Abstract
The TP53 is important in functions of cell cycle control, apoptosis, and maintenance of DNA integrity. Studies on the association between p53 codon 72 polymorphism and primary open-angle glaucoma (POAG) risk have yielded conflicting results. Published literature from PubMed and Web of Science databases was retrieved. All studies evaluating the association between p53 codon 72 polymorphisms and POAG were included. Pooled odds ratio (OR) and 95% confidence interval (CI) were calculated. Eleven separate studies including 2541 cases and 1844 controls were pooled in the meta-analysis. We did not detect a significant association between POAG risk and p53 codon 72 polymorphism overall population except allele genetic model (C vs. G: OR = 0.961, 95% CI = 0.961–0.820, P = 0.622). In the stratified analysis for Asians and Caucasians, there was an association between p53 codon 72 polymorphism and POAG. In the dominant model in the overall population and by ethnicity subgroups, the highest elevated POAG risk was presented. In summary, these results indicate that p53 codon 72 polymorphism is likely an important genetic factor contributing to susceptibility of POAG. However, more case–controls studies based on larger sample size and stratified by ethnicity are suggested to further clarify the relationship between p53 codon 72 polymorphism and POAG.
Keywords: Codon 72, meta-analysis, p53, polymorphism, primary open-angle glaucoma
Glaucoma is one of the leading causes of irreversible blindness worldwide, affecting 60 million people worldwide, with 8.4 million people with the disease being bilaterally blind.[1] The most common form is primary open-angle glaucoma (POAG), which is characterized by retinal ganglion cell apoptosis and visual field changes corresponding with the excavation of the optic disk and an open anterior chamber angle.[2,3]
It is suggested that glaucoma influenced by a combination of genetic and environmental risk factors.[4] This condition is attributed to several genes, the majority of which are yet unidentified.[5] Until now, linkage analysis in large affected families has yielded 25 chromosomal loci linked to POAG. The POAG-associated genes include atrial natriuretic peptide, apolipoprotein E, optic atrophy 1, p53, glutathione S-transferase (GST), interleukins, and tumor necrosis factor-α.[6] However, the role of these genes in the etiology of POAG is still controversial.[7] Recently, several genome-wide association studies have been performed for POAG, which revealed several genetic variants associated with the disease. Several chromosomal loci have now been reported as linked to POAG, such as p53 (MIM 602432).[6] However, the association of p53 with POAG has been a source of controversy as reports published recently showed conflicting results.[8]
TP53 is a tumor suppressor that plays an important role in cell cycle regulation and the maintenance of genome integrity.[9] TP53 mediates the cellular response to DNA damage through effects on gene transcription, DNA synthesis and repair, genomic plasticity, and apoptosis. Functional polymorphisms of the TP53 gene which influence the above activities of TP53 protein might be associated with human susceptibility to cancer.[10] The codon 72 polymorphism (rs1042522) is located in exon 4 of TP53 gene and involves a CCC → CGC transition leading to a proline (Pro) → arginine (Arg) amino acid substitution at position 72 (Pro72Arg), and the 72Arg allele shows more efficient in inducing apoptosis and lower ability in inducing cell cycle arrest and DNA repair.[10,11]
Several previous studies have explored the association between the p53 codon 72 polymorphism and POAG susceptibility; however, existing results are inconsistent. Therefore, we conducted this meta-analysis to obtain accurate and up-to-date estimates of the association between the TP53 codon 72 polymorphism and POAG.
Materials and Methods
Search strategy
We performed a comprehensive search of the PubMed, Web of Science, and SID databases to identify potentially relevant studies on the association between p53 codon 72 Arg/Pro polymorphism and POAG risk up to January 2015. The following terms were used in the literature search: “POAG,” “p53,” “codon 72,” “rs1042522,” “Arg72Pro,” “polymorphism,” “variant,” and “SNP.” The search was restricted to humans with language exclusions. The references of retrieved publication, published reviews, letters, and comments were scanned for additional relevant studies. Additional studies were identified by a manual search of the references from the original or review articles on this topic.
Inclusion criteria
Studies included in our meta-analysis had to meet the following inclusion criteria: (1) evaluated the association between the TP53 codon 72 polymorphism and POAG susceptibility; (2) a case–control study; (3) provided the number of individual genotypes in both the case and control groups, or enabled the genotypes to be calculated from available published data; (4) published only in English; and (5) when multiple publications reported on the same or overlapping data, we chose the most recent or largest population. Studies were excluded if one of the following existed: (1) none-case–control studies; (2) studies that contained overlapping data; (3) not offering the source of cases and controls or other essential information; (4) reviews and repeated literature were also excluded; (5) no usable data reported.
