Abstract
Actinoalloteichus hymeniacidonis HPA 177T is a Gram-positive, strictly aerobic, black pigment producing and spore-forming actinomycete, which forms branching vegetative hyphae and was isolated from the marine sponge Hymeniacidon perlevis.
Actinomycete bacteria are prolific producers of secondary metabolites, some of which have been developed into anti-microbial, anti-tumor and immunosuppressive drugs currently used in human therapy. Considering this and the growing interest in natural products as sources of new drugs, actinomycete bacteria from the hitherto poorly explored marine environments may represent promising sources for drug discovery.
As A. hymeniacidonis, isolated from the marine sponge, is a type strain of the recently described and rare genus Actinoalloteichus, knowledge of the complete genome sequence enables genome analyses to identify genetic loci for novel bioactive compounds. This project, describing the 6.31 Mbp long chromosome, with its 5346 protein-coding and 73 RNA genes, will aid the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: Actinoalloteichus, Strictly aerobic, Non-motile, Gram-positive, Non-acid-fast, Branching vegetative hyphae, Spore forming, Secondary metabolite biosynthesis gene clusters
Introduction
Strain HPA 177T is the type strain of the species Actinoalloteichus hymeniacidonis, it was isolated from the marine sponge Hymeniacidon perlevis at the intertidal beach of Dalian, Yellow Sea, North-China, during investigation of its actinomycete diversity [1].
Members of the diverse order Actinomycetales are a major source of a variety of novel bioactive and possibly pharmaceutically important compounds and drugs, such as anticancer agents [2–4], antibiotics [5, 6] and also other industrially relevant molecules and enzymes with diverse biological activities [5, 7]. Especially marine actinomycetes became a focus of research since they have evolved the greatest genomic and metabolic diversity and are auspicious sources of novel secondary metabolites and enzymes [5, 7–9].
The comparison of the complete genome sequences of members of the rare genus Actinoalloteichus might unravel unknown gene clusters dedicated to the biosynthesis of such molecules as bioactive secondary metabolites and enzymes. This has already been demonstrated for the genomes of strains belonging to closely related genera, such as Kutzneria, Saccharomonospora, Crossiella , Kibdelosporangium, and Streptoalloteichus [10–19].
Organism information
Classification and features
The genus Actinoalloteichus was established by Tamura et al. (2000) on the basis of morphological, physiological, chemotaxonomic and phylogenetic criteria. The genus contains Gram-positive, non-acid-fast, aerobic organisms with branching vegetative hyphae [20]. The aerial mycelium of Actinoalloteichus develops straight spore chains [20]. According to 16S rDNA gene sequence analysis Actinoalloteichus is part of the family Pseudonocardiaceae, suborder Pseudonocardineae, order Actinomycetales, class Actinobacteria [20, 21] (Table 1). It differs from other genera of its family by its morphological characteristics, fatty acid components and its non-motility [20].
Table 1.
Classification and general features of Actinoalloteichus hymeniacidonis HPA 177T according to the MIGS recommendations [46]
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [47] | |
| Phylum ‘Actinobacteria’ | TAS [48] | ||
| Class Actinobacteria | TAS [21] | ||
| Order Actinomycetales | TAS [49, 50] | ||
| Suborder Pseudonocardianeae | TAS [51] | ||
| Family Pseudonocardiaceae | TAS [51, 52] | ||
| Genus Actinoalloteichus | TAS [20] | ||
| Species Actinoalloteichus hymeniacidonis | TAS [1] | ||
| Type-strain HPA177T (DSM 45092 = CGMCC 4.2500 = JCM 13436) | TAS [1] | ||
| Gram stain | positive | TAS [1] | |
| Cell shape | branching hyphae | TAS [1] | |
| Motility | non-motile | NAS | |
| Sporulation | straight spores in aerial mycelia | TAS [1] | |
| Temperature range | mesophile (15–45 °C) | TAS [1] | |
| Optimum temperature | not reported | ||
| pH range, optimum | not reported | ||
| Carbon source | fructose, glucose, maltose, mannitol, mannose, xylose, rhamnose, sucrose, sorbitol, citrate | TAS [1] | |
| MIGS-6 | Habitat | Microbiological community of the intertidal marine sponge Hymeniacidon perlevis | TAS [1] |
| MIGS-6.3 | Salinity | not reported | |
| MIGS-22 | Oxygen requirement | Aerobic | TAS [1] |
| MIGS-15 | Biotic relationship | Commensal | TAS [1] |
| MIGS-14 | Pathogenicity | non-pathogen | NAS |
| MIGS-4 | Geographic location | China: inter-tidal beach of Dalian, Yellow Sea | TAS [1] |
| MIGS-5 | Sample collection time | not reported | |
| MIGS-4.1 | Latitude | 38°52′ N | TAS [1] |
| MIGS-4.2 | Longitude | 121°41′ E | TAS [1] |
| MIGS-4.4 | Altitude | not reported |
aEvidence codes - TAS Traceable Author Statement (i.e., a direct report exists in the literature), NAS Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [53]
The genus Actinoalloteichus currently contains only five known species. Besides Actinoalloteichus hymeniacidonis HPA 177T the other currently known members are the halophilic Actinoalloteichus hoggarensis [22], Actinoalloteichus nanshanensis , isolated from the rhizosphere of a fig tree [23], the soil bacterium Actinoalloteichus spitiensis [24] and Actinoalloteichus cyanogriseus , the type species of the genus isolated from a soil sample collected from the Yunnan province of China [20].
