Abstract
Background.
People living with human immunodeficiency virus (PLWH) have been disproportionally affected by methicillin-resistant Staphylococcus aureus (MRSA) colonization and infection, in particular by clones USA300 and USA500. However, the contribution of epidemiological, bacterial, and immunological risk factors to the excess of S aureus in PLWH remain incompletely understood.
Methods.
In this cross-sectional study, we determined the prevalence and molecular epidemiology of S aureus colonization in 93 PLWH attending an urban human immunodeficiency virus (HIV) clinic. Participants completed a structured interview assessing demographic information and risk factors for MRSA. Swabs were obtained from the nose, throat, and groin and cultured for S aureus and Staphylococcus epidermidis.
Results.
Most participants had well controlled HIV infection (89, 96% CD4 >200). Thirty-six (39%) individuals were colonized with S aureus at 1 or more body sites, including 6 (6%) with MRSA. Regular gym use was a risk factor for S aureus but not MRSA carriage. In contrast, S epidermidis was present in almost all individuals (n = 84, 90%), predominantly in the nares (n = 66, 71%). Using generalized estimating equation models, we observed that the odds of S aureus colonization were significantly and drastically reduced when S epidermidis was detected (P = .0001). After controlling for site, gender, and age, we identified that the odds of S aureus colonization were 80% less if S epidermidis was present (adjusted odds ratio, 0.20; 95% confidence interval, .09–.45; P < .0001).
Conclusions.
Taken together, we observed a lower prevalence of S aureus and MRSA colonization than has been previously reported in PLWH. In this cohort, colonization with S epidermidis was protective against S aureus colonization.
Keywords. HIV, immune dysregulation, MRSA, Staphylococcus aureus, Staphylococcus epidermidis.
Over the past 15 years, community-associated methicillin-resistant Staphylococcus aureus infections (CA-MRSA) have emerged sequential to healthcare-associated (HA) MRSA infections [1]. Community-associated MRSA account for the majority of skin and soft tissue infections (SSTIs) in the United States [2]. People living with human immunodeficiency virus (PLWH) have been disproportionally affected by both HA- and CA-MRSA as evidenced by their increased frequency of S aureus colonization, skin infections, and invasive blood stream infections [3–6]. Some studies have suggested that PLWH have a 6–18 times higher incident rate of S aureus infection when compared with healthy human immunodeficiency virus (HIV)-negative controls [3, 7].
The increased incidence of S aureus infections is likely multifactorial and includes behavioral, host immune, and pathogen factors [5, 7]. It has been shown that injection drug use, homelessness, high-risk sexual activity, or extended hospital stays can contribute to this increased burden [8, 9]. Moreover, severe immunodeficiency as manifested by low CD4 counts significantly contribute to worse S aureus outcomes [10], but even in PLWH on antiretroviral therapy the overall incidence of S aureus colonization and disease remains significantly elevated [5, 11].
In the healthcare setting, nasal carriage of MRSA has been associated with subsequent MRSA infections [12]. The role of S aureus and MRSA colonization in subsequent infections is less clear in the community setting [13]. More recently, colonization of body sites other than the nares have been recognized as potential reservoirs for infecting S aureus strains [14, 15], including in PLWH [6]. These studies have also suggested that certain clonal types such as USA300 and USA500 preferentially colonize certain body sites such as the groin, in particular in patients infected with HIV [16, 17]. This suggests possible specific interactions between the impaired immune system after HIV infection and the molecular make-up of distinct S aureus clones. Patients with HIV infection, even when on antiretroviral therapy, appear to have persistent defects in Th17-mediated immune responses, which are critical in controlling S aureus infections [18, 19]. Moreover, concomitant increased Th2 response and chronic immune activation can lead to the downregulation of antimicrobial peptides human β-defensin (hBD)2 and hBD3, which are also important in the keratinocyte response to S aureus [20].
In addition to host factors, S aureus colonization is also determined by interaction with local microbiota. It has been suggested that the frequent commensal Staphylococcus epidermidis in particular has the ability to directly inhibit S aureus colonization by secretion of a serine protease, Esp1, or by activation of Toll-like receptor-2 on keratinocytes, triggering the release of antimicrobial peptides [20, 21]. The importance of this interaction in patients with HIV remains unknown.
In this study, we aimed to determine the prevalence and molecular epidemiology of S aureus, MRSA, and S epidermidis colonization in an inner-city population of HIV-infected individuals.
METHODS
Study Population
This cross-sectional study was reviewed and approved by the Columbia University Medical Center (New York, NY) Institutional Review Board. The study took place in January and February 2013 at the New York Presbyterian’s Comprehensive HIV Program clinic. Patients were informed about the study by their primary care provider and, after giving verbal consent to be contacted, were approached by the study team. After providing written informed consent, patients were recruited into the study. In total, 96 patients met the inclusion criteria of being HIV positive and ≥18 years of age; 93 patients completed the survey and provided all body site swabs. Patients attending the clinic were ineligible to participate if their HIV status was negative or unknown (n = 1); or if they had inflammatory bowel disease (n = 1). Other predefined exclusion criteria of active or acute acquired immune deficiency syndrome-defining opportunistic infections within 4 weeks before study entry or the current use of systemic immunosuppressive medications (eg, corticosteroids) within 14 days before study entry were not encountered. Participants were compensated with a $10 gift card to a CVS Pharmacy.