Data extraction
Two authors independently extracted the following trial data from included studies: The following items were collected from each study:first author's surname, year of publication, ethnicity, countries of origin, total number of cases and controls, and genotype frequencies of cases and controls. Different descents were categorized as Caucasians, Asians, and Mixed populations which included more than one ethnic descent. For case–control studies, data were extracted separately for each group whenever possible.
Statistical analysis
Deviation from the Hardy–Weinberg equilibrium (HWE) among the controls was evaluated for each single study using online calculator named HWE calculator for 2 alleles (http://www.had2know.com). Dichotomous data were presented as odds ratio (OR) with 95% confidence interval (CI). Statistical heterogeneity was measured using the Q statistic test (P < 0.10 was considered statistically significant heterogeneity). An I2 value of 25% indicates low heterogeneity, 50% indicates moderate heterogeneity and 75% indicates high heterogeneity. The heterogeneity was considered statistically significant at I2 >50% or P < 0.10. Either a random-effects model (DerSimonian–Laird method) or fixed-effects model (Mantel–Haenszel method) was used to calculate pooled effect estimates in the presence or absence of heterogeneity, respectively. To establish the effect of heterogeneity among the studies on the conclusions of this meta-analysis, subgroup analyses were conducted based on ethnicity. Several methods were used to assess the potential for publication bias. Visual inspection of funnel plot asymmetry was conducted. The Begg's rank correlation method and the Egger's weighted regression method were used to statistically assess publication bias. P < 0.05 was considered statistically significant. All the statistical tests were performed with Comprehensive Meta Analysis software (CMA) version 2.0 (Biostat, USA).
Results
Study characteristics
After comprehensive searching, a total of 21 articles were retrieved, but only 11 full-text publications[8,11,12,13,14,15,16,17,18,19,20] which catered to the inclusion criteria were finally included in our meta-analysis. The characteristics of all studies are summarized in Table 1. The 11 studies were published from 2002 to 2015 with 5 were carried out in Asian countries, 6 in Europe countries, and 8 in America. Of these 54 studies, the number of cases in the included studies for GSTM1 deletion varied from 58 to 523 patients. The genotype distribution in controls of the included studies all agreed with HWE. The genotype frequencies for p3 polymorphism of cases and controls are presented in Table 2 in detail.
Table 1.
Distribution of p53 codon 72 genotypes among primary open-angle glaucoma cases and controls included in the meta-analysis
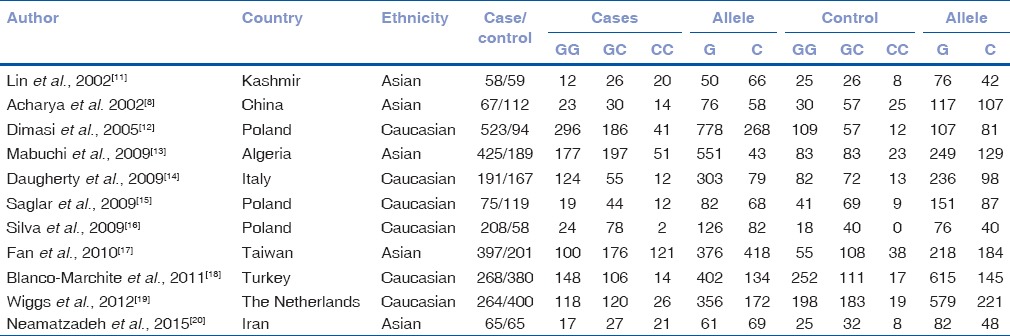
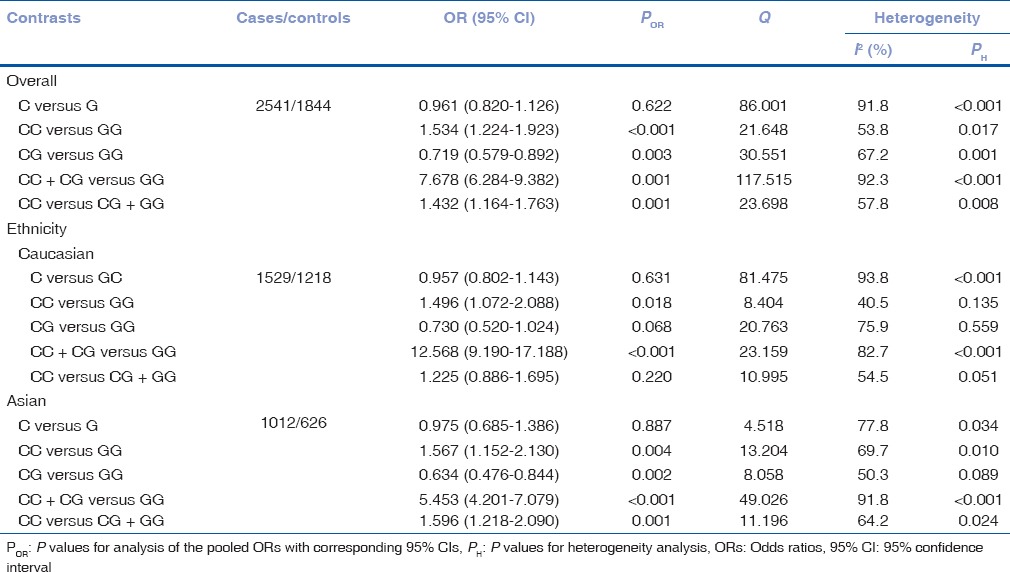
Table 2.