A representative 16S rRNA sequence of A. hymeniacidonis HPA 177T was compared to the Ribosomal Database Project database [25] confirming the initial taxonomic classification. On the basis of the 16S rDNA, A. hymeniacidonis shows highest similarity to A. hoggarensis AH97T (99.2%) and A. nanshanensis NEAU119T (98.3%). Together with A. spitiensis DSM 44848 T (96.8%) and A. cyanogriseus IFO 14455T (96.4%), they form a distinct clade within the family Pseudonocardiaceae. Figure 1 shows the phylogenetic neighborhood of A. hymeniacidonis in a 16S rRNA gene based tree.
Fig. 1.

Phylogenetic tree highlighting the position of A. hymeniacidonis HPA 177T (given in bold) relative to type strains of other species within the genus Actinoalloteichus and related genera of the family Pseudonocardiaceae. The tree uses sequences aligned by the RDP aligner. Using the Jukes-Cantor corrected distance model, a distance matrix is constructed based on alignment model positions without the use of alignment inserts, using a minimum comparable position of 200. The tree is built with RDP Tree Builder, which utilizes Weighbor [54] with an alphabet size of 4 and length size of 1000. The building of the tree also involves a bootstrapping process repeated 100 times to generate a majority consensus tree [55]. Streptomyces albus DSM 40313T was used as the root organism. Species for which a complete or draft genome sequence is available are underlined
A. hymeniacidonis HPA 177T forms branching vegetative hyphae (Fig. 2), which are grey to black in color and tend to fragment after 3 weeks of cultivation (1). The aerial hyphae develop spores of a dimension of 0.6 × 0.8 μm [1]. HPA 177T is strictly aerobic and non-motile [1]. Growth of A. hymeniacidonis was shown at temperatures between 15 and 45 °C (optimal growth between 20 and 37 °C) [1]. HPA 177T can utilize fructose, glucose, maltose, mannitol, mannose, xylose, rhamnose, sucrose, sorbitol, sodium citrate, casein, or starch as carbon sources, but not arabinose, inositol, and raffinose [1] (Table 1). It grows well on yeast extract/malt extract agar or oatmeal agar and produces a black soluble pigment when growing on yeast extract/malt extract agar as well as on peptone/yeast extract/iron agar [1]. It has been shown that the strain grows faster on ISP2 agar media prepared with 50% of artificial sea water, which, considering the source of isolation, probably reflects an adaptation to the marine environment. Urea is not decomposed by A. hymeniacidonis, and this strain shows neither hydrolysis of aesculin or hippurate, nor utilization of calcium malate, sodium oxalate, or sodium succinate nor reduction of nitrate [1].
Fig. 2.
Colony of A. hymeniacidonis HPA 177T grown at 28 °C for 8 days on ISP2 agar medium prepared with artificial sea water
Chemotaxonomic data
The cell wall of A. hymeniacidonis contains diaminopimelic acids (A2pm) [1]. The major menaquinone is MK-9(H4) (64%), followed by MK-9(H6) (23%) and MK-9(H8) (12%).
The phospholipids were shown to be mainly composed of phosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol, phosphatidylinositol mannoside as well as of some other glucosamine containing phospholipids of unknown structure as diagnostic polar lipids [1]. A. hymeniacidonis does not contain mycolic acids [1].