Survey
Patients completed structured interviews using audio computer-assisted self-interviewing software. Questions assessed demographic information and risk factors for MRSA, including personal care habits, as well as pertinent aspects of medical, social, and sexual histories. In addition, retrospective reviews of patient medical records were undertaken to ascertain relevant clinical and laboratory information. This also included assessment of underlying skin diseases (eczema, psoriasis, seborrheic dermatitis, lichenoid dermatitis, skin allergies, acne, tinea, basal cell carcinoma, and zoster) or skin and soft tissue infections and opportunistic infections and antibiotic exposures over the 3 months before enrollment.
Microbiological Sample Collection and Molecular Studies
After completing the survey, the nose, throat, and groin of participants were sampled using sterile premoistened swabs (BD BBL CultureSwab; BD Diagnostic Systems, Sparks, MD). Additional skin sites were sampled if study participants reported possible infected skin lesions. Samples were processed as previously described [22]. In brief, culture swabs were incubated overnight at 37°C in 6% salt-supplemented Tryptic soy broth and plated onto Mannitol salt agar (Becton Dickinson, Sparks, MD). Positive, mannitol-fermenting yellow colonies were isolated onto 5% sheep blood/Tryptic soy agar plates (blood/TSA) (Becton Dickinson). Staphylococcus aureus was identified from blood/TSA by coagulase and Protein A detection kit (Murex StaphAurex). In addition, all nonmannitol-fermenting and Staphaurex-negative colonies were isolated onto TSA (Becton Dickinson, Sparks, MD). Staphylococcus epidermidis was identified from TSA by species-specific polymerase chain reaction (PCR) as previously described [23].
All S aureus isolates were genotyped by staphylococcal protein A (spa) repeat-region sequencing and analysis (Ridom-staphsoftware). Strain relatedness was further evaluated using integrated based-upon repeat pattern (BURP) algorithms for spa Clonal Complex clustering (spa-CCs) [22]. Presence and type of Staphylococcal Chromosomal Cassette (SCC)mec, determined by multiplex PCR, was used to evaluate methicillin resistance [24]. Isolates were further genotyped by testing for the presence of the arginine-catabolic mobile element (ACME) gene by PCR [22].
All S epidermidis isolates were tested for the presence of the serine protease esp gene by PCR [21]. For antibiotic susceptibility testing, we randomly selected 1 S epidermidis isolate per participant from half of the S epidermidis-colonized individuals, due to cost restraints. Isolates were tested for resistance to penicillin, levofloxacin, gentamicin, erythromycin, linezolid, tetracycline, cefoxitin, and rifampin using the Kirby-Bauer method and Clinical and Laboratory Standards Institute (CLSI) standards [25].
Statistical Analyses
All statistical analyses were performed using SAS 9.4 (SAS Institute Inc., Cary, NC). We tested 2 separate outcomes against each hypothesized risk factor: colonization with S aureus (methicillin-sensitive S aureus [MSSA] or MRSA) at any body site or colonization with MRSA at any body site. Comparisons of colonized to noncolonized participants on dichotomous variables were carried out using χ2 or Fisher’s exact tests where appropriate. Bivariate analyses with continuous predictors were evaluated using unpaired Student t tests. Generalized estimating equations (GEEs) were used to evaluate the relationship between S aureus and S epidermidis colonization. This method allowed us to control for the multiple body site swabs taken per individual. All statistical tests were 2-sided, with P < .05 considered significant.
RESULTS
Study Demographics and Colonization Prevalence
This cross-sectional study included 93 HIV-positive participants with a mean age of 50 (interquartile range, 44–60). Approximately one third of the population was female (n = 32, 34%), two thirds were male (n = 60, 65%), and 1 individual was transgender (n = 1, 1%; Table 1). Hispanics (n = 44, 48%) and African Americans (n = 34, 37%) comprised the largest ethnic groups in our study, whereas whites (n = 9, 10%) and other races (n = 5, 5%) were less frequently represented. The majority of participants self-identified as heterosexual (n = 55, 59%); one third (n = 31, 33%) self-identified as men who have sex with men (50% of men). Most participants had well controlled HIV infection because 89 individuals (96%) had a recent CD4 count >200 and an undetectable viral load at their most recent visit. Only 4 patients had a recent viral load >1000. Few participants (n = 5, 5%) were hospitalized in the 3 months preceding study participation.
Table 1.
