Abstract
Gene expression programs are largely regulated by the tissue-specific expression of lineage-defining transcription factors or by the inducible expression of transcription factors in response to specific stimuli. Here I will review our own work over the last 20 years to show how specific activation signals also lead to the wide-spread re-distribution of pre-existing constitutive transcription factors to sites undergoing chromatin reorganization. I will summarize studies showing that activation of kinase signaling pathways creates open chromatin regions that recruit pre-existing factors which were previously unable to bind to closed chromatin. As models I will draw upon genes activated or primed by receptor signaling in memory T cells, and genes activated by cytokine receptor mutations in acute myeloid leukemia. I also summarize a hit-and-run model of stable epigenetic reprograming in memory T cells, mediated by transient Activator Protein 1 (AP-1) binding, which enables the accelerated activation of inducible enhancers.
Keywords: Transcription, epigenetics, immunological memory, leukemia
Introduction
Vertebrate development requires the progressive differentiation of stem cells into all the tissues that make up the whole animal. This is accompanied by many consecutive lineage choices at branch points where cells choose alternate fates. Cellular differentiation is typically accompanied by the activation of a new program of gene expression dictated by the activation of lineage-defining transcription factor genes. Cells also have the capacity to express inducible transcription factors that enable responses to specific extra-cellular signals. These responses are initiated by a wide variety surface receptors that allow cells to respond to regulatory molecules controlling cell growth, differentiation and survival, or to signals that trigger specific reactions such as immune responses. In this review I will focus primarily on (a) T Cell Receptor (TCR) signals that not only induce immune response genes but also prime them for subsequent responses [1], and (b) The receptor FLT3 which maintains myeloid progenitor cells and is frequently mutated in Acute Myeloid Leukemia (AML) [2]. In both cases I will describe how receptor activation opens up newly accessible regions of chromatin and enables the binding of pre-existing factors such as RUNX1 that cannot otherwise bind to these sites when they are occupied by nucleosomes.
Chromatin Remodeling Directed by Inducible Factors
The vast bulk of the genome is occupied by regularly spaced nucleosomes that assemble as highly condensed chromatin fibers. Most nucleosomes comprise ~ 146 bp of DNA wrapped around a histone protein octamer made up of two molecules each of histones H2A, H2B, H3 and H4 [3]. Nucleosomes within chromatin are on average spaced ~ 185 to 195 bp apart and typically exist as a highly compacted fiber which at the lowest level of compaction is ~ 30 nm in diameter. Significantly, much of the DNA occupied by nucleosomes is inaccessible to many transcription factors (TF) under normal conditions. Tightly regulated transcriptional enhancers (such as the GM-CSF enhancer [4]) are often encompassed by nucleosomes, and are dependent upon the activation of specific TFs that recruit remodelers which either disrupt or reposition nucleosomes [5,6]. The mechanisms regulating this process involve a wide variety of histone modifying enzymes and chromatin remodelers and these have been described in detail previously [3,5,7-10].
