang H, Geng S. et al. Analysis of whole genomic expression profiles and screening of the key signaling pathways associated with pancreatic cancer. Int J Clin Exp Pathol. 2012;5(6):537–546. [PMC free article] [PubMed] [Google Scholar]
- Bodoky G, Timcheva C, Spigel DR. et al. A phase II open-label randomized study to assess the efficacy and safety of selumetinib (AZD6244 [ARRY-142886]) versus capecitabine in patients with advanced or metastatic pancreatic cancer who have failed first-line gemcitabine therapy. Invest New Drugs. 2012;30(3):1216–1223. doi: 10.1007/s10637-011-9687-4. [DOI] [PubMed] [Google Scholar]
- Cascinu S, Berardi R, Sobrero A. et al. Sorafenib does not improve efficacy of chemotherapy in advanced pancreatic cancer: A GISCAD randomized phase II study. Dig Liver Dis. 2014;46(2):182–186. doi: 10.1016/j.dld.2013.09.020. [DOI] [PubMed] [Google Scholar]
- Hartley ML, Bade NA, Prins PA. et al. Pancreatic cancer, treatment options, and GI-4000. Hum Vaccin Immunother. 2015;11(4):931–937. doi: 10.1080/21645515.2015.1011017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heestand GM, Kurzrock R. Molecular landscape of pancreatic cancer: implications for current clinical trials. Oncotarget. 2015;6(7):4553–4561. doi: 10.18632/oncotarget.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante JR, Somer BG, Park JO. et al. A randomised, double-blind, placebo-controlled trial of trametinib, an oral MEK inhibitor, in combination with gemcitabine for patients with untreated metastatic adenocarcinoma of the pancreas. Eur J Cancer. 2014;50(12):2072–2081. doi: 10.1016/j.ejca.2014.04.024. [DOI] [PubMed] [Google Scholar]
- Javles M, Golan T, Maitra A. et al. Changing the course of pancreatic cancer--Focus on recent translational advances. Cancer Treat Rev. 2016;44:17–25. doi: 10.1016/j.ctrv.2016.01.004. [DOI] [PubMed] [Google Scholar]
- Mann KM KM, Ying H, Juan J. et al. KRAS-related proteins in pancreatic cancer. Pharmacol Ther. 2016;168:29–32. doi: 10.1016/j.pharmthera.2016.09.003. [DOI] [PubMed] [Google Scholar]
- Gutierrez ML, Corchete L, Teodosio C. et al. Identification and characterization of the gene expression profiles for protein coding and non-coding RNAs of pancreatic ductal adenocarcinomas. Oncotarget. 2015;6(22):19070–19086. doi: 10.18632/oncotarget.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. The epigenotype. 1942. Int J Epidemiol. 2012;41(1):10–13. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- Sharma S, Kelly TK, Jones PA. et al. Epigenetics in cancer. Carcinogenesis. 2010;31(1):27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomberk GA, Urrutia R. The Triple-Code Model for Pancreatic Cancer: Cross Talk Among Genetics, Epigenetics, and Nuclear Structure. Surg Clin North Am. 2015;95(5):935–952. doi: 10.1016/j.suc.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheishvili D, Boureau L, Szyf M. DNA demethylation and invasive cancer: implications for therapeutics. Br J Pharmacol. 2015;172(11):2705–2715. doi: 10.1111/bph.12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B, Robertson KD. DNA methylation and human disease. Adv Exp Med Biol. 2013;754:3–29. doi: 10.1007/978-1-4419-9967-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6(8):597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan S, Van Emburgh BO, Robertson KD. DNA methylation in development and human disease. Mutat Res. 2008;647(1-2):30–38. doi: 10.1016/j.mrfmmm.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klutstein M, Nejman D, Greenfield R. et al. DNA Methylation in Cancer and Aging. Cancer Res. 2016;76(12):3446–3450. doi: 10.1158/0008-5472.CAN-15-3278. [DOI] [PubMed] [Google Scholar]
- Schutte M, Hruban RH, Geradts J. et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57(15):3126–3130. [PubMed] [Google Scholar]
- Fernandez-Zapico ME, Gonzalez-Paz NC, Weiss E. et al. Ectopic expression of VAV1 reveals an unexpected role in pancreatic cancer tumorigenesis. Cancer Cell. 2005;7(1):39–49. doi: 10.1016/j.ccr.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Huang PH, Lu PJ, Ding LY. et al. TGFbeta promotes mesenchymal phenotype of pancreatic cancer cells, in part, through epigenetic activation of VAV1. Oncogene. 2016 doi: 10.1038/onc.2016.378. [DOI] [PubMed] [Google Scholar]