Meta-analysis results for the p53 codon 72 polymorphism and primary open-angle glaucoma risk
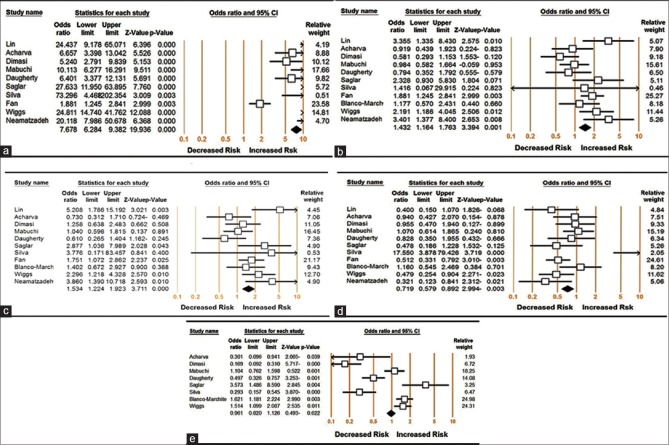
Evaluation of heterogeneity
To analyze heterogeneity among the selected studies, Q-test and I2 statistics were employed, and heterogeneity was noticed in all the five genetic models. Therefore, random-effects model was applied to synthesize the data [Table 2]. Then, we assessed the source of heterogeneity for dominant comparison (CC + CG vs. GG) by ethnicity. As a result, ethnicity (χ2 = 14.41; df = 2; P = 0.001) was found to contribute to substantial heterogeneity. In addition, subgroup analyses revealed that the heterogeneity was significantly reduced in the small sample size group and large sample size group in all genetic models, suggesting that the total sample size was the source of heterogeneity [Table 2 and Fig. 1].
Figure 1.
Meta-analysis of the odds ratio for p53 codon 72 polymorphism associated with primary open-angle glaucoma. (a) Dominant model: CC + CG versus GG; (b) recessive model: CC versus CG + GG; (c) additive model: CC versus GG; (d) heterozygous model: CG versus GG; and (e) allelic model: C versus G
Meta-analysis results
Table 2 lists the main results of the meta-analysis. The association of the p53 codon 72 polymorphism with POAG risk under different genetic models is shown in Table 2. The overall data comprising 2541 cases and 1844 controls exhibited significant increased POAG risk under the dominant (CC + CG vs. GG: OR = 7.678, 95% CI = 6.284–9.382, P < 0.001) [Fig. 1a], recessive (CC vs. CG + GG: OR = 1.432, 95% CI = 1.164–1.763, P = 0.001) [Fig. 1b], additive (CC vs. GG: OR = 1.534, 95% CI = 1.224–1.923, P < 0.001) [Fig. 1c], heterozygous genetic models (CG vs. GG: OR = 0.719, 95% CI = 0.579–0.892, P = 0.003) [Fig. 1d], except allelic model (C vs. G: OR = 0.961, 95% CI = 0.961–0.820, P = 0.622) [Fig. 1e].