Inducible Disruption of Nucleosomes by TCR-inducible Factors
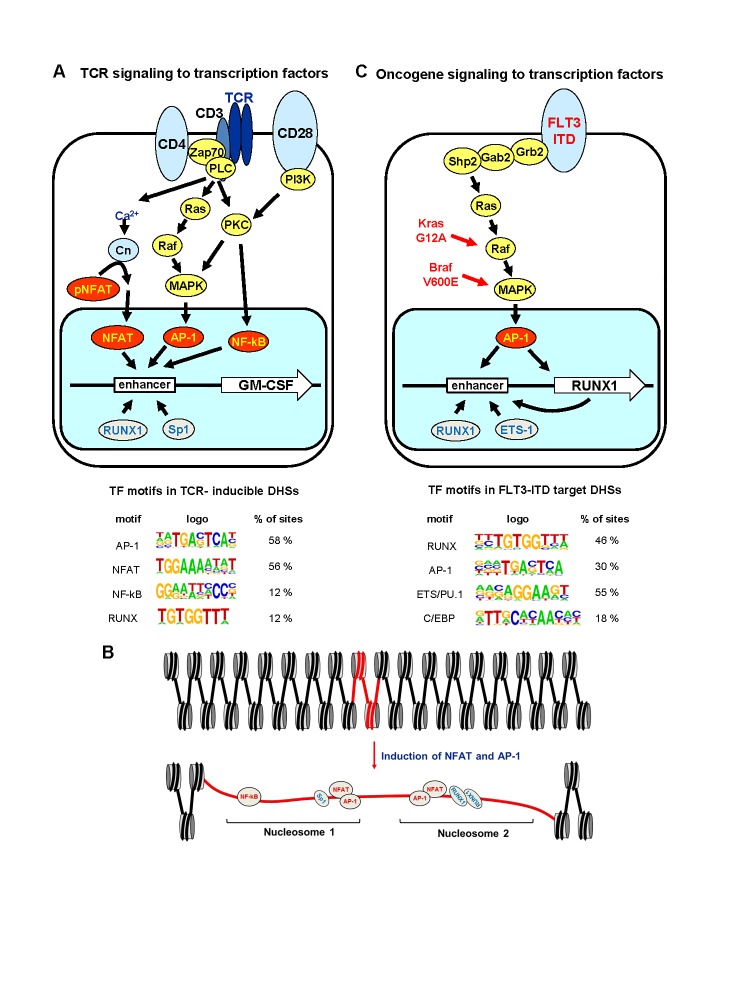
Inducible genes for cytokines such as IL-2, IL-3 and GM-CSF are activated in T cells primarily in response to TCR signaling to the NFAT, AP-1, and NF-κB families of inducible TFs (Figure 1A) [11,12]. NFAT represents a major target of Ca2+ signaling and AP-1 and NF-κB represent major targets of kinase signaling pathways, such as PKC and MAPK [12-15]. We have used the human GM-CSF locus extensively as a model for studying mechanisms of locus activation by TCR signaling. We demonstrated that the GM-CSF gene is regulated by a highly inducible enhancer 3 kb upstream of the gene which encompasses two essential composite NFAT/AP-1 elements and rapidly forms an inducible DNaseI Hypersensitive Site (DHS) via mechanisms dependent on both NFAT and AP-1 [4,16-18]. The activation of this enhancer involves the displacement of two positioned nucleosomes that normally occupy most of the known TF binding sites within the enhancer (Figure 1B) [4]. These sites include binding sites for the constitutively expressed factors Sp1 and RUNX1 [18,19]. Significantly, we used in vivo footprinting to show that these pre-existing TFs can only occupy the enhancer in T cells and mast cells after the DHS has been induced by NFAT and AP-1 [4,20]. This is consistent with a model whereby the binding of TFs to their recognition sequences is in many cases tightly controlled at the level of chromatin accessibility.
Figure 1.
Global redistribution of TF binding in T cells and AML cells in response to activation of receptor signaling. A. TCR signaling pathways linked to the activation of inducible genes such as GM-CSF in T cells. Shown underneath are TF motifs that were found to be enriched in a population of ~1000 inducible DHSs identified in a global analysis of stimulated mouse T cells [1]; B. Scale model of the chromatin architecture and TF occupancy at the human GM-CSFenhancer in T cells before and after activation of TCR signaling; C. Gene regulatory network activated via MAPK signaling pathways by FLT3, Ras and Raf gene mutations in AML. Shown underneath are TF motifs that were found to be enriched in a population of ~1000 DHSs that are specifically enriched in AML carrying FLT-ITD mutations [2].
Genome-wide studies of inducible regulatory elements further confirmed the general principle whereby there is global redistribution of the binding of constitutively expressed TFs to inducible DHSs in T cells in response to TCR signaling [1]. RUNX1 chromatin immunoprecipitation (ChIP-Seq) analyses of inducible DHSs revealed that many of these sites exhibit de novo binding of RUNX1 in parallel with AP-1 in response to stimulation of Ca2+ and MAPK signaling pathways. Furthermore, binding motifs for RUNX factors are enriched in both inducible and constitutive DHSs in T cells [1]. RUNX1 binding sites were present in 12 percent of the inducible DHSs detected in T cells (Figure 1A) [1]. This is consistent with a model whereby inducible activation of transcription is mediated by inducible factors working in close cooperation with constitutively expressed factors.