The cellular fatty acids are mainly composed of anteiso pentadecanoic acid (C15:0 anteiso) (20%), cis-8-heptadecenoic acid (C17:1 ω8c) (19%), isopalmitic acid (C16:0 iso) (16%), heptadecanoic acid (C17:0) (11%) and other fatty acids occurring in lower amounts [1]. Galactose, glucose, mannose, and ribose are whole cell sugars of HPA 177T [1].
Genome sequencing information
Genome project history
Due to the increasing interest in exploiting new and rare actinomycetes as new sources of novel secondary metabolites [5], Actinoalloteichus hymeniacidonis HPA 177T, a member of the rare genus Actinoalloteichus [20], was selected for sequencing. While not being part of the GEBA project [26], sequencing of the type strain will aid the GEBA effort. The genome project is deposited in the Genomes OnLine Database [27] and the complete genome sequence is deposited in GenBank. A summary of the project information is shown in Table 2.
Table 2.
Genome sequencing project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Nextera DNA Sample Prep Kit, Nextera Mate Pair Sample Prep Kit |
| MIGS-29 | Sequencing platforms | Illumina MiSeq |
| MIGS-31.2 | Fold coverage | 159.00× |
| MIGS-30 | Assemblers | Newbler version 2.8 |
| MIGS-32 | Gene calling method | GeneMark, Glimmer |
| Locus Tag | TL08 | |
| GenBank ID | CP014859 | |
| GenBank Date of Release | September 28, 2016 | |
| GOLD ID | Gp01114707 | |
| NCBI project ID | PRJNA273752 | |
| MIGS-13 | Source material identifier | DSM 45092 |
| Project relevance | Industrial, GEBA |
Growth conditions and DNA isolation
A. hymeniacidonis HPA 177T was grown aerobically in 50 ml 3% TSB medium (Oxoid, UK) in 250 mL baffled flasks at 28 °C, 250 rpm. Genomic DNA was isolated using Wizard Genomic DNA Purification Kit (Promega, USA) from ~2 g of mycelium (wet weight) using the manufacturer’s protocol with the following modification. The clarified lysate prior to precipitation of DNA with isopropanol was extracted once with ½ volume of a 1:1 mixture of phenol/chloroform (pH 8.0).
Genome sequencing and assembly
Two libraries were prepared: a WGS library using the Illumina-Compatible Nextera DNA Sample Prep Kit (Epicentre, WI, U.S.A.) and a 6 k MatePair library using the Nextera Mate Pair Sample Preparation Kit, both according to the manufacturer’s protocol. Both libraries were sequenced in a 2× 250 bp paired read run on the MiSeq platform, yielding 4,594,541 total reads, providing 159.00× coverage of the genome. Reads were assembled using the Newbler assembler v2.8 (Roche). The initial Newbler assembly consisted of 31 contigs in five scaffolds, with a total of 50 contigs larger than 100 bp. Analysis of the five scaffolds revealed three to make up the chromosome and the remaining two containing the three copies of the RRN operon.
The Phred/Phrap/Consed software package [28–31] was used for sequence assembly and quality assessment in the subsequent finishing process, gaps between contigs were closed by manual editing in Consed (for repetitive elements).
Genome annotation
Gene prediction and primary annotation were done using the IMG ER pipeline [32]. Additionally, genes were identified using GeneMark [33], GLIMMER [34], and Prodigal [35]. For annotation, BLAST searches against the NCBI Protein Clusters Database [36] were performed and the annotation was enriched by searches against the Conserved Domain Database [37] and subsequent assignment of coding sequences to COGs. Non-coding genes and miscellaneous features were predicted using tRNAscan-SE [38], Infernal [39], RNAMMer [40], Rfam [41], TMHMM [42], and SignalP [43].
Genome properties
The genome includes one circular chromosome of 6,306,386 bp (68.08% G+C content) (Fig. 3). Among a total of 5425 predicted genes, 5346 are protein coding genes. 4068 (74.90%) of the protein coding genes were assigned a putative function, the remaining were annotated as hypothetical proteins. The properties and the statistics of the genome are summarized in Tables 3 and 4, and the circular plot is shown in Fig. 3.
Fig. 3.