- Ilan L, Katzav S. Human Vav1 expression in hematopoietic and cancer cell lines is regulated by c-Myb and by CpG methylation. PLoS ONE. 2012;7(1):e29939. doi: 10.1371/journal.pone.0029939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent A, Omura N, Hong SM. et al. Genome-wide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clin Cancer Res. 2011;17(13):4341–4354. doi: 10.1158/1078-0432.CCR-10-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nones K, Waddell N, Song S. et al. Genome-wide DNA methylation patterns in pancreatic ductal adenocarcinoma reveal epigenetic deregulation of SLIT-ROBO, ITGA2 and MET signaling. Int J Cancer. 2014;135(5):1110–1118. doi: 10.1002/ijc.28765. [DOI] [PubMed] [Google Scholar]
- Blockus H, Chedotal A. Slit-Robo signaling. Development. 2016;143(17):3037–3044. doi: 10.1242/dev.132829. [DOI] [PubMed] [Google Scholar]
- Gara RK, Kumari S, Ganju A. et al. Slit/Robo pathway: a promising therapeutic target for cancer. Drug Discov Today. 2015;20(1):156–164. doi: 10.1016/j.drudis.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohrig A, Detjen KM, Hilfenhaus G. et al. Axon guidance factor SLIT2 inhibits neural invasion and metastasis in pancreatic cancer. Cancer Res. 2014;74(5):1529–1540. doi: 10.1158/0008-5472.CAN-13-1012. [DOI] [PubMed] [Google Scholar]
- Huang T, Kang W, Cheng AS. et al. The emerging role of Slit-Robo pathway in gastric and other gastro intestinal cancers. BMC Cancer. 2015;15:950. doi: 10.1186/s12885-015-1984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutruel C, Bergmann F, Rooman I. et al. Early epigenetic downregulation of WNK2 kinase during pancreatic ductal adenocarcinoma development. Oncogene. 2014;33(26):3401–3410. doi: 10.1038/onc.2013.312. [DOI] [PubMed] [Google Scholar]
- Tan AC, Jimeno A, Lin SH. et al. Characterizing DNA methylation patterns in pancreatic cancer genome. Mol Oncol. 2009;3(5-6):425–438. doi: 10.1016/j.molonc.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng DF, Kanai Y, Sawada M. et al. DNA methylation of multiple tumor-related genes in association with overexpression of DNA methyltransferase 1 (DNMT1) during multistage carcinogenesis of the pancreas. Carcinogenesis. 2006;27(6):1160–1168. doi: 10.1093/carcin/bgi361. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Uzvolgyi E, Liang G. et al. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27(11):2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang L, Xu J. et al. Aberrant DNA methyltransferase expression in pancreatic ductal adenocarcinoma development and progression. J Exp Clin Cancer Res. 2013;32:86. doi: 10.1186/1756-9966-32-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Wang F, Yang L. et al. Expression of DNMT1 and DNMT3a are regulated by GLI1 in human pancreatic cancer. PLoS ONE. 2011;6(11):e27684. doi: 10.1371/journal.pone.0027684. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Peng DF, Kanai Y, Sawada M. et al. Increased DNA methyltransferase 1 (DNMT1) protein expression in precancerous conditions and ductal carcinomas of the pancreas. Cancer Sci. 2005;96(7):403–408. doi: 10.1111/j.1349-7006.2005.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simo-Riudalbas L, Esteller M. Targeting the histone orthography of cancer: drugs for writers, erasers and readers. Br J Pharmacol. 2015;172(11):2716–2732. doi: 10.1111/bph.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazienza V, Tavano F, Benegiamo G. et al. Correlations among PPARgamma, DNMT1, and DNMT3B Expression Levels and Pancreatic Cancer. PPAR Res. 2012;2012:461784. doi: 10.1155/2012/461784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RK, Wang YC. Dysregulated transcriptional and post-translational control of DNA methyltransferases in cancer. Cell Biosci. 2014;4:46. doi: 10.1186/2045-3701-4-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso D, Bintz J, Lomberk G. et al. Pivotal Role of the Chromatin Protein Nupr1 in Kras-Induced Senescence and Transformation. Sci Rep. 2015;5:17549. doi: 10.1038/srep17549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Fukushima N, Hruban RH. et al. CpG island methylation profile of pancreatic intraepithelial neoplasia. Mod Pathol. 2008;21(3):238–244. doi: 10.1038/modpathol.3800991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Zhang JJ, Zhu R. et al. The increase in the expression and hypomethylation of MUC4 gene with the progression of pancreatic ductal adenocarcinoma. Med Oncol. 2011;28(Suppl 1):S175–S184. doi: 10.1007/s12032-010-9683-0. [DOI] [PubMed] [Google Scholar]