Others have shown that binding of specific TFs to nucleosomal DNA becomes unfavorable once the site is positioned more than about 20 bp from the DNA exit point on the nucleosome [21]. In the case of the GM-CSF enhancer, the binding sites for Sp1 and RUNX1 are located deep inside the nucleosome [4], which is why binding to these sites requires nucleosome disruption by other factors. These data are summarized in the model shown in Figure 1B. However, there are exceptions to these general observations such as a group of TFs referred to as pioneer factors. These include the FOX forkhead family of TFs which are specialists at binding to nucleosomal DNA [22]. These TFs function as pioneer factors by mimicking histone H1 and binding to the nucleosomal dyad to open up condensed chromatin fibers. In contrast, AP-1 would appear to be a factor which primarily disrupts nucleosomes within relatively accessible chromatin [1,4] by recruiting remodelers such as Brg1, and histone modifiers such as CBP [23-25]. In this context, inducible TFs such as AP-1 function by very different mechanisms than pioneer factors which function at a much higher level of chromatin structure. However, when a DHS is drawn to scale, as in Figure 1B, it reveals the extent to which DNA is made accessible (i.e. hypersensitive) by the unraveling of nucleosomes. In this example, the unraveling of two nucleosomes is expected to free up ~ 400 bp of DNA which is equivalent to ~ 15 nucleosome diameters [3].
Receptor Mutations in Leukemia Lead to Redistribution of Constitutive TFs
Acute Myeloid Leukemia (AML) provides another context whereby activation of receptor signaling leads to the de novo binding of both inducible and constitutive factors. FLT3 is a cytokine receptor that is frequently mutated via internal tandem duplications (ITD), leading to constitutive activation of multiple signaling pathways, including MAPK (Figure 1C) [26-28]. Genome-wide analyses of DHSs (DNase-Seq) of AML samples carrying different mutations revealed the existence of a subset of over a thousand DHSs specifically enriched in FLT3-ITD-positive AML [2]. This subset of FLT3-ITD-specific DHSs was also highly enriched for binding sites for constitutively expressed ETS and RUNX factors in addition to inducible AP-1 factors (Figure 1C). RUNX1 ChIP-Seq confirmed that many of these de novo DHSs recruited RUNX1 [2]. These data implied that AP-1 is a major target of FLT3-ITD and are consistent with the above model derived from T cells whereby induction of AP-1 leads to destabilization of nucleosomes and de novo binding of constitutive TFs to sites that were previously inaccessible. As depicted in Figure 1C, it is likely that a similar global redistribution of pre-existing TFs will be also found in leukemia carrying activating mutations in Ras and Raf family members such as Kras G12A, Nras G12V and Braf V600E [29,30].
Transient Receptor Signaling in T cells Leads to Stable Maintenance of Epigenetic Priming and Constitutive TF Binding in Memory T cells
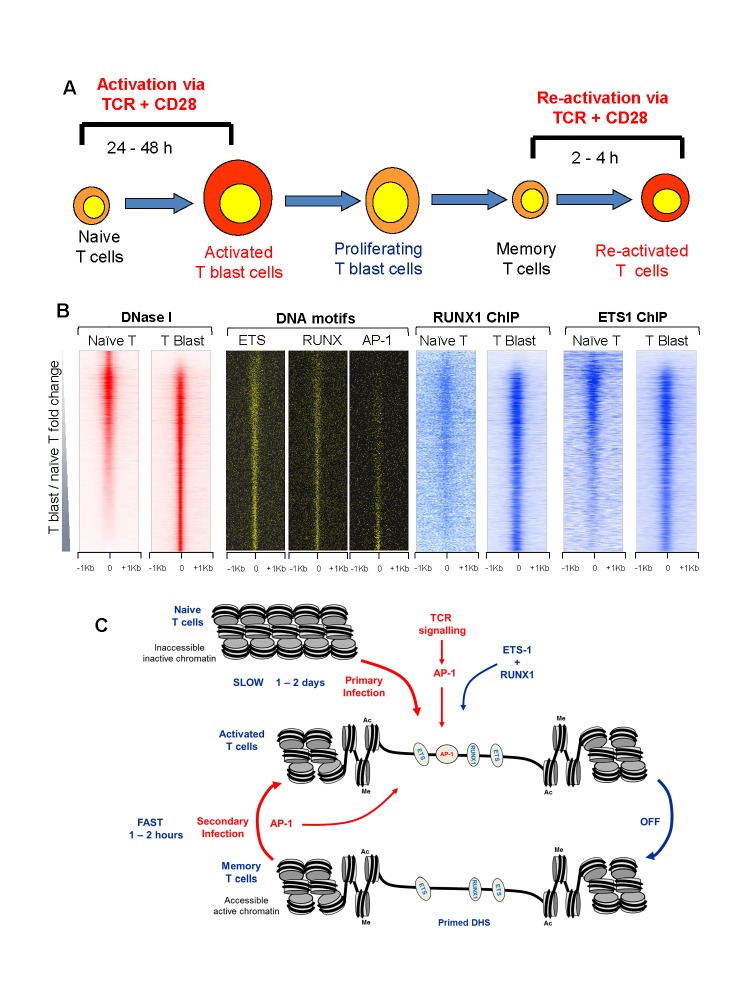
When naïve T cells are stimulated for the first time they undergo an extensive program of chromatin remodeling that takes place over a period of 24 to 48 hours as they become transformed to rapidly dividing T blast cells [31] (Figure 2A). Many of the inducible genes expressed by activated effector T cells remain highly unresponsive in naïve T cells until they have completed the process of blast cell transformation [1]. However, these genes can be induced in a few hours in T blast cells, and this property is stably retained in memory T cells which have returned to the quiescent state [1,32-35].