Demographics, HIV History, and Staphylococcus aureus Risk Factors of the Study Population
| S aureus Colonization | MRSA Colonization | ||||||
|---|---|---|---|---|---|---|---|
| Characteristic | Total N (%) | Yes (N = 36) | No (N = 57) | P a | Yes (N = 6) | No (N = 87) | P a |
| Demographics | |||||||
| Gender (n = 92) | |||||||
| Male | 60 (65) | 25 | 35 | .21 | 5 | 55 | .68 |
| Female | 32 (34) | 11 | 21 | 1 | 31 | ||
| Ethnicity (n = 92) | .36 | .09 | |||||
| Hispanic | 44 (48) | 18 | 26 | 2 | 42 | ||
| African American | 34 (37) | 10 | 24 | 1 | 33 | ||
| White | 9 (10) | 5 | 4 | 2 | 7 | ||
| Other | 5 (5) | 3 | 2 | 1 | 4 | ||
| Residence | .76 | 1 | |||||
| Apartment/house | 81 (87) | 32 | 49 | 6 | 75 | ||
| Otherb | 12 (13) | 4 | 8 | 0 | 12 | ||
| Sexual orientation | .65 | .40 | |||||
| MSM | 31 (33) | 11 | 20 | 3 | 28 | ||
| Other | 62 (67) | 25 | 37 | 3 | 59 | ||
| Work | .083 | .12 | |||||
| Unemployed | 55 (59) | 21 | 34 | 2 | 53 | ||
| Employed | 31 (33) | 13 | 18 | 3 | 28 | ||
| Retired | 5 (5) | 0 | 5 | 0 | 5 | ||
| Student | 2 (2) | 2 | 0 | 1 | 1 | ||
| Mean age (SD) | 50 (11) | 51 (11) | 50 (12) | .55 | 47 (12) | 50 (12) | .51 |
| HIV History | |||||||
| HIV risk factor (n = 89) | .94 | .90 | |||||
| Sex (male-female) | 44 (49) | 19 | 25 | 3 | 41 | ||
| Sex (male-male) | 30 (34) | 11 | 19 | 3 | 27 | ||
| Healthcare-associated | 7 (8) | 2 | 5 | 0 | 7 | ||
| Multiple risks | 6 (7) | 2 | 4 | 0 | 6 | ||
| Injection drug use | 2 (2) | 1 | 1 | 0 | 2 | ||
| Current CD4 count | .94 | ||||||
| <200 | 4 (4) | 2 | 2 | 1 | 3 | .07 | |
| 200 to 500 | 31 (33) | 12 | 19 | 0 | 31 | ||
| ≥500 | 58 (62) | 22 | 36 | 5 | 53 | ||
| Mean current CD4 % (SD) (n = 92) | 28 (10) | 27 (10) | 28 (11) | .69 | 31 (8) | 27 (11) | .38 |
| CD4 nadir (n = 89) | .78 | 1 | |||||
| <100 | 29 (33) | 12 | 17 | 2 | 27 | ||
| ≥100 | 60 (67) | 23 | 37 | 4 | 56 | ||
| Current* viral load | .14 | .71 | |||||
| Nondetectable | 69 (74) | 28 | 41 | 4 | 65 | ||
| 20–1000 | 20 (22) | 5 | 15 | 2 | 18 | ||
| >1000 | 4 (4) | 3 | 1 | 0 | 4 | ||
| S aureus risk factors | |||||||
| Lives with <5-year-olds | 2 (2) | 1 | 1 | 1 | 0 | 2 | 1 |
| Lives with a pet | 36 (39) | 14 | 22 | .98 | 3 | 33 | .67 |
| Lives alone | 39 (42) | 18 | 21 | .21 | 3 | 36 | .39 |
| Uses a gym* (n = 92) | 28 (30) | 16 | 12 | .019 | 4 | 24 | .07 |
| Shares towels | 6 (6) | 4 | 2 | .20 | 0 | 6 | 1 |
| Shares clothes | 2 (2) | 1 | 1 | 1 | 0 | 2 | 1 |
| Shaving, any site | 85 (91) | 33 | 52 | 1 | 4 | 81 | .08 |
| S aureus infections | |||||||
| Ever | 20 (22) | 9 | 11 | .51 | 2 | 18 | .61 |
| MSSA | 12 (13) | 3 | 9 | .36 | 0 | 12 | 1 |
| MRSA | 8 (9) | 6 | 2 | .052 | 2 | 6 | .08 |
| Within 1 year | 3 (3) | 2 | 1 | .56 | 1 | 2 | .18 |
| MSSA | 2 (2) | 1 | 1 | 1 | 0 | 2 | 1 |
| MRSA | 1 (1) | 1 | 0 | .39 | 1 | 0 | .06 |
| History of skin condition | 24 (26) | 10 | 14 | .73 | 2 | 22 | .65 |
| Previous skin infections | |||||||
| Ever | 15 (16) | 4 | 11 | .35 | 0 | 15 | .58 |
| Recent* | 11 (12) | 4 | 7 | 1 | 0 | 11 | 1 |
| Any sexual partners* (n = 89) | 49 (55) | 16 | 33 | .097 | 4 | 2 | .69 |
| Sexual partner with skin wounds | 2 (2) | 1 | 1 | 1 | 0 | 2 | 1 |
| Hospital admission* (n = 92) | 5 (5) | 0 | 5 | .15 | 0 | 5 | 1 |
| Past opportunistic infections | |||||||
| Any | 25 (27) | 8 | 17 | .42 | 0 | 25 | .19 |
| Thrush | 15 (16) | 4 | 11 | .30 | 0 | 15 | .58 |
| PCP | 8 (9) | 2 | 6 | .48 | 0 | 8 | 1 |
| Kaposi’s sarcoma | 3 (3) | 0 | 3 | .28 | 0 | 3 | 1 |
| Antimicrobial exposure* | |||||||
| Trimethoprim-sulfamethoxazole | 7 (8) | 2 | 5 | .70 | 0 | 7 | 1 |
| Mupirocin | 2 (2) | 1 | 1 | 1 | 0 | 2 | 1 |
| Dapsone | 3 (3) | 1 | 2 | 1 | 0 | 3 | 1 |
| Other antimicrobials | 10 (11) | 3 | 7 | .74 | 0 | 10 | 1 |
Abbreviations: HIV, human immunodeficiency virus; MSM, men whom have sex with men; MRSA, methicillin-resistant S aureus; MSSA, methicillin-sensitive S aureus; PCP, pneumocystis pneumonia; SD, standard deviation; SRO, single-room occupancy.