- Bigey P, Ramchandani S, Theberge J. et al. Transcriptional regulation of the human DNA Methyltransferase (dnmt1) gene. Gene. 2000;242(1-2):407–418. doi: 10.1016/s0378-1119(99)00501-6. [DOI] [PubMed] [Google Scholar]
- DesJarlais R, Tummino PJ. Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology. Biochemistry (Mosc) 2016;55(11):1584–1599. doi: 10.1021/acs.biochem.5b01210. [DOI] [PubMed] [Google Scholar]
- McGinty RK, Tan S. Nucleosome structure and function. Chem Rev. 2015;115(6):2255–2273. doi: 10.1021/cr500373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16(3):178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- Cutter AR, Hayes JJ. A brief review of nucleosome structure. FEBS Lett. 2015;589(20 Pt A):2914–2922. doi: 10.1016/j.febslet.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann T, Schneider R. Targeting histone modifications--epigenetics in cancer. Curr Opin Cell Biol. 2013;25(2):184–189. doi: 10.1016/j.ceb.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Ebert A, Lein S, Schotta G. et al. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14(4):377–392. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- Gabor Miklos GL, Maleszka R. Epigenomic communication systems in humans and honey bees: from molecules to behavior. Horm Behav. 2011;59(3):399–406. doi: 10.1016/j.yhbeh.2010.05.016. [DOI] [PubMed] [Google Scholar]
- Wenzel D, Palladino F, Jedrusik-Bode M. Epigenetics in C. elegans: facts and challenges. Genesis. 2011;49(8):647–661. doi: 10.1002/dvg.20762. [DOI] [PubMed] [Google Scholar]
- Cosgrove MS. Writers and readers: deconvoluting the harmonic complexity of the histone code. Nat Struct Mol Biol. 2012;19(8):739–740. doi: 10.1038/nsmb.2350. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Caldas C. Chromatin modifier enzymes, the histone code and cancer. Eur J Cancer. 2005;41(16):2381–2402. doi: 10.1016/j.ejca.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications - writers that read. EMBO Rep. 2015;16(11):1467–1481. doi: 10.15252/embr.201540945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosogane M, Funayama R, Nishida Y. et al. Ras-induced changes in H3K27me3 occur after those in transcriptional activity. PLoS Genet. 2013;9(8):e1003698. doi: 10.1371/journal.pgen.1003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JC, Whetstine JR. Chromatin landscape: methylation beyond transcription. Epigenetics. Epigenetics. 2011;6(1):9–15. doi: 10.4161/epi.6.1.13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AK, Barton MC. Bromodomain histone readers and cancer. J Mol Biol. 2016 doi: 10.1016/j.jmb.2016.11.020. [DOI] [PubMed] [Google Scholar]
- Lomberk G, Wallrath L, Urrutia R. et al. The Heterochromatin Protein 1 family. Genome Biol. 2006;7(7):228. doi: 10.1186/gb-2006-7-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7(3):299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crea F, Paolicchi E, Marquez VE. et al. Polycomb genes and cancer: time for clinical application? Crit Rev Oncol Hematol. 2012;83(2):184–193. doi: 10.1016/j.critrevonc.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Cao Q, Yu J, Dhanasekaran SM. et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27(58):7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Karuturi RK, Sun F. et al. CDKN1C (p57) is a direct target of EZH2 and suppressed by multiple epigenetic mechanisms in breast cancer cells. PLoS ONE. 2009;4(4):e5011. doi: 10.1371/journal.pone.0005011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez ME, Li X, Toy K. et al. Downregulation of EZH2 decreases growth of estrogen receptor-negative invasive breast carcinoma and requires BRCA1. Oncogene. 2009;28(6):843–853. doi: 10.1038/onc.2008.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ougolkov AV, Bilim VN, Billadeau DD. Regulation of pancreatic tumor cell proliferation and chemoresistance by the histone methyltransferase enhancer of zeste homologue 2. Clin Cancer Res. 2008;14(21):6790–6796. doi: 10.1158/1078-0432.CCR-08-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Chen J, Zhan Q. et al. H2AK119Ub1 and H3K27Me3 in molecular staging for survival prediction of patients with pancreatic ductal adenocarcinoma. Oncotarget. 2014;5(21):10421–10433. doi: 10.18632/oncotarget.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Fukamachi K, Tsuda H. et al. RAS oncogenic signal upregulates EZH2 in pancreatic cancer. Biochem Biophys Res Commun. 2012;417(3):1074–1079. doi: 10.1016/j.bbrc.2011.12.099. [DOI] [PubMed] [Google Scholar]