Figure 2.
Mechanisms of establishing and maintaining immunological memory in T cells. A. Stages of T cell activation and differentiation; B. DNase-Seq and TF ChIP-Seq, plus motif locations, for 2 Kb segments of the mouse genome spanning the ~17,000 strongest DHSs in naïve CD4 T cells and CD4 T blast cells, ranked in order of increasing signal in T blast cells relative to naïve T cells [2]; C. Scale model depicting mechanisms of TF binding and epigenetic re-programming of activated and memory T cells.
In order to understand the molecular basis of the establishment of immunological memory in T cells, we performed an extensive series of DNA-Seq and ChIP-Seq analyses in naïve T cells, T blast cells, and memory T cells [1]. These studies revealed ~ 3000 DHSs that were first acquired by T blast cells during the transformation process and then stably maintained in memory T cells. Reminiscent of the inducible DHSs and the FLT3-ITD-AML-specific DHSs, these memory T cell-specific DHSs were also enriched for ETS, RUNX1 and AP-1 motifs. However, in this case AP-1 was only bound by these DHSs in direct response to ongoing TCR signaling, whereas ETS-1 and RUNX1 remained stably bound to these sites for many cell cycles after the removal of TCR signaling stimuli (Figure 2B). Hence, it appears that even transient activation of AP-1 can lead to the creation of de novo DHSs which are then stably maintained by newly recruited constitutively expressed TFs once the AP-1 stimulus is removed. This simple hit-and-run mechanism provides an elegant mechanisms that can account for both the establishment and the maintenance of immunological memory in T cells (Figure 2C).
The memory T cell-specific DHSs functioned as locus priming elements by maintaining islands of active chromatin marked by histone H3 K4 me2 and H3 K4 Ac27 modifications in the vicinity of inducible enhancers [1]. This pairing of priming elements, which do not typically have intrinsic enhancer activity of their own, with inducible enhancers has the additional potential to explain (i) why the kinetics of inducible gene activation is so much faster in memory T cells and effector T cells than in naive T cells, and (ii) how chromatin priming can be maintained without also activating steady state levels of transcription.
Conclusions and Outlook
It is to be expected that the general principles outlined above will be reiterated in essentially all other model systems whereby inducible factors invoke a specific program of gene expression in response to specific stimuli. What is fascinating is the concept that in some cases the need for a specific factor is transient. Once specific loci have been reprogrammed by one factor, they can in some cases be maintained as open chromatin by additional TFs recruited in response to the first factor. This may also be the case for RUNX1 in the reprogramming of haemopoietic lineage cells [36].
In the case of memory T cells, we have established that ongoing TCR signaling is not required to maintain regions of active chromatin which were initially reprogrammed by TCR signaling. However, it will be important for future studies to investigate the role of signaling from other types of receptors, such as cytokine and TNF super family receptors (TNFRSF), in the long term reinforcement of epigenetic priming at inducible loci in memory T cells. There is already an indication that this is likely to be the case as immunological memory is deficient in cells lacking TNFRSF4/OX40 [37,38] or IL-7 [39].