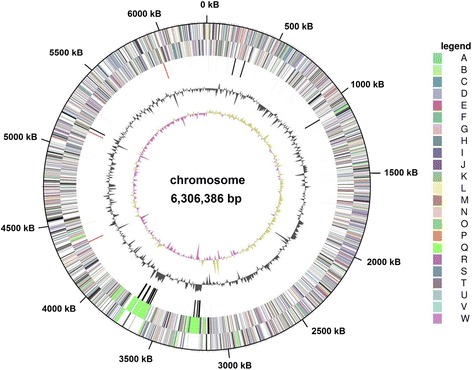
Graphical map of the chromosome of A. hymeniacidonis HPA 177T. From the outside to the center: Genes on forward strand (colored by COG categories), genes on reverse strand (colored by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), G+C content, G+C skew
Table 3.
Genome Statistics
| Attribute | Value | % of totala |
|---|---|---|
| Genome size (bp) | 6,306,386 | 100.00 |
| DNA coding (bp) | 5,516,402 | 87.47 |
| DNA G+C (bp) | 4,293,157 | 68.08 |
| DNA scaffolds | 1 | 100.00 |
| Total genes | 5425 | 100.00 |
| Protein-coding genes | 5346 | 98.54 |
| RNA genes | 73 | 1.34 |
| Pseudo genes | 6 | 0.11 |
| Genes with internal clusters | 753 | 13.86 |
| Genes with function prediction | 4068 | 74.90 |
| Genes assigned to COGs | 3329 | 61.30 |
| Genes with Pfam domains | 4327 | 79.67 |
| Genes with signal peptides | 381 | 7.02 |
| Genes with transmembrane helices | 1271 | 23.40 |
| CRISPR repeats | 15 |
aThe total is based on either the size of the genome in base pairs or the total number of total genes in the annotated genome
Table 4.
Number of genes associated with the general COG functional categories
| Code | value | % age | Description |
|---|---|---|---|
| J | 206 | 5.33 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.03 | RNA processing and modification |
| K | 439 | 11.36 | Transcription |
| L | 109 | 2.82 | Replication, recombination and repair |
| B | 1 | 0.03 | Chromatin structure and dynamics |
| D | 33 | 0.85 | Cell cycle control, cell division, chromosome partitioning |
| V | 150 | 3.88 | Defense mechanisms |
| T | 184 | 4.76 | Signal transduction mechanisms |
| M | 159 | 4.11 | Cell wall/membrane biogenesis |
| N | 7 | 0.18 | Cell motility |
| U | 29 | 0.75 | Intracellular trafficking and secretion, and vesicular transport |
| O | 136 | 3.52 | Posttranslational modification, protein turnover, chaperones |
| Z | Cytoskeleton | ||
| W | 4 | 0.1 | Extracellular structures |
| C | 213 | 5.51 | Energy production and conversion |
| G | 348 | 9 | Carbohydrate transport and metabolism |
| E | 334 | 8.64 | Amino acid transport and metabolism |
| F | 94 | 2.43 | Nucleotide transport and metabolism |
| H | 255 | 6.6 | Coenzyme transport and metabolism |
| I | 181 | 4.68 | Lipid transport and metabolism |
| P | 204 | 5.28 | Inorganic ion transport and metabolism |
| Q | 190 | 4.91 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 450 | 11.64 | General function prediction only |
| S | 135 | 3.49 | Function unknown |
| X | 4 | 0.1 | Mobilome: prophages, transposons |
| - | 2102 | 38.7 | Not in COGs |
Insights from the genome sequence
Gene clusters for biosynthesis of secondary metabolites
So far, there have been no reports on isolation of secondary metabolites from A. hymeniacidonis HPA 177T. However, keeping in mind that all actinomycete genomes sequenced so far contain SMBGCs, the genome of strain HPA 177T was analyzed for their presence using the online version of software antiSMASH 3.0.4 [44]. The results of the analysis were manually curated to confirm or edit borders of the clusters, identify closest homologues in the databases based on BLAST search (Table 5), and to gain a more detailed insight into the biosynthesis of the corresponding compound. In total, 25 SMBGCs were identified, 11 of which appeared to be unique at the time of analysis and based on the public database searches. This conclusion was based on the unique composition of the core genes in the clusters encoding scaffold-building enzymes, and in some cases, such as stand-alone terpene cyclase or type III polyketide synthase genes, on low (below 60%) identity of their products to proteins in the NCBI database. Based on this analysis, it seems possible that A. hymeniacidonis HPA 177T has the genetic capacity to produce novel compounds some of which, e.g. peptide-polyketide hybrids, terpenoids, and unique lassopeptides, may represent bioactive metabolites suitable for drug development. Given its habitat, A. hymeniacidonis might be the real source of secondary metabolites that are thought to originate from its host sponge, comparable to. e.g. Theonella swinhoi and Entotheonella sp. [45]. The knowledge on the SMBGCs and their putative products will assist in identification of the corresponding compounds, and may pave the way to biosynthetic engineering toward generation of new analogues.