*Variable was assessed for the 3 months preceding study enrollment.
aχ2 or Fisher’s exact test was used for analyses of dichotomous variables. Unpaired Student t test was used for analyses of continuous variables.
bSROs, transitional, and shelters.
The self-reported burden of prior S aureus infections was low. Approximately one fifth (n = 20, 22%) of the study population reported ever having been diagnosed with an S aureus infection, and very few of these infections (n = 3, 3%) had occurred within the past year. Eleven participants (12%) had a skin infection in the past 3 months. Twenty-two participants reported using antibiotics within 3 months before study participation, which was confirmed for 20 patients by clinical chart review. Other common risk factors for S aureus infections, such as receiving tattoos (n = 3), recent incarceration (n = 1), or injection drug use (n = 1), were infrequently reported.
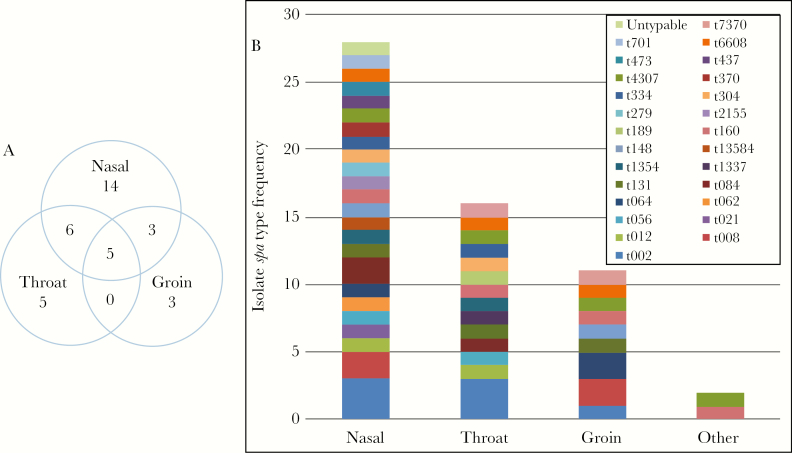
By culturing the nares, throat, and groin, we identified 36 individuals colonized with S aureus at 1 or more body sites (39%). Of these, 6 were colonized with MRSA (6%). Colonization with S aureus was most prevalent at the nares (n = 28, 30%), followed by throat (n = 16, 17%), groin (n = 11, 12%), and other sites (n = 2, 2%). Colonization occurred at multiple sites in 14 individuals, including 5 with colonization at all 3 sites. Eight individuals were colonized only at the throat or groin without concomitant nasal colonization (Figure 1A).
Figure 1.
Staphylococcus aureus colonization (A) by body site and (B) by frequency of spa types, stratified on body site. Data are expressed as absolute frequencies. Genotyping of isolates from all body sites yielded 26 unique spa-types.
We detected S epidermidis colonization in a majority of individuals at 1 or more body sites (n = 84, 90%). Colonization with S epidermidis was most prevalent at the nares (n = 66, 71%), followed by throat (n = 50, 54%), groin (n = 37, 40%), and other sites (n = 11, 12%). Nearly two thirds of S epidermidis colonized individuals were colonized at multiple body sites (n = 53, 57%); 18 were colonized at all 3 sites. Colonization with any S aureus, MRSA alone, or S epidermidis at each body site did not differ between males and females (Table 2).
Table 2.
Staphylococcus aureus and Staphylococcus epidermidis Colonization Prevalence by Body Site and Gender
| N = 92* | S aureus | MRSA | S epidermidis (esp-Positive) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Male | Female | P a | Male | Female | P a | Male | Female | P a | |
| Nasal | 19 | 9 | .73 | 3 | 0 | .55 | 41 | 24 | .50 |
| Throat | 12 | 4 | .37 | 2 | 0 | .54 | 36 | 13 | .08 |
| Inguinal | 7 | 4 | 1 | 2 | 1 | 1 | 23 | 13 | .83 |
Abbreviations: MRSA, methicillin-resistant S aureus.
*Transgender individuals were excluded.
aχ2 or Fisher’s exact test was used for analyses.