Subgroup analyses of the different ethnic groups were performed. The results are shown in Table 2 and significant increased POSG risk was found in Caucasians under dominant (CC + CG vs. GG: OR = 12.568, 95% CI = 9.190–17.188, P < 0.001) and additive genetic models (CC vs. GG: OR = 1.496, 95% CI = 1.072–2.088, P = 0.018). Similarly, increased POAG risks were shown among Asians under dominant (CC + CG vs. GG: OR = 12.568, 95% CI = 9.190–17.188, P < 0.001), additive (CC vs. GG: OR = 1.496, 95% CI = 1.072–2.088, P = 0.018), recessive (CC vs. CG + GG: OR = 1.596, 95% CI = 1.218–2.090, P < 0.001), heterozygous genetic models (CG vs. GG: OR = 0.634, 95% CI = 0.476–0.844, P = 0.002), except allele model (C vs. G: OR = 0.975, 95% CI = 0.685–1.386, P = 0.887).
In the dominant model in the overall population and by ethnicity subgroups, the highest elevated POAG risk was presented. The results indicated that individuals who carry variant G allele might have an increased POAG risk compared with those who bear wild-type C allele [Table 2].
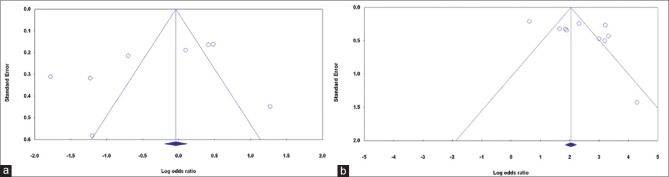
Publication bias
Publication bias of the selected articles was assessed by the Begg's funnel plot and Egger's test. The shape of the funnel plot did not show obvious publication bias [Fig. 2]. Similarly, no evidence of publication bias was observed by Egger's test (P = 0.173 for allelic genetic model; P = 0.299 and for dominant genetic model; P = 0.115).
Figure 2.
Begg's funnel plot of the meta-analysis of prostate cancer risk and p53 codon 72 polymorphism. (a) Allelic model: C versus G, and (b) dominant model: CC + CG versus GG
Discussion
In the present study, we systemically reviewed all available published studies and performed a meta-analysis to derive a more precise estimation of the association between p53 gene polymorphism and susceptibility to POAG. Our meta-analysis included 15 separate studies involving 2700 cases and 2365 controls. In Asians, we detected an association of the ε4 genotype with elevated risk for glaucoma, mainly for POAG. Thus, the Arg genotype may be associated with elevated risk for POAG in Asians.
The p53 gene is located on the short arm of chromosome 17, and its protein product is related to the regulation of the cell cycle.[1,2] It is well known that p53 are important components involved in the apoptosis pathway. A number of studies have investigated the genetic effects of p53 codon 72 polymorphism on POAG susceptibility with conflicting results.[21] Meta-analysis across sufficiently homogeneous studies can further enhance the sample size and power. Meta-analysis is, therefore, becoming a popular method for resolving discrepancies in genetic association studies.[22] Thus, to explain these contradictory results, as well as to decrease uncertainty about the effect size of estimated risk, a meta-analysis was conducted, examining all available studies related to p53 codon 72 polymorphism and its associations with POAG.
In the present study, we systemically reviewed all available published studies and performed a meta-analysis to derive a more precise estimation of the association between p53 codon 72 polymorphism and susceptibility to POAG. Our meta-analysis included 11 separate studies involving 2700 cases and 2365 controls. The results have shown a significant association between p53 codon 72 and POAG except allele model. By ethnicity-pooled analysis, we detected an association of the p53 gene genotype with elevated risk for POAG, in Asians and Caucasians. However, the number of studies and the number of subjects in the studies included in the meta-analysis were small, especially for the Asian population.
To the best of our knowledge, there is only a meta-analysis that assessed the association between p53 codon 72 polymorphism and POAG. The study of Guo et al. has 9 case–control studies with 1930 cases and 1463 controls, their conclusion indicate that it provided evidence that the p53 codon 72 polymorphism is an association with the risk of POAG. Guo et al. concluded that a significant association was found between the p53 codon 72 polymorphism and POAG risk when all the eligible studies were pooled into the meta-analysis; however, significant risk of POAG was observed in recessive model (OR = 1.31, 95% CI = 1.05–1.64, P = 0.017).[21] In subgroup analyses for ethnicity, they have found the association between codon 72 polymorphism and risk for POAG in Asian populations (recessive model: OR = 1.36, 95% CI = 1.03–1.80, P = 0.026) but not in Caucasian populations,[21] suggesting that the genetic background or environment they live in may influence the p53 codon 72 polymorphism on POAG susceptibility. The previous studies have identified that the ethnic differences may affect genetic predisposition to POAG.[8,16,17,23] However, the results of their meta-analysis should be interpreted with caution due to some limitations. However, in the stratified analysis by ethnicity, we have found significant increased risks in Asian and Caucasians for some genetic models.