- Toll AD, Dasgupta A, Potoczek M. et al. Implications of enhancer of zeste homologue 2 expression in pancreatic ductal adenocarcinoma. Hum Pathol. 2010;41(9):1205–1209. doi: 10.1016/j.humpath.2010.03.004. [DOI] [PubMed] [Google Scholar]
- van Vlerken LE, Kiefer CM, Morehouse C. et al. EZH2 is required for breast and pancreatic cancer stem cell maintenance and can be used as a functional cancer stem cell reporter. Stem Cells Transl Med. 2013;2(1):43–52. doi: 10.5966/sctm.2012-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallen-St Clair J, Soydaner-Azeloglu R, Lee KE. et al. EZH2 couples pancreatic regeneration to neoplastic progression. Genes Dev. 2012;26(5):439–444. doi: 10.1101/gad.181800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Xia W, Zhang Z. et al. Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Mol Carcinog. 2008;47(9):701–706. doi: 10.1002/mc.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N. et al. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25(5):485–498. doi: 10.1101/gad.2019811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K. et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32(4):503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Romero C, Rooman I, Skoudy A. et al. The epigenetic regulators Bmi1 and Ring1B are differentially regulated in pancreatitis and pancreatic ductal adenocarcinoma. J Pathol. 2009;219(2):205–213. doi: 10.1002/path.2585. [DOI] [PubMed] [Google Scholar]
- Bednar F, Schofield HK, Collins MA. et al. Bmi1 is required for the initiation of pancreatic cancer through an Ink4a-independent mechanism. Carcinogenesis. 2015;36(7):730–738. doi: 10.1093/carcin/bgv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RC, Dou Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat Rev Cancer. 2015;15(6):334–346. doi: 10.1038/nrc3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S, Hofemeister H, Marks H. et al. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development. 2014;141(3):526–537. doi: 10.1242/dev.102681. [DOI] [PubMed] [Google Scholar]
- Hu D, Gao X, Morgan MA. et al. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol. 2013;33(23):4745–4754. doi: 10.1128/MCB.01181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lin C, Smith ER. et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29(22):6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matkar S, Thiel A, Hua X. Menin: a scaffold protein that controls gene expression and cell signaling. Trends Biochem Sci. 2013;38(8):394–402. doi: 10.1016/j.tibs.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nuland R, Smits AH, Pallaki P. et al. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol Cell Biol. 2013;33(10):2067–2077. doi: 10.1128/MCB.01742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slany RK. The molecular biology of mixed lineage leukemia. Haematologica. 2009;94(7):984–993. doi: 10.3324/haematol.2008.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sausen M, Phallen J, Adleff V. et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun. 2015;6:7686. doi: 10.1038/ncomms8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntsman DG, Chin SF, Muleris M. et al. MLL2, the second human homolog of the Drosophila trithorax gene, maps to 19q13.1 and is amplified in solid tumor cell lines. Oncogene. 1999;18(56):7975–7984. doi: 10.1038/sj.onc.1203291. [DOI] [PubMed] [Google Scholar]
- Dawkins JB, Wang J, Maniati E. et al. Reduced Expression of Histone Methyltransferases KMT2C and KMT2D Correlates with Improved Outcome in Pancreatic Ductal Adenocarcinoma. Cancer Res. 2016;76(16):4861–4871. doi: 10.1158/0008-5472.CAN-16-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedi U, Mishra VK, Wasilewski D. et al. Epigenetic plasticity: a central regulator of epithelial-to-mesenchymal transition in cancer. Oncotarget. 2014;5(8):2016–2029. doi: 10.18632/oncotarget.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon RP, Robaa D, Alhalabi Z. et al. KATching-Up on Small Molecule Modulators of Lysine Acetyltransferases. J Med Chem. 2016;59(4):1249–1270. doi: 10.1021/acs.jmedchem.5b01502. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389(6649):349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Marmorstein R, Zhou MM. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol. 2014;6(7):a018762. doi: 10.1101/cshperspect.a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti R, Conte M, Altucci L. Targeting Histone Deacetylases in Diseases: Where Are We? Antioxid Redox Signal. 2015;23(1):99–126. doi: 10.1089/ars.2013.5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano CN, Izetti P, Pereira MP. et al. H4K12 and H3K18 Acetylation Associates With Poor Prognosis in Pancreatic Cancer. Appl Immunohistochem Mol Morphol. 2016;24(5):337–344. doi: 10.1097/PAI.0000000000000194. [DOI] [PubMed] [Google Scholar]