Molecular and Phenotypic Characterization
Among the 36 colonized individuals, we observed 26 different spa-types. Of the 14 individuals colonized at multiple body sites, only 1 had different spa-types at the 3 tested body sites. The vast majority of colonizing isolates were MSSA (86%) and belonged to a diversity of spa-types (Figure 1B). The most frequent spa-type was t002, accounting for 15% of MSSA isolates. Half of the MRSA isolates were spa-type t008, consistent with USA300. Six of the 8 MRSA isolates (75%) were ACME positive, consistent with USA300. We observed spa-type t064 in both the MSSA and MRSA group.
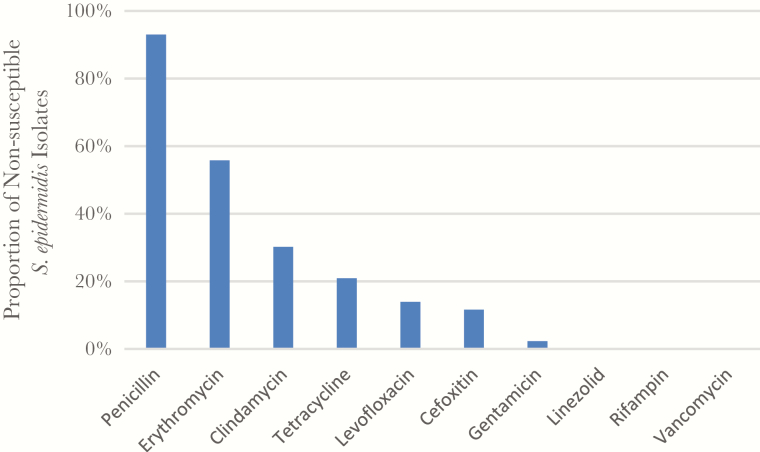
Staphylococcus epidermidis have been associated with substantial antibiotic resistance [26]. Antibiotic susceptibility testing on a subset of the S epidermidis collection showed that nearly all isolates were resistant to penicillin (n = 40, 93%; Figure 2). Only 12% (n = 5) were resistant to cefoxitin, consistent with methicillin-resistant S epidermidis (Figure 2). Methicillin-resistant S epidermidis strains were more likely to be nonsusceptible to tetracycline compared with the methicillin-sensitive S epidermidis group (80% vs 13%, respectively; Fisher’s exact test, P = .005). The 2 groups had no other significant differences in antibiotic susceptibilities, and all isolates were susceptible to linezolid, rifampin, and vancomycin. All S epidermidis isolates were typed for the esp gene and were positive.
Figure 2.
Staphylococcus epidermidis antibiograms. Susceptibilities were determined using the Kirby-Bauer disk diffusion method and compared against Clinical and Laboratory Standards Institute standards.
Risk Factors for Staphylococcus aureus Colonization
To assess for risk factors of S aureus colonization, we carried out bivariate analyses. Participants colonized with S aureus at any body site were more likely to have regularly used a gym within the last 3 months (44%) compared with those who were not colonized (21%; P = .019; Table 1). However, this association did not hold for carriage of MRSA (P = .067). A reported history of S aureus infections within 1 year of the interview was not associated with carriage of S aureus or MRSA. No other demographic characteristics or healthcare risk factors were significantly different when comparing carriers to noncarriers.
Almost all participants (90%) were colonized with S epidermidis at 1 or more body sites. The esp gene was ubiquitous throughout this sample collection, with all isolates displaying positive PCR results. On the individual level, we found that 5 of the 6 (83%) MRSA-colonized individuals and 27 of the 30 (90%) MSSA-colonized individuals were also colonized with S epidermidis at at least 1 body site. However, collapsing the data onto the individual level does not take into account variation across body sites. Hence, we chose to further investigate this relationship by stratifying our observations by swabbed body site. We found that when the nose or throat was colonized with S epidermidis, S aureus colonization was less likely to co-occur at the same site. This was most apparent in the throat, where only 2 participants were cocolonized, compared with 14 who were colonized with S aureus alone (Table 3). Because swabs from the same individual cannot be considered independent observations, we used GEE models to assess the statistical significance of S epidermidis colonization on S aureus colonization. In the GEE model, an interaction term between body site and S epidermidis colonization was not significant. We found that the odds of S aureus colonization were significantly and drastically reduced when S epidermidis was detected (P = .0001; Table 3). After controlling for swab site, gender, and age, we identified that the odds of S aureus colonization were 80% less if a person was colonized with S epidermidis (adjusted odds ratio [aOR], 0.20; 95% confidence interval, .09–.45; P < .0001) (Table 3). More importantly, this dramatic protective effect did not differ significantly across body sites (interaction term nonsignificant; P = .10). We were unable to perform a similar analysis on MRSA alone due to low numbers.
Table 3.