Table 5.
Secondary metabolite biosynthesis gene clusters identified in the genome of Actinoalloteichus hymeniacidonis DSM 45092 using antiSMASH 3.0.4 software followed by manual curation
| No | Cluster type | Presence in another bacterium# | Putative product |
|---|---|---|---|
| 1 | Ectoine | Saccharopolyspora rectivirgula DSM 43113 | Ectoine |
| 2 | NRPS-PKSI | Nonomuraea candida DSM 45086 | NRS peptide-polyketide hybrid |
| 3 | Ladderane | Saccharomonospora viridis DSM 43017 | Ladderane |
| 4 | NRPS-PKSI | - | NRS peptide-polyketide hybrid |
| 5 | Ectoine | multiple Actinoalloteichus spp. | Ectoine |
| 6 | Lassopeptide | - | Lassopeptide |
| 7 | Terpene | Kribbella flavida DSM 17836 | Terpenoid |
| 8 | PKSII | - | Aromatic polyketide |
| 9 | Terpene | - | Terpenoid |
| 10 | Siderophore | Saccharomonospora paurometabolica YIM 90007 | Siderophore |
| 11 | Terpene | Actinosynnema mirum DSM 43827 | Carotenoid |
| 12 | PKSIII | - | Stilbene-like polyketide |
| 13 | NRPS-PKSI | Streptomyces sp. NTK 937 | Polycyclic tetramate macrolactam |
| 14 | NRPS | Streptomyces sp. SirexAA-E | Coelibactin |
| 15 | PKSI | - | 34-membered macrocyclic lactone |
| 16 | NRPS-PKSI | Streptomyces bingchenggensis BCW-1 | NRS peptide-polyketide hybrid |
| 17 | Terpene | - | Terpenoid |
| 18 | NRPS | - | NRS peptide |
| 19 | PKSI | Saccharomonospora xinjiangensis XJ-54 | Glycosylated polyene macrolide |
| 20 | NRPS | - | Mannopeptimycin-like NRS peptide |
| 21 | PKSI | Amycolatopsis nigrescens CSC17Ta-90 | Hygrocin-like polyketide |
| 22 | Oligosaccharide | Nocardiopsis kunsanensis DSM 44524 | Oligosaccharide |
| 23 | Butyrolactone | - | Butyrolactone |
| 24 | Siderophore | - | Siderophore |
| 25 | PKSII | Microbispora sp. ATCC PTA-5024 | Aromatic polyketide |
Notes: NRS non-ribosomally synthesized. Shaded cells show potentially unique gene clusters. #Presence in other bacteria based on the publically available data as of January 27, 2016
Conclusion
The genome sequence of A. hymeniacidonis HPA 177T represents the first genome of the A. hoggarensis/A. hymeniacidonis/A. nanshanensis subgroup, the first complete genome of this genus as well as the first of a marine species of this genus. As such, it will be a useful basis for future genome comparisons. The presence of 25 SMBGCs indicates a great potential for secondary metabolite production, either by heterologous expression in suitable hosts or by activating the clusters by genetic engineering.
Funding
Christian Rückert acknowledges funding through a grant by the Federal Ministry for Education and Research (0316017A) within the BioIndustry2021 initiative. SZ acknowledges support of the University of Vienna.
We acknowledge support of the publication fee by the Deutsche Forschungsgemeinschaft and the Open Access Publication Funds of Bielefeld University Library.