- Gayther SA, Batley SJ, Linger L. et al. Mutations truncating the EP300 acetylase in human cancers. Nat Genet. 2000;24(3):300–303. doi: 10.1038/73536. [DOI] [PubMed] [Google Scholar]
- Stenzinger A, Endris V, Klauschen F. et al. High SIRT1 expression is a negative prognosticator in pancreatic ductal adenocarcinoma. BMC Cancer. 2013;13:450. doi: 10.1186/1471-2407-13-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouaissi M, Silvy F, Loncle C. et al. Further characterization of HDAC and SIRT gene expression patterns in pancreatic cancer and their relation to disease outcome. PLoS ONE. 2014;9(9):e108520. doi: 10.1371/journal.pone.0108520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Copeland RA, Keilhack H. PRC2 and SWI/SNF Chromatin Remodeling Complexes in Health and Disease. Biochemistry (Mosc) 2016;55(11):1600–1614. doi: 10.1021/acs.biochem.5b01191. [DOI] [PubMed] [Google Scholar]
- Lusser A, Kadonaga JT. Chromatin remodeling by ATP-dependent molecular machines. Bioessays. 2003;25(12):1192–1200. doi: 10.1002/bies.10359. [DOI] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11(7):481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- Hang CT, Yang J, Han P. et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466(7302):62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Yun R, Datta A. et al. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol. 2003;23(21):7475–7487. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khursheed M, Kolla JN, Kotapalli V. et al. ARID1B, a member of the human SWI/SNF chromatin remodeling complex, exhibits tumour-suppressor activities in pancreatic cancer cell lines. Br J Cancer. 2013;108(10):2056–2062. doi: 10.1038/bjc.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Tian X, Wang F. et al. BRG1 promotes chemoresistance of pancreatic cancer cells through crosstalking with Akt signalling. Eur J Cancer. 2014;50(13):2251–2262. doi: 10.1016/j.ejca.2014.05.017. [DOI] [PubMed] [Google Scholar]
- Numata M, Morinaga S, Watanabe T. et al. The clinical significance of SWI/SNF complex in pancreatic cancer. Int J Oncol. 2013;42(2):403–410. doi: 10.3892/ijo.2012.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Figura G, Fukuda A, Roy N. et al. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol. 2014;16(3):255–267. doi: 10.1038/ncb2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Molin M, Hong SM, Hebbar S. et al. Loss of expression of the SWI/SNF chromatin remodeling subunit BRG1/SMARCA4 is frequently observed in intraductal papillary mucinous neoplasms of the pancreas. Hum Pathol. 2012;43(4):585–591. doi: 10.1016/j.humpath.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagl NG Jr., Zweitzig DR, Thimmapaya B. et al. The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer Res. 2006;66(3):1289–1293. doi: 10.1158/0008-5472.CAN-05-3427. [DOI] [PubMed] [Google Scholar]
- Romero OA, Setien F, John S. et al. The tumour suppressor and chromatin-remodelling factor BRG1 antagonizes Myc activity and promotes cell differentiation in human cancer. EMBO Mol Med. 2012;4(7):603–616. doi: 10.1002/emmm.201200236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessmann E, Schneider G, Ellenrieder V. et al. MYC in pancreatic cancer: novel mechanistic insights and their translation into therapeutic strategies. Oncogene. 2016;35(13):1609–1618. doi: 10.1038/onc.2015.216. [DOI] [PubMed] [Google Scholar]
- Schleger C, Verbeke C, Hildenbrand R. et al. c-MYC activation in primary and metastatic ductal adenocarcinoma of the pancreas: incidence, mechanisms, and clinical significance. Mod Pathol. 2002;15(4):462–469. doi: 10.1038/modpathol.3880547. [DOI] [PubMed] [Google Scholar]