GEE Models Assessing the Association of Staphylococcus epidermidis With Staphylococcus aureus Colonization
| SA Colonization (MSSA or MRSA) | SE Colonized | Not SE Colonized |
|---|---|---|
| Nasal | N = 66 | N = 27 |
| SA colonized | 11 (17) | 17 (63) |
| Not SA colonized | 55 (83) | 10 (37) |
| Throat | N = 50 | N = 43 |
| SA colonized | 2 (4) | 14 (33) |
| Not SA colonized | 48 (96) | 29 (67) |
| Inguinal | N = 37 | N = 56 |
| SA colonized | 5 (14) | 6 (11) |
| Not SA colonized | 32 (86) | 50 (89) |
| Variables in GEE Model | OR of SA Colonization (95% CI) | SE Parameter P Value |
| SE | 0.32 (0.18–0.57) | .0001 |
| SE, site* | 0.20 (0.09–0.44) | <.0001 |
| SE, site, gender, age | 0.20 (0.09–0.45) | <.0001 |
Abbreviations: CI, confidence interval; CLSI, Clinical and Laboratory Standards Institute; GEE, generalized estimating equation; MRSA, methicillin-resistant S aureus; MSSA, methicillin-sensitive S aureus; OR, odds ratio; SA, S aureus; SE, S epidermidis.
*Swab site was not a significant effect measure modifier of the association between SE and SA colonization.
DISCUSSION
In this population of HIV-positive individuals attending an urban comprehensive HIV clinic, we observed a lower prevalence of MRSA colonization (6.5%) compared with 15%–20% reported in previous studies on individuals infected with HIV [6, 27]. However, the prevalence of MRSA colonization was still higher than the 1.5% reported nationally from the general population [28] and what we previously observed in healthy individuals attending the hospital’s dental clinic (2%) [22]. Moreover, the overall prevalence of S aureus colonization by 3 body-site screening (39%) was relatively low compared with prior studies in other HIV study populations [29]. More importantly, GEE aOR indicated a protective effect of S epidermidis against S aureus colonization. It appears unlikely that this effect was mediated by Esp1 protease because all S epidermidis isolates carried the corresponding gene. It has been suggested that this enzyme harbors direct activity against S aureus [21], although Fredheim et al [30] also found no association between presence of the esp gene and S aureus carriage. These differences might reflect clonal variability of S aureus isolates between studies and S aureus resistance to Esp-mediated killing in some samples. Although USA300 and USA500 were predominant amongst MRSA isolates consistent with prior studies [16, 17], MSSA were highly diverse in our study population.
Alternative means of S epidermidis conferring protection to S aureus might involve other direct bacterial effectors such as the release of phenol soluble modulins, lantibiotics, or an indirect effect via modulating the local immune response. This includes S epidermidis-mediated induction of interleukin (IL)-17A(+) CD8(+) T cells that migrate to the epidermis and skin [31]. It has been shown that clearance of nasal S aureus carriage requires T cells and IL-17A, which leads to recruitment of neutrophils. Even after initiation of antiretroviral treatment and recovery of CD4 cells, the T-cell homeostasis in infected patients remains disturbed and includes a Th2 polarization away from Th-17, upregulation of regulatory T cells, and persistently elevated CD8 T cell counts. Future studies are needed to address how polarization of the T-cell response in HIV patients contributes to colonization with S aureus and S epidermidis and potential competition between these organisms.
Competition between S aureus and S epidermidis is likely not restricted to PLWH [31] and might also differ at particular sites. Few studies have assessed the prevalence of S epidermidis colonization in healthy individuals [32]. Our observations here are comparable to early investigations where colonization ranged from 62% at the legs and 78% at the nares to 92% in the axillae [33, 34]. More recently, Staphylococcus lugdunensis has been suggested as capable of inhibiting S aureus colonization at the nares via release of lugdunin [35]. A series of investigations has also documented the contribution of the innate immune response to mediate competition between bacteria such as Haemophilus influenza and Streptococcus pneumoniae at mucosal sites [36].
In a recent meta-analysis of 6558 PLWH from 32 studies, 6.9% were MRSA carriers. A history of hospitalization over the past year conferred a 3.1 times higher risk and recent incarceration a 1.7 times higher risk of MRSA colonization [37]. Several important differences between our study and others need to be considered. High-risk social behaviors such as drug use or recent incarceration as well as crowding were low in our study population as were previous S aureus infections or recent hospital admission [38, 39]. Our patient population was slightly older with an average age of 50 [5, 27], and we enrolled a higher proportion of heterosexual men and overall more women (34%) than others [5, 27, 39, 40]. The majority of our study population was Hispanic, which has previously been associated with lower MRSA colonization in PLWH [27]. Although we did not observe significant gender differences between S aureus carriage in the current study, we recently observed that men attending a sexually transmitted disease clinic were almost 4 times more likely to harbor S aureus in the anterior nares [15]. Gym use was significantly associated with S aureus colonization (P = .019), but it was not associated with MRSA colonization (P = .067), which might be explained by our relatively small sample size. In a recent study, Crum-Cianflone et al [38] also identified public gym use as a risk factor for S aureus colonization and concluded that specific behaviors, rather than HIV-related risk factors, predicted S aureus colonization and SSTIs.
Low CD4 counts (<100) have been associated with an increased risk of MRSA colonization in previous studies from approximately 10 years ago [41–43]. Participants in our study were under better control and, on average, had high CD4 counts. Although approximately one third of participants had a documented CD4 nadir of <100, CD4 nadir was not associated with MRSA or S aureus colonization. Antibiotic exposure over the past 3 months was overall low (~20%) compared with other studies in this community (~50%) [22], despite the use of prophylactic antibiotics to prevent opportunistic infections, and might have contributed to the lower MRSA prevalence. Antibiotic resistance in S epidermidis was also low.