Authors’ contributions
LS prepared and wrote the manuscript, AA and AW performed library preparation and sequencing, JK coordinated the study, SZ isolated genomic DNA, analyzed genome for the presence of secondary metabolite biosynthesis gene clusters, and contributed to writing the manuscript, and CR assembled and analyzed the genome sequence. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- CeBiTec
Center for Biotechnology
- GEBA
Genomic Encyclopedia of Bacteria and Archaea
- SMBGC
Secondary metabolite biosynthesis gene cluster
References
- 1.Zhang H, Zheng W, Huang J, Luo H, Jin Y, Zhang W, Liu Z, Huang Y. Actinoalloteichus hymeniacidonis sp. nov., an actinomycete isolated from the marine sponge Hymeniacidon perleve. Int J Syst Evol Microbiol. 2006;56:2309–2312. doi: 10.1099/ijs.0.64217-0. [DOI] [PubMed] [Google Scholar]
- 2.Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinispora. Angew Chem Int Ed Engl. 2003;42:355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- 3.Prudhomme J, McDaniel E, Ponts N, Bertani S, Fenical W, Jensen P, Le Roch K. Marine actinomycetes: a new source of compounds against the human malaria parasite. PLoS One. 2008;3:e2335. doi: 10.1371/journal.pone.0002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhatnagar I, Kim S. Immense essence of excellence: marine microbial bioactive compounds. Mar Drugs. 2010;8:2673–2701. doi: 10.3390/md8102673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramani R, Aalbersberg W. Marine actinomycetes: an ongoing source of novel bioactive metabolites. Microbiol Res. 2012;167:571–580. doi: 10.1016/j.micres.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Manivasagan P, Venkatesan J, Sivakumar K, Kim S. Marine actinobacterial metabolites: current status and future perspectives. Microbiol Res. 2013;168:311–332. doi: 10.1016/j.micres.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Ramesh S, Mathivanan N. Screening of marine actinomycetes isolated from the Bay of Bengal, India for antimicrobial activity and industrial enzymes. World J Microbiol Biotechnol. 2009;25:2103–2111. doi: 10.1007/s11274-009-0113-4. [DOI] [Google Scholar]
- 8.Manivasagan P, Kang K, Sivakumar K, Li-Chan, Eunice CY, Oh H, Kim S. Marine actinobacteria: an important source of bioactive natural products. Environ Toxicol Pharmacol. 2014;38:172–188. doi: 10.1016/j.etap.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 9.Jensen PR, Mincer TJ, Williams PG, Fenical W. Marine actinomycete diversity and natural product discovery. Antonie Van Leeuwenhoek. 2005;87:43–48. doi: 10.1007/s10482-004-6540-1. [DOI] [PubMed] [Google Scholar]
- 10.Tao M, Wang L, Wendt-Pienkowski E, Zhang N, Yang D, Galm U, Coughlin JM, Xu Z, Shen B. Functional characterization of tlmH in Streptoalloteichus hindustanus E465-94 ATCC 31158 unveiling new insight into tallysomycin biosynthesis and affording a novel bleomycin analog. Mol Biosyst. 2010;6:349–356. doi: 10.1039/B918106G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rebets Y, Tokovenko B, Lushchyk I, Rückert C, Zaburannyi N, Bechthold A, Kalinowski J, Luzhetskyy A. Complete genome sequence of producer of the glycopeptide antibiotic Aculeximycin Kutzneria albida DSM 43870T, a representative of minor genus of Pseudonocardiaceae. BMC Genomics. 2014;15:885. doi: 10.1186/1471-2164-15-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikemoto T, Matsunaga H, Konishi K, Okazaki T, Enokita R, Torikata A. Aculeximycin, a new antibiotic from Streptosporangium albidum. I. taxonomy of producing organism and fermentation. J Antibiot. 1983;36:1093–1096. doi: 10.7164/antibiotics.36.1093. [DOI] [PubMed] [Google Scholar]
- 13.Ohkuma H, Tenmyo O, Konishi M, Oki T, Kawaguchi H. BMY-28190, a novel antiviral antibiotic complex. J Antibiot. 1988;41:849–854. doi: 10.7164/antibiotics.41.849. [DOI] [PubMed] [Google Scholar]