- Sharma T, Bansal R, Haokip DT. et al. SMARCAL1 Negatively Regulates C-Myc Transcription By Altering The Conformation Of The Promoter Region. Sci Rep. 2015;5:17910. doi: 10.1038/srep17910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Whyte WA, Zepeda-Mendoza CJ. et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27(24):2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Amaral PP, Schlesinger FJ. et al. The reality of pervasive transcription. PLoS Biol. 2011;9(7):e1000625. doi: 10.1371/journal.pbio.1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2(11):986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859(1):169–176. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- Han T, Jiao F, Hu H. et al. EZH2 promotes cell migration and invasion but not alters cell proliferation by suppressing E-cadherin, partly through association with MALAT-1 in pancreatic cancer. Oncotarget. 2016;7(10):11194–11207. doi: 10.18632/oncotarget.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Burstin J, Eser S, Paul MC. et al. E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology. 2009;137(1):361–371. doi: 10.1053/j.gastro.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Jutooru I, Chadalapaka G. et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32(13):1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo R, Shimamura T, Mimori K. et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71(20):6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li Z, Zheng S. et al. Expression profile of long non-coding RNAs in pancreatic cancer and their clinical significance as biomarkers. Oncotarget. 2015;6(34):35684–35698. doi: 10.18632/oncotarget.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavorgna G, Vago R, Sarmini M. et al. Long non-coding RNAs as novel therapeutic targets in cancer. Pharmacol Res. 2016;110:131–138. doi: 10.1016/j.phrs.2016.05.018. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang J. Therapeutic Potentials of Noncoding RNAs: Targeted Delivery of ncRNAs in Cancer Cells. Adv Exp Med Biol. 2016;927:429–458. doi: 10.1007/978-981-10-1498-7_16. [DOI] [PubMed] [Google Scholar]
- Taucher V, Mangge H, Haybaeck J. Non-coding RNAs in pancreatic cancer: challenges and opportunities for clinical application. Cell Oncol (Dordr) 2016;39(4):295–318. doi: 10.1007/s13402-016-0275-7. [DOI] [PubMed] [Google Scholar]
- Kowalczyk MS, Higgs DR, Gingeras TR. Molecular biology: RNA discrimination. Nature. 2012;482(7385):310–311. doi: 10.1038/482310a. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F. et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Tang YT, Xu XH, Yang XD. et al. Role of non-coding RNAs in pancreatic cancer: the bane of the microworld. World J Gastroenterol. 2014;20(28):9405–9417. doi: 10.3748/wjg.v20.i28.9405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Malumbres M. miRNAs and cancer: an epigenetics view. Mol Aspects Med. 2013;34(4):863–874. doi: 10.1016/j.mam.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive V, Minella AC, He L. Outside the coding genome, mammalian microRNAs confer structural and functional complexity. Sci Signal. 2015;8(368):re2. doi: 10.1126/scisignal.2005813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar SL, Lehmann U. DNA methylation, microRNAs, and their crosstalk as potential biomarkers in hepatocellular carcinoma. World J Gastroenterol. 2014;20(24):7894–7913. doi: 10.3748/wjg.v20.i24.7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macha MA, Seshacharyulu P, Krishn SR. et al. MicroRNAs (miRNAs) as biomarker(s) for prognosis and diagnosis of gastrointestinal (GI) cancers. Curr Pharm Des. 2014;20(33):5287–5297. doi: 10.2174/1381612820666140128213117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawa Z, Haque I, Ghosh A. et al. The miRacle in Pancreatic Cancer by miRNAs: Tiny Angels or Devils in Disease Progression. Int J Mol Sci. 2016;17(6) doi: 10.3390/ijms17060809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Li P, Chen X. et al. miR-221/222 induces pancreatic cancer progression through the regulation of matrix metalloproteinases. Oncotarget. 2015;6(16):14153–14164. doi: 10.18632/oncotarget.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma MZ, Kong X, Weng MZ. et al. Candidate microRNA biomarkers of pancreatic ductal adenocarcinoma: meta-analysis, experimental validation and clinical significance. J Exp Clin Cancer Res. 2013;32:71. doi: 10.1186/1756-9966-32-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JK, Hong SM, Karikari CA. et al. Aberrant MicroRNA-155 expression is an early event in the multistep progression of pancreatic adenocarcinoma. Pancreatology. 2010;10(1):66–73. doi: 10.1159/000231984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gironella M, Seux M, Xie MJ. et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci U S A. 2007;104(41):16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Chen J, Wang J. et al. Putative tumor suppressor gene SEL1L was downregulated by aberrantly upregulated hsa-mir-155 in human pancreatic ductal adenocarcinoma. Mol Carcinog. 2014;53(9):711–721. doi: 10.1002/mc.22023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WJ, Zhao YP, Zhang TP. et al. MLH1 as a direct target of MiR-155 and a potential predictor of favorable prognosis in pancreatic cancer. J Gastrointest Surg. 2013;17(8):1399–1405. doi: 10.1007/s11605-013-2230-5. [DOI] [PubMed] [Google Scholar]