Several limitations to our study need to be considered. This was a relatively small sample, with low MRSA prevalence and surveyed over a short time period. The study might have lacked power to detect small differences between colonized and noncolonized individuals. The study population reflects a single clinic in an urban environment with low self-reported behavioral risk factors for S aureus carriage. We did not measure immune markers such as soluble CD14 or IL-17 and their potential impact on S aureus colonization. We used standard culture techniques for isolation of S aureus [22]. We cannot exclude that this approach may have interfered with isolation of S epidermidis. However, given the high prevalence of S epidermidis colonization at the different body sites, this appears less likely.
CONCLUSIONS
Taken together, our results of a relatively low MRSA prevalence likely reflect a combination of PLWH with well controlled HIV and lack of high-risk behaviors previously associated with S aureus colonization and infection. Further research should explore whether the observed protective effect of S epidermidis on S aureus colonization extends beyond PLWH and what the molecular mediators of these interactions are.
Acknowledgments
We thank the patients for their participation.
Finanical support. This study was funded by National Institutes of Health/National Institute of Allergy and Infectious Diseases (grant K08 AI090013; to A.-C. U.) and a Columbia University Provost grant (to A.-C. U.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Moran GJ, Talan DA. Community-associated methicillin-resistant Staphylococcus aureus: is it in your community and should it change practice? Ann Emerg Med 2005; 45:321–2. [DOI] [PubMed] [Google Scholar]
- 2. Moran GJ, Krishnadasan A, Gorwitz RJ, et al. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med 2006; 355:666–74. [DOI] [PubMed] [Google Scholar]
- 3. Crum-Cianflone NF, Burgi AA, Hale BR. Increasing rates of community-acquired methicillin-resistant Staphylococcus aureus infections among HIV-infected persons. Int J STD AIDS 2007; 18:521–6. [DOI] [PubMed] [Google Scholar]
- 4. Miller M, Cespedes C, Vavagiakis P, et al. Staphylococcus aureus colonization in a community sample of HIV-infected and HIV-uninfected drug users. Eur J Clin Microbiol Infect Dis 2003; 22:463–9. [DOI] [PubMed] [Google Scholar]
- 5. Shet A, Mathema B, Mediavilla JR, et al. Colonization and subsequent skin and soft tissue infection due to methicillin-resistant Staphylococcus aureus in a cohort of otherwise healthy adults infected with HIV type 1. J Infect Dis 2009; 200:88–93. [DOI] [PubMed] [Google Scholar]
- 6. Peters PJ, Brooks JT, Limbago B, et al. Methicillin-resistant Staphylococcus aureus colonization in HIV-infected outpatients is common and detection is enhanced by groin culture. Epidemiol Infect 2011; 139:998–1008. [DOI] [PubMed] [Google Scholar]
- 7. Popovich KJ, Weinstein RA, Aroutcheva A, et al. Community-associated methicillin-resistant Staphylococcus aureus and HIV: intersecting epidemics. Clin Infect Dis 2010; 50:979–87. [DOI] [PubMed] [Google Scholar]
- 8. Shadyab AH, Crum-Cianflone NF. Methicillin-resistant Staphylococcus aureus (MRSA) infections among HIV-infected persons in the era of highly active antiretroviral therapy: a review of the literature. HIV Med 2012; 13:319–32. [DOI] [PubMed] [Google Scholar]
- 9. Miller M, Cespedes C, Bhat M, et al. Incidence and persistence of Staphylococcus aureus nasal colonization in a community sample of HIV-infected and -uninfected drug users. Clin Infect Dis 2007; 45:343–6. [DOI] [PubMed] [Google Scholar]
- 10. Jacobson MA, Gellermann H, Chambers H. Staphylococcus aureus bacteremia and recurrent staphylococcal infection in patients with acquired immunodeficiency syndrome and AIDS-related complex. Am J Med 1988; 85:172–6. [DOI] [PubMed] [Google Scholar]
- 11. Ramsetty SK, Stuart LL, Blake RT, et al. Risks for methicillin-resistant Staphylococcus aureus colonization or infection among patients with HIV infection. HIV Med 2010; 11:389–94. [DOI] [PubMed] [Google Scholar]
- 12. von Eiff C, Becker K, Machka K, et al. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N Engl J Med 2001; 344:11–6. [DOI] [PubMed] [Google Scholar]
- 13. Miller M, Cook HA, Furuya EY, et al. Staphylococcus aureus in the community: colonization versus infection. PLoS One 2009; 4:e6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang ES, Tan J, Eells S, et al. Body site colonization in patients with community-associated methicillin-resistant Staphylococcus aureus and other types of S. aureus skin infections. Clin Microbiol Infect 2010; 16:425–31. [DOI] [PubMed] [Google Scholar]
- 15. Miko BA, Uhlemann AC, Gelman A, et al. High prevalence of colonization with Staphylococcus aureus clone USA300 at multiple body sites among sexually transmitted disease clinic patients: an unrecognized reservoir. Microbes Infect 2012; 14:1040–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gordon RJ, Quagliarello B, Cespedes C, et al. A molecular epidemiological analysis of 2 Staphylococcus aureus clonal types colonizing and infecting patients with AIDS. Clin Infect Dis 2005; 40:1028–36. [DOI] [PubMed] [Google Scholar]
