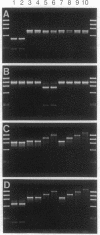
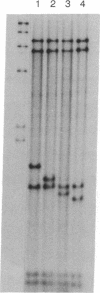
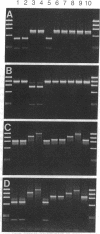
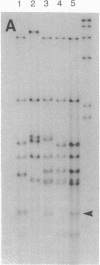
Abstract
Previous DNA studies have revealed that feral neotropical African bees have largely retained an African genetic integrity. Additional DNA testing is needed to confirm these findings, to understand the processes responsible, and to follow African bee spread into the temperate United States. To facilitate surveys, the polymerase chain reaction was utilized. African and European honeybee mitochondrial DNA (mtDNA) was identified through amplified segments that carry informative restriction site and length polymorphisms. The ability to discriminate among honeybee subspecies was established by testing a total of 129 colonies from Africa and Europe. Matriline identities could thus be determined for imported New World bees. Among 41 managed and feral colonies in the United States and north Mexico, two European lineages (west and east) were distinguished. From neotropical regions, 72 feral colonies had African mtDNA and 4 had European mtDNA. The results support earlier conclusions that neotropical African bees have spread as unbroken African maternal lineages. Old and New World African honeybee populations exhibit different frequencies of a mtDNA length polymorphism. Through standard analyses, a north African mtDNA type that may have been imported previously from Spain or Portugal was not detected among neotropical African bees.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barton N. H., Hewitt G. M. Adaptation, speciation and hybrid zones. Nature. 1989 Oct 12;341(6242):497–503. doi: 10.1038/341497a0. [DOI] [PubMed] [Google Scholar]
- Bee D. J. Clinical diagnosis of ovarian dysfunction. Vet Rec. 1991 Mar 2;128(9):214–214. doi: 10.1136/vr.128.9.214-a. [DOI] [PubMed] [Google Scholar]
- Crozier R. H., Crozier Y. C., Mackinlay A. G. The CO-I and CO-II region of honeybee mitochondrial DNA: evidence for variation in insect mitochondrial evolutionary rates. Mol Biol Evol. 1989 Jul;6(4):399–411. doi: 10.1093/oxfordjournals.molbev.a040553. [DOI] [PubMed] [Google Scholar]
- Hall H. G. DNA differences found between Africanized and European honeybees. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4874–4877. doi: 10.1073/pnas.83.13.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall H. G., Muralidharan K. Evidence from mitochondrial DNA that African honey bees spread as continuous maternal lineages. Nature. 1989 May 18;339(6221):211–213. doi: 10.1038/339211a0. [DOI] [PubMed] [Google Scholar]
- Hall H. G. Parental analysis of introgressive hybridization between African and European honeybees using nuclear DNA RFLPs. Genetics. 1990 Jul;125(3):611–621. doi: 10.1093/genetics/125.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michener C. D. The Brazilian bee problem. Annu Rev Entomol. 1975;20:399–416. doi: 10.1146/annurev.en.20.010175.002151. [DOI] [PubMed] [Google Scholar]
- Reiss Y., Goldstein J. L., Seabra M. C., Casey P. J., Brown M. S. Inhibition of purified p21ras farnesyl:protein transferase by Cys-AAX tetrapeptides. Cell. 1990 Jul 13;62(1):81–88. doi: 10.1016/0092-8674(90)90242-7. [DOI] [PubMed] [Google Scholar]
- Rinderer T. E., Hellmich R. L., 2nd, Danka R. G., Collins A. M. Male reproductive parasitism: a factor in the africanization of European honey-bee populations. Science. 1985 May 31;228(4703):1119–1121. doi: 10.1126/science.228.4703.1119. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Smith D. R., Brown W. M. Polymorphisms in mitochondrial DNA of European and Africanized honeybees (Apis mellifera). Experientia. 1988 Mar 15;44(3):257–260. doi: 10.1007/BF01941730. [DOI] [PubMed] [Google Scholar]
- Smith D. R., Taylor O. R., Brown W. M. Neotropical Africanized honey bees have African mitochondrial DNA. Nature. 1989 May 18;339(6221):213–215. doi: 10.1038/339213a0. [DOI] [PubMed] [Google Scholar]
- Vlasak I., Burgschwaiger S., Kreil G. Nucleotide sequence of the large ribosomal RNA of honeybee mitochondria. Nucleic Acids Res. 1987 Mar 11;15(5):2388–2388. doi: 10.1093/nar/15.5.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]