Heterogeneity is a potential problem when interpreting the results of a meta-analysis, and finding the sources of heterogeneity is one of the most important goals of meta-analysis.[24] In the present meta-analysis, significant between-study moderate- to high-level heterogeneity in the pooled analyses of total eligible studies was observed in all genetic models [Table 2]. To find the sources of heterogeneity, we performed meta-regression and subgroup analyses. From the omitting studies and subgroups analysis, we found that ethnicity and source of control might not be the source of heterogeneity [Table 2].
Some potential limitations in our study should be considered. First, our meta-analysis was based on unadjusted estimates, and the confounding factors such as gender and age could not be controlled because most studies did not provide these data. Second, this meta-analysis is limited to language and database restrictions. The PubMed database is the only search source and included published studies were in English. Third, gene-gene and gene-environment interactions were not addressed in our meta-analysis. Fourth, the overall sample sizes of included literatures are quite small. Hence, we should have larger sample size, rigorous design approach, perfect retrieval strategy, and reasonable inclusion and exclusion criteria in the future studies.
Conclusion
This meta-analysis suggests that the p53 codon 72 polymorphism may be associated with increased risk of POAG, especially among Asians. However, it is necessary to conduct large sample studies and well-matched controls. Moreover, gene-gene and gene-environment interactions should also be considered in the analysis. Such studies taking these factors into account may eventually lead to our better, comprehensive understanding of the association between the p53 polymorphism and POAG risk.
Financial support and sponsorship
This study was supported in part by the Shahid Sadoughi University of Medical Sciences, Yazd, Iran.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
The authors would like to thank the Department of Ophthalmology and Medical Genetics, Shahid Sadoughi University of Medical Sciences, Yazd, Iran.
References
- 1.Kyari F, Abdull MM, Bastawrous A, Gilbert CE, Faal H. Epidemiology of glaucoma in Sub-Saharan Africa: Prevalence, incidence and risk factors. Middle East Afr J Ophthalmol. 2013;20:111–25. doi: 10.4103/0974-9233.110605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu T, Xie L, Ye J, He X. Family-based analysis identified CD2 as a susceptibility gene for primary open angle glaucoma in Chinese Han population. J Cell Mol Med. 2014;18:600–9. doi: 10.1111/jcmm.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao X, Yang C, Tong Y, Zhang X, Xu L, Li Y. Identification a novel MYOC gene mutation in a Chinese family with juvenile-onset open angle glaucoma. Mol Vis. 2010;16:1728–35. [PMC free article] [PubMed] [Google Scholar]
- 4.Fingert JH. Primary open-angle glaucoma genes. Eye (Lond) 2011;25:587–95. doi: 10.1038/eye.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Micheal S, Ayub H, Khan MI, Bakker B, Schoenmaker-Koller FE, Ali M, et al. Association of known common genetic variants with primary open angle, primary angle closure, and pseudoexfoliation glaucoma in Pakistani cohorts. Mol Vis. 2014;20:1471–9. [PMC free article] [PubMed] [Google Scholar]
- 6.Rao KN, Nagireddy S, Chakrabarti S. Complex genetic mechanisms in glaucoma: An overview. Indian J Ophthalmol. 2011;59(Suppl 1):S31–42. doi: 10.4103/0301-4738.73685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahdy MA. Gene therapy in glaucoma-part 2: Genetic etiology and gene mapping. Oman J Ophthalmol. 2010;3:51–9. doi: 10.4103/0974-620X.64227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acharya M, Mitra S, Mukhopadhyay A, Khan M, Roychoudhury S, Ray K. Distribution of p53 codon 72 polymorphism in Indian primary open angle glaucoma patients. Mol Vis. 2002;8:367–71. [PubMed] [Google Scholar]