Acknowledgments
This work was supported by the BBSRC and by Bloodwise. I would like to especially thank Constance Bonifer, Sarah Bevington, Pierre Cauchy, Sally James, Fernando J. Calero-Nieto, and Andrew Bert for all their work and help that went into the research upon which this article is based.
Glossary
- AML
Acute myeloid leukemia
- AP-1
Activator Protein 1
- bp
base pairs
- ChIP
Chromatin immune-precipitation
- DHS
DNase I Hypersensitive Site
- GM-CSF
Granulocyte-Macrophage Colony-Stimulating Factor
- ITD
Internal Tandem Duplication
- Kb
Kilobases
- MAPK
Mitogen Activated Protein Kinase
- NFAT
Nuclear Factor of Activated T cells
- PKC
Protein Kinase C
- TCR
T cell Receptor
- TF
Transcription factor
References
- Bevington SL, Cauchy P, Piper J. et al. Inducible chromatin priming is associated with the establishment of immunological memory in T cells. EMBO J. 2016;35(5):515–535. doi: 10.15252/embj.201592534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchy P, James SR, Zacarias-Cabeza J. et al. Chronic FLT3-ITD Signaling in Acute Myeloid Leukemia Is Connected to a Specific Chromatin Signature. Cell Rep. 2015;12(5):821–836. doi: 10.1016/j.celrep.2015.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill PN. Structure and function of active chromatin and DNase I hypersensitive sites. FEBS J. 2011;278(13):2182–2210. doi: 10.1111/j.1742-4658.2011.08128.x. [DOI] [PubMed] [Google Scholar]
- Johnson BV, Bert AG, Ryan GR. et al. GM-CSF enhancer activation requires cooperation between NFAT and AP-1 elements and is associated with extensive nucleosome reorganization. Mol Cell Biol. 2004;24(18):7914–7930. doi: 10.1128/MCB.24.18.7914-7930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461(7261):193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- Field Y, Fondufe-Mittendorf Y, Moore IK. et al. Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat Genet. 2009;41(4):438–445. doi: 10.1038/ng.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9(1):15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet. 2010;11(6):426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- Macian F, Lopez-Rodriguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20(19):2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- Shannon MF, Coles LS, Vadas MA. et al. Signals for activation of the GM-CSF promoter and enhancer in T cells. Crit Rev Immunol. 1997;17(3-4):301–323. doi: 10.1615/critrevimmunol.v17.i3-4.30. [DOI] [PubMed] [Google Scholar]
- Hogan PG, Chen L, Nardone J. et al. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17(18):2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- Serfling E, Berberich-Siebelt F, Chuvpilo S. et al. The role of NF-AT transcription factors in T cell activation and differentiation. Biochim Biophys Acta. 2000;1498(1):1–18. doi: 10.1016/s0167-4889(00)00082-3. [DOI] [PubMed] [Google Scholar]
- Brownlie RJ, Zamoyska R. T cell receptor signalling networks: branched, diversified and bounded. Nat Rev Immunol. 2013;13(4):257–269. doi: 10.1038/nri3403. [DOI] [PubMed] [Google Scholar]
- Cockerill PN, Bert AG, Jenkins F. et al. Human GM-CSF enhancer function is associated with cooperative interactions between AP-1 and NFATp/c. Mol Cell Biol. 1995;15(4):2071–2079. doi: 10.1128/mcb.15.4.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill PN, Shannon MF, Bert AG. et al. The granulocyte-macrophage colony-stimulating factor/interleukin 3 locus is regulated by an inducible cyclosporin A-sensitive enhancer. Proc Natl Acad Sci U S A. 1993;90(6):2466–2470. doi: 10.1073/pnas.90.6.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bert AG, Johnson BV, Baxter EW. et al. A modular enhancer is differentially regulated by GATA and NFAT elements that direct different tissue-specific patterns of nucleosome positioning and inducible chromatin remodeling. Mol Cell Biol. 2007;27(8):2870–2885. doi: 10.1128/MCB.02323-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill PN, Osborne CS, Bert AG. et al. Regulation of GM-CSF gene transcription by core-binding factor. Cell Growth Differ. 1996;7(7):917–922. [PubMed] [Google Scholar]