- Yu J, Li A, Hong SM. et al. MicroRNA alterations of pancreatic intraepithelial neoplasias. Clin Cancer Res. 2012;18(4):981–992. doi: 10.1158/1078-0432.CCR-11-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S, Saleh H, Sethi S. et al. MicroRNA profiling of diagnostic needle aspirates from patients with pancreatic cancer. Br J Cancer. 2012;107(8):1354–1360. doi: 10.1038/bjc.2012.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Raulefs S, Bruns P. et al. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol Cancer. 2015;14:94. doi: 10.1186/s12943-015-0358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Abou Tayoun AN, Abo KM. et al. MicroRNAs as diagnostic markers for pancreatic ductal adenocarcinoma and its precursor, pancreatic intraepithelial neoplasm. Cancer Genet. 2013;206(6):217–221. doi: 10.1016/j.cancergen.2013.05.020. [DOI] [PubMed] [Google Scholar]
- Vila-Navarro E, Vila-Casadesus M, Moreira L. et al. MicroRNAs for Detection of Pancreatic Neoplasia: Biomarker Discovery by Next-generation Sequencing and Validation in 2 Independent Cohorts. Ann Surg. 2016 doi: 10.1097/SLA.0000000000001809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C, Shen Y, Yang T. et al. Diagnostic value of microRNA for pancreatic cancer: a meta-analysis. Arch Med Sci. 2012;8(5):749–755. doi: 10.5114/aoms.2012.31609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YY, Zheng JY, Liu J. et al. miR-183 induces cell proliferation, migration, and invasion by regulating PDCD4 expression in the SW1990 pancreatic cancer cell line. Biomed Pharmacother. 2015;70:151–157. doi: 10.1016/j.biopha.2015.01.016. [DOI] [PubMed] [Google Scholar]
- Miao F, Zhu J, Chen Y. et al. MicroRNA-183-5p promotes the proliferation, invasion and metastasis of human pancreatic adenocarcinoma cells. Oncol Lett. 2016;11(1):134–140. doi: 10.3892/ol.2015.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JK, Henry JC, Jiang J. et al. miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem Biophys Res Commun. 2011;406(4):518–523. doi: 10.1016/j.bbrc.2011.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Hao J, Xie F. et al. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis. 2011;32(8):1183–1189. doi: 10.1093/carcin/bgr105. [DOI] [PubMed] [Google Scholar]
- Zhou L, Zhang WG, Wang DS. et al. MicroRNA-183 is involved in cell proliferation, survival and poor prognosis in pancreatic ductal adenocarcinoma by regulating Bmi-1. Oncol Rep. 2014;32(4):1734–1740. doi: 10.3892/or.2014.3374. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A. et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi M, Teimoori-Toolabi L, Arzanani MK. et al. MicroRNA-148b and microRNA-152 reactivate tumor suppressor genes through suppression of DNA methyltransferase-1 gene in pancreatic cancer cell lines. Cancer Biol Ther. 2014;15(4):419–427. doi: 10.4161/cbt.27630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Zhang JG, Liu Y. et al. miR-148b functions as a tumor suppressor in pancreatic cancer by targeting AMPKalpha1. Mol Cancer Ther. 2013;12(1):83–93. doi: 10.1158/1535-7163.MCT-12-0534-T. [DOI] [PubMed] [Google Scholar]
- Qazi AM, Gruzdyn O, Semaan A. et al. Restoration of E-cadherin expression in pancreatic ductal adenocarcinoma treated with microRNA-101. Surgery. 2012;152(4):704–711. doi: 10.1016/j.surg.2012.07.020. [DOI] [PubMed] [Google Scholar]