- 9.Lin HY, Huang CH, Wu WJ, Chang LC, Lung FW. TP53 codon 72 gene polymorphism paradox in associated with various carcinoma incidences, invasiveness and chemotherapy responses. Int J Biomed Sci. 2008;4:248–54. [PMC free article] [PubMed] [Google Scholar]
- 10.Peng Q, Lao X, Chen Z, Lai H, Deng Y, Wang J, et al. TP53 and MDM2 gene polymorphisms, gene-gene interaction, and hepatocellular carcinoma risk: Evidence from an updated meta-analysis. PLoS One. 2013;8:e82773. doi: 10.1371/journal.pone.0082773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin HJ, Chen WC, Tsai FJ, Tsai SW. Distributions of p53 codon 72 polymorphism in primary open angle glaucoma. Br J Ophthalmol. 2002;86:767–70. doi: 10.1136/bjo.86.7.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimasi DP, Hewitt AW, Green CM, Mackey DA, Craig JE. Lack of association of p53 polymorphisms and haplotypes in high and normal tension open angle glaucoma. J Med Genet. 2005;42:e55. doi: 10.1136/jmg.2005.032458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mabuchi F, Sakurada Y, Kashiwagi K, Yamagata Z, Iijima H, Tsukahara S. Lack of association between p53 gene polymorphisms and primary open angle glaucoma in the Japanese population. Mol Vis. 2009;15:1045–9. [PMC free article] [PubMed] [Google Scholar]
- 14.Daugherty CL, Curtis H, Realini T, Charlton JF, Zareparsi S. Primary open angle glaucoma in a Caucasian population is associated with the p53 codon 72 polymorphism. Mol Vis. 2009;15:1939–44. [PMC free article] [PubMed] [Google Scholar]
- 15.Saglar E, Yucel D, Bozkurt B, Ozgul RK, Irkec M, Ogus A. Association of polymorphisms in APOE, p53, and p21 with primary open-angle glaucoma in Turkish patients. Mol Vis. 2009;15:1270–6. [PMC free article] [PubMed] [Google Scholar]
- 16.Silva RE, Arruda JT, Rodrigues FW, Moura KK. Primary open angle glaucoma was not found to be associated with p53 codon 72 polymorphism in a Brazilian cohort. Genet Mol Res. 2009;8:268–72. doi: 10.4238/vol8-1gmr578. [DOI] [PubMed] [Google Scholar]
- 17.Fan BJ, Liu K, Wang DY, Tham CC, Tam PO, Lam DS, et al. Association of polymorphisms of tumor necrosis factor and tumor protein p53 with primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2010;51:4110–6. doi: 10.1167/iovs.09-4974. [DOI] [PubMed] [Google Scholar]
- 18.Blanco-Marchite C, Sánchez-Sánchez F, López-Garrido MP, Iñigez-de-Onzoño M, López-Martínez F, López-Sánchez E, et al. WDR36 and P53 gene variants and susceptibility to primary open-angle glaucoma: Analysis of gene-gene interactions. Invest Ophthalmol Vis Sci. 2011;52:8467–78. doi: 10.1167/iovs.11-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiggs JL, Hewitt AW, Fan BJ, Wang DY, Figueiredo Sena DR, O’Brien C, et al. The p53 codon 72 PRO/PRO genotype may be associated with initial central visual field defects in Caucasians with primary open angle glaucoma. PLoS One. 2012;7:e45613. doi: 10.1371/journal.pone.0045613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neamatzadeh H, Soleimanizad R, Zare-Shehneh M, Gharibi S, Shekari A, Rahimzadeh AB. Association between p53 codon 72 (Arg72Pro) polymorphism and primary open-angle glaucoma in Iranian patients. Iran Biomed J. 2015;19:51–6. doi: 10.6091/ibj.1379.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Y, Zhang H, Chen X, Yang X, Cheng W, Zhao K. Association of TP53 polymorphisms with primary open-angle glaucoma: A meta-analysis. Invest Ophthalmol Vis Sci. 2012;53:3756–63. doi: 10.1167/iovs.12-9818. [DOI] [PubMed] [Google Scholar]
- 22.Munafò MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20:439–44. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Ressiniotis T, Griffiths PG, Birch M, Keers S, Chinnery PF. Primary open angle glaucoma is associated with a specific p53 gene haplotype. J Med Genet. 2004;41:296–8. doi: 10.1136/jmg.2003.016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ioannidis JP, Patsopoulos NA, Evangelou E. Uncertainty in heterogeneity estimates in meta-analyses. BMJ. 2007;335:914–6. doi: 10.1136/bmj.39343.408449.80. [DOI] [PMC free article] [PubMed] [Google Scholar]