- Bowers SR, Calero-Nieto FJ, Valeaux S. et al. Runx1 binds as a dimeric complex to overlapping Runx1 sites within a palindromic element in the human GM-CSF enhancer. Nucleic Acids Res. 2010;38(18):6124–6134. doi: 10.1093/nar/gkq356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams CC, Workman JL. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol Cell Biol. 1995;15(3):1405–1421. doi: 10.1128/mcb.15.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I. et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9(2):279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Ndlovu MN, Van Lint C, Van Wesemael K. et al. Hyperactivated NF-{kappa}B and AP-1 transcription factors promote highly accessible chromatin and constitutive transcription across the interleukin-6 gene promoter in metastatic breast cancer cells. Mol Cell Biol. 2009;29(20):5488–5504. doi: 10.1128/MCB.01657-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Yamauchi M, Nishina M. et al. Identification of SWI.SNF complex subunit BAF60a as a determinant of the transactivation potential of Fos/Jun dimers. J Biol Chem. 2001;276(4):2852–2857. doi: 10.1074/jbc.M009633200. [DOI] [PubMed] [Google Scholar]
- Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114(Pt 13):2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- Stirewalt DL, Radich JP. The role of FLT3 in haematopoietic malignancies. Nat Rev Cancer. 2003;3(9):650–665. doi: 10.1038/nrc1169. [DOI] [PubMed] [Google Scholar]
- Thiede C, Steudel C, Mohr B. et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99(12):4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- Masson K, Ronnstrand L. Oncogenic signaling from the hematopoietic growth factor receptors c-Kit and Flt3. Cell Signal. 2009;21(12):1717–1726. doi: 10.1016/j.cellsig.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Falini B, Martelli MP, Tiacci E. BRAF-V600E mutation in hairy cell leukemia: from bench to bedside. Blood. 2016;128(15):1918–1927. doi: 10.1182/blood-2016-07-418434. [DOI] [PubMed] [Google Scholar]
- Renneville A, Roumier C, Biggio V. et al. Cooperating gene mutations in acute myeloid leukemia: a review of the literature. Leukemia. 2008;22(5):915–931. doi: 10.1038/leu.2008.19. [DOI] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ. et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95(5):625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- Zediak VP, Johnnidis JB, Wherry EJ. et al. Cutting edge: persistently open chromatin at effector gene loci in resting memory CD8+ T cells independent of transcriptional status. J Immunol. 2011;186(5):2705–2709. doi: 10.4049/jimmunol.1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zediak VP, Wherry EJ, Berger SL. The contribution of epigenetic memory to immunologic memory. Curr Opin Genet Dev. 2011;21(2):154–159. doi: 10.1016/j.gde.2011.01.016. [DOI] [PubMed] [Google Scholar]
- Rogers PR, Dubey C, Swain SL. Qualitative changes accompany memory T cell generation: faster, more effective responses at lower doses of antigen. J Immunol. 2000;164(5):2338–2346. doi: 10.4049/jimmunol.164.5.2338. [DOI] [PubMed] [Google Scholar]
- Sprent J, Surh CD. T cell memory. Annu Rev Immunol. 2002;20:551–579. doi: 10.1146/annurev.immunol.20.100101.151926. [DOI] [PubMed] [Google Scholar]
- Chen MJ, Yokomizo T, Zeigler BM. et al. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457(7231):887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun M, Ma CS, Akcay A. et al. Inherited human OX40 deficiency underlying classic Kaposi sarcoma of childhood. J Exp Med. 2013;210(9):1743–1759. doi: 10.1084/jem.20130592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspal FM, Kim MY, McConnell FM. et al. Mice deficient in OX40 and CD30 signals lack memory antibody responses because of deficient CD4 T cell memory. J Immunol. 2005;174(7):3891–3896. doi: 10.4049/jimmunol.174.7.3891. [DOI] [PubMed] [Google Scholar]
- Seddon B, Tomlinson P, Zamoyska R. Interleukin 7 and T cell receptor signals regulate homeostasis of CD4 memory cells. Nat Immunol. 2003;4(7):680–686. doi: 10.1038/ni946. [DOI] [PubMed] [Google Scholar]