- Bao B, Ali S, Banerjee S. et al. Curcumin analogue CDF inhibits pancreatic tumor growth by switching on suppressor microRNAs and attenuating EZH2 expression. Cancer Res. 2012;72(1):335–345. doi: 10.1158/0008-5472.CAN-11-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao B, Wang Z, Ali S. et al. Metformin inhibits cell proliferation, migration and invasion by attenuating CSC function mediated by deregulating miRNAs in pancreatic cancer cells. Cancer Prev Res (Phila) 2012;5(3):355–364. doi: 10.1158/1940-6207.CAPR-11-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor E, Waghray M, Lee CJ. et al. Bmi1 enhances tumorigenicity and cancer stem cell function in pancreatic adenocarcinoma. PLoS ONE. 2013;8(2):e55820. doi: 10.1371/journal.pone.0055820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali SR, Humphreys KJ, McKinnon RA. et al. Impact of Histone Deacetylase Inhibitors on microRNA Expression and Cancer Therapy: A Review. Drug Dev Res. 2015;76(6):296–317. doi: 10.1002/ddr.21268. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Maruyama R, Yamamoto E. et al. Epigenetic alteration and microRNA dysregulation in cancer. Front Genet. 2013;4:258. doi: 10.3389/fgene.2013.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Gu Y, Li Z. et al. miR-615-5p is epigenetically inactivated and functions as a tumor suppressor in pancreatic ductal adenocarcinoma. Oncogene. 2015;34(13):1629–1640. doi: 10.1038/onc.2014.101. [DOI] [PubMed] [Google Scholar]
- Hanoun N, Delpu Y, Suriawinata AA. et al. The silencing of microRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis. Clin Chem. 2010;56(7):1107–1118. doi: 10.1373/clinchem.2010.144709. [DOI] [PubMed] [Google Scholar]
- Wang P, Chen L, Zhang J. et al. Methylation-mediated silencing of the miR-124 genes facilitates pancreatic cancer progression and metastasis by targeting Rac1. Oncogene. 2014;33(4):514–524. doi: 10.1038/onc.2012.598. [DOI] [PubMed] [Google Scholar]
- Feng H, Wang Y, Su J. et al. MicroRNA-148a Suppresses the Proliferation and Migration of Pancreatic Cancer Cells by Down-regulating ErbB3. Pancreas. 2016;45(9) Suppl 1:1263–1271. doi: 10.1097/MPA.0000000000000677. [DOI] [PubMed] [Google Scholar]
- Zhang R, Li M, Zang W. et al. MiR-148a regulates the growth and apoptosis in pancreatic cancer by targeting CCKBR and Bcl-2. Tumour Biol. 2014;35(1):837–844. doi: 10.1007/s13277-013-1115-2. [DOI] [PubMed] [Google Scholar]
- Li A, Omura N, Hong SM. et al. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 2010;70(13):5226–5237. doi: 10.1158/0008-5472.CAN-09-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, He C, Deng S. et al. MiR-548an, Transcriptionally Downregulated by HIF1alpha/HDAC1, Suppresses Tumorigenesis of Pancreatic Cancer by Targeting Vimentin Expression. Mol Cancer Ther. 2016;15(9):2209–2219. doi: 10.1158/1535-7163.MCT-15-0877. [DOI] [PubMed] [Google Scholar]
- Li BS, Liu H, Yang WL. Reduced miRNA-218 expression in pancreatic cancer patients as a predictor of poor prognosis. Genet Mol Res. 2015;14(4):16372–16378. doi: 10.4238/2015.December.9.5. [DOI] [PubMed] [Google Scholar]
- Li CH, To KF, Tong JH. et al. Enhancer of zeste homolog 2 silences microRNA-218 in human pancreatic ductal adenocarcinoma cells by inducing formation of heterochromatin. Gastroenterology. 2013;144(5):1086–1097. doi: 10.1053/j.gastro.2013.01.058. [DOI] [PubMed] [Google Scholar]
- Hessmann E, Johnsen SA, Siveke JT. et al. Epigenetic treatment of pancreatic cancer: is there a therapeutic perspective on the horizon? Gut. 2016 doi: 10.1136/gutjnl-2016-312539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klieser E, Swierczynski S, Mayr C. et al. Role of histone deacetylases in pancreas: Implications for pathogenesis and therapy. World J Gastrointest Oncol. 2015;7(12):473–483. doi: 10.4251/wjgo.v7.i12.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kampen JG, Marijnissen-van Zanten MA, Simmer F. et al. Epigenetic targeting in pancreatic cancer. Cancer Treat Rev. 2014;40(5):656–664. doi: 10.1016/j.ctrv.2013.12.002. [DOI] [PubMed] [Google Scholar]
- Marks DL, Olson RL, Fernandez-Zapico ME. Epigenetic control of the tumor microenvironment. Epigenomics. 2016;8(12):1671–1687. doi: 10.2217/epi-2016-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillmore CM, Xu C, Desai PT. et al. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature. 2015;520(546):239–242. doi: 10.1038/nature14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Wang X, Shen X. et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18(4):316–328. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitler BG, Aird KM, Garipov A. et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21(3):231–238. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SK, Warholic NM, Wigle TJ. et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A. 2013;110(19):7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]