- 14.Tamura T, Ishida Y, Otoguro M, Hatano K, Suzuki K. Classification of ‘Streptomyce tenebrarius’ Higgins and Kastner as Streptoalloteichus tenebrarius nom. rev., comb. nov., and emended description of the genus Streptoalloteichus. Int J Syst Evol Microbiol. 2008;58:688–691. doi: 10.1099/ijs.0.65272-0. [DOI] [PubMed] [Google Scholar]
- 15.Yu W, Liu W, Huang H, Zheng F, Wang X, Wu Y, Li K, Xie X, Jin Y. Application of a novel alkali-tolerant thermostable DyP-type peroxidase from Saccharomonospora viridis DSM 43017 in biobleaching of eucalyptus kraft pulp. PLoS One. 2014;9:e110319. doi: 10.1371/journal.pone.0110319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawai F, Oda M, Tamashiro T, Waku T, Tanaka N, Yamamoto M, Mizushima H, Miyakawa T, Tanokura M. A novel Ca2+-activated, thermostabilized polyesterase capable of hydrolyzing polyethylene terephthalate from Saccharomonospora viridis AHK190. Appl Microbiol Biotechnol. 2014;98:10053–10064. doi: 10.1007/s00253-014-5860-y. [DOI] [PubMed] [Google Scholar]
- 17.Labeda DP. Crossiella gen. nov., a new genus related to Streptoalloteichus. Int J Syst Evol Microbiol. 2001;51:1575–1579. doi: 10.1099/00207713-51-4-1575. [DOI] [PubMed] [Google Scholar]
- 18.Tomita K, Hoshino Y, Miyaki T. Kibdelosporangium albatum sp. nov., producer of the antiviral antibiotics cycloviracins. Int J Syst Bact. 1993;43:297–301. doi: 10.1099/00207713-43-2-297. [DOI] [PubMed] [Google Scholar]
- 19.Jarerat A, Tokiwa Y, Tanaka H. Poly(L-lactide) degradation by Kibdelosporangium aridum. Biotechnol Lett. 2003;25:2035–2038. doi: 10.1023/B:BILE.0000004398.38799.29. [DOI] [PubMed] [Google Scholar]
- 20.Tamura T, Zhiheng L, Yamei Z, Hatano K. Actinoalloteichus cyanogriseus gen. nov., sp. nov. Int J Syst Evol Microbiol. 2000;50:1435–1440. doi: 10.1099/00207713-50-4-1435. [DOI] [PubMed] [Google Scholar]
- 21.Stackebrandt E, Rainy FA, Ward-Rainy NL. Proposal for a New Hierarchic Classification System, Actinobacteria classis nov. Int J Syst Evol Microbiol. 1997;47:479–491. [Google Scholar]
- 22.Boudjelal F, Zitouni A, Bouras N, Schumann P, Spröer C, Sabaou N, Klenk H. Actinoalloteichus hoggarensis sp. nov., an actinomycete isolated from Saharan soil. Int J Syst Evol Microbiol. 2015;65:2006–2010. doi: 10.1099/ijs.0.000216. [DOI] [PubMed] [Google Scholar]
- 23.Xiang W, Liu C, Wang X, Du J, Xi L, Huang Y. Actinoalloteichus nanshanensis sp. nov., isolated from the rhizosphere of a fig tree (Ficus religiosa) Int J Syst Evol Microbiol. 2011;61:1165–1169. doi: 10.1099/ijs.0.023283-0. [DOI] [PubMed] [Google Scholar]
- 24.Singla AK, Mayilraj S, Kudo T, Krishnamurthi S, Prasad GS, Vohra RM. Actinoalloteichus spitiensis sp. nov., a novel actinobacterium isolated from a cold desert of the Indian Himalayas. Int J Syst Evol Microbiol. 2005;55:2561–2564. doi: 10.1099/ijs.0.63720-0. [DOI] [PubMed] [Google Scholar]
- 25.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, Hooper SD, Pati A, Lykidis A, Spring S, Anderson IJ, D’haeseleer P, Zemla A, Singer M, Lapidus A, Nolan M, Copeland A, Han C, Chen F, Cheng J, Lucas S, Kerfeld C, Lang E, Gronow S, Chain P, Bruce D, Rubin EM, Kyrpides NC, Klenk H, Eisen JA. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liolios K, Chen IA, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2010;38:D346–D354. doi: 10.1093/nar/gkp848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. doi: 10.1101/gr.8.3.186. [DOI] [PubMed] [Google Scholar]
- 29.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 30.Gordon D. Viewing and editing assembled sequences using Consed. Curr Protoc Bioinformatics. 2003;2:11.2.1–11.2.43. doi: 10.1002/0471250953.bi1102s02. [DOI] [PubMed] [Google Scholar]
- 31.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 32.Markowitz VM, Mavromatis K, Ivanova NN, Chen IA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics. 2009;25:2271–2278. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- 33.Borodovsky M, Mills R, Besemer J, Lomsadze A. Prokaryotic gene prediction using GeneMark and GeneMark.hmm. Curr Protoc Bioinformatics. 2003;1:4.5.1–4.5.16. doi: 10.1002/0471250953.bi0405s01. [DOI] [PubMed] [Google Scholar]
- 34.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hyatt D, Chen G, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klimke W, Agarwala R, Badretdin A, Chetvernin S, Ciufo S, Fedorov B, Kiryutin B, O’Neill K, Resch W, Resenchuk S, Schafer S, Tolstoy I, Tatusova T. The National Center for Biotechnology Information’s Protein Clusters Database. Nucleic Acids Res. 2009;37:D216–D223. doi: 10.1093/nar/gkn734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eddy SR. A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMC Bioinformatics. 2002;3:18. doi: 10.1186/1471-2105-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lagesen K, Hallin P, Rødland EA, Staerfeldt H, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 43.Bendtsen JD, Nielsen H, Heijne G, von Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 44.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W, Breitling R, Takano E, Medema MH. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson MC, Mori T, Rückert C, Uria AR, Helf MJ, Takada K, Gernert C, Steffens UA, Heycke N, Schmitt S, Rinke C, Helfrich EJN, Brachmann AO, Gurgui C, Wakimoto T, Kracht M, Crüsemann M, Hentschel U, Abe I, Matsunaga S, Kalinowski J, Takeyama H, Piel J. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506:58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- 46.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, Ashburner M, Axelrod N, Baldauf S, Ballard S, Boore J, Cochrane G, Cole J, Dawyndt P, Vos P de, DePamphilis C, Edwards R, Faruque N, Feldman R, Gilbert J, Gilna P, Glöckner FO, Goldstein P, Guralnick R, Haft D, Hancock D, Hermjakob H, Hertz-Fowler C, Hugenholtz P, Joint I, Kagan L, Kane M, Kennedy J, Kowalchuk G, Kottmann R, Kolker E, Kravitz S, Kyrpides N, Leebens-Mack J, Lewis SE, Li K, Lister AL, Lord P, Maltsev N, Markowitz V, Martiny J, Methe B, Mizrachi I, Moxon R, Nelson K, Parkhill J, Proctor L, White O, Sansone S, Spiers A, Stevens R, Swift P, Taylor C, Tateno Y, Tett A, Turner S, Ussery D, Vaughan B, Ward N, Whetzel T, San Gil I, Wilson G, Wipat A. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26:541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains and Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodfellow M. Phylum XXVI. Actinobacteria phyl. nov. Bergey’s Manual of Systematic Bacteriology 2012;5, Part A:33.
- 49.Zhi XY, Li WJ, Stackebrandt E. An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int J Syst Evol Microbiol. 2009;59:589–608. doi: 10.1099/ijs.0.65780-0. [DOI] [PubMed] [Google Scholar]
- 50.Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Med J Aust. 1980;2:3–4. doi: 10.5694/j.1326-5377.1980.tb131800.x. [DOI] [PubMed] [Google Scholar]
- 51.Labeda DP, Goodfellow M, Chun J, Zhi X-Y, Li W-J. Reassessment of the systematics of the suborder Pseudonocardianeae: transfer of the genera within the family Actinosynnemataceae Labeda and Kroppenstedt 2000 emend. Zhi et al. 2009 into an emended family Pseudonocardiaceae Embley et al. 1989 emend. Zhi et al. 2009. Int J Syst Evol Microbiol. 2011;61:1259–1264. doi: 10.1099/ijs.0.024984-0. [DOI] [PubMed] [Google Scholar]
- 52.Embley MT, Smida J, Stackebrandt E. The phylogeny of mycolate-less wall chemotype IV Actinomycetes and description of Pseudonocardiaceae fam. nov. Syst Appl Microbiol. 1988;1:44–52. doi: 10.1016/S0723-2020(88)80047-X. [DOI] [Google Scholar]
- 53.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bruno WJ, Socci ND, Halpern AL. Weighted neighbor joining: a likelihood-based approach to distance-based phylogeny reconstruction. Mol Biol Evol. 2000;17:189–197. doi: 10.1093/oxfordjournals.molbev.a026231. [DOI] [PubMed] [Google Scholar]
- 55.Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–D172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]