- 17. Mermel LA, Eells SJ, Acharya MK, et al. Quantitative analysis and molecular fingerprinting of methicillin-resistant Staphylococcus aureus nasal colonization in different patient populations: a prospective, multicenter study. Infect Control Hosp Epidemiol 2010; 31:592–7. [DOI] [PubMed] [Google Scholar]
- 18. Ishigame H, Kakuta S, Nagai T, et al. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity 2009; 30:108–19. [DOI] [PubMed] [Google Scholar]
- 19. Cho JS, Pietras EM, Garcia NC, et al. IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J Clin Invest 2010; 120:1762–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lai Y, Cogen AL, Radek KA, et al. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J Invest Dermatol 2010; 130:2211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iwase T, Uehara Y, Shinji H, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 2010; 465:346–9. [DOI] [PubMed] [Google Scholar]
- 22. Uhlemann AC, Knox J, Miller M, et al. The environment as an unrecognized reservoir for community-associated methicillin resistant Staphylococcus aureus USA300: a case-control study. PLoS One 2011; 6:e22407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hirotaki S, Sasaki T, Kuwahara-Arai K, Hiramatsu K. Rapid and accurate identification of human-associated staphylococci by use of multiplex PCR. J Clin Microbiol 2011; 49:3627–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Milheiriço C, Oliveira DC, de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus . Antimicrob Agents Chemother 2007; 51:3374–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard—Ninth Edition. CLSI document M2-A9. Wayne, PA; Clinical Laboratory Standards Institute; 2006. [Google Scholar]
- 26. Miragaia M, Thomas JC, Couto I, et al. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J Bacteriol 2007; 189:2540–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Popovich KJ, Hota B, Aroutcheva A, et al. Community-associated methicillin-resistant Staphylococcus aureus colonization burden in HIV-infected patients. Clin Infect Dis 2013; 56:1067–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gorwitz RJ, Kruszon-Moran D, McAllister SK, et al. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001-2004. J Infect Dis 2008; 197:1226–34. [DOI] [PubMed] [Google Scholar]
- 29. Lee CJ, Sankaran S, Mukherjee DV, et al. Staphylococcus aureus oropharyngeal carriage in a prison population. Clin Infect Dis 2011; 52:775–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fredheim EG, Flægstad T, Askarian F, Klingenberg C. Colonisation and interaction between S. epidermidis and S. aureus in the nose and throat of healthy adolescents. Eur J Clin Microbiol Infect Dis 2015; 34:123–9. [DOI] [PubMed] [Google Scholar]
- 31. Naik S, Bouladoux N, Wilhelm C, et al. Compartmentalized control of skin immunity by resident commensals. Science 2012; 337:1115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev 2014; 27:870–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kloos WE, Schleifer KH. Isolation and characterization of staphylococci from human skin. Int J System Bacteriol 1975; 25:62–79. [Google Scholar]
- 34. Kloos WE, Musselwhite MS. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl Microbiol 1975; 30:381–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zipperer A, Konnerth MC, Laux C, et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 2016; 535:511–6. [DOI] [PubMed] [Google Scholar]
- 36. Lysenko ES, Ratner AJ, Nelson AL, Weiser JN. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog 2005; 1:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zervou FN, Zacharioudakis IM, Ziakas PD, et al. Prevalence of and risk factors for methicillin-resistant Staphylococcus aureus colonization in HIV infection: a meta-analysis. Clin Infect Dis 2014; 59:1302–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Crum-Cianflone NF, Wang X, Weintrob A, et al. Specific behaviors predict Staphylococcus aureus colonization and skin and soft tissue infections among human immunodeficiency virus-infected persons. Open Forum Infect Dis 2015; 2:ofv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vieira MT, Marlow MA, Aguiar-Alves F, et al. Living conditions as a driving factor in persistent methicillin-resistant Staphylococcus aureus colonization among HIV-infected youth. Pediatr Infect Dis J 2016; 35:1126–31. [DOI] [PubMed] [Google Scholar]
- 40. Imaz A, Camoez M, Di Yacovo S, et al. Prevalence of methicillin-resistant Staphylococcus aureus colonization in HIV-infected patients in Barcelona, Spain: a cross-sectional study. BMC Infect Dis 2015; 15:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cenizal MJ, Hardy RD, Anderson M, et al. Prevalence of and risk factors for methicillin-resistant Staphylococcus aureus (MRSA) nasal colonization in HIV-infected ambulatory patients. J Acquir Immune Defic Syndr 2008; 48:567–71. [DOI] [PubMed] [Google Scholar]
- 42. McDonald LC, Lauderdale TL, Lo HJ, et al. Colonization of HIV-infected outpatients in Taiwan with methicillin-resistant and methicillin-susceptible Staphylococcus aureus . Int J STD AIDS 2003; 14:473–7. [DOI] [PubMed] [Google Scholar]
- 43. Villacian JS, Barkham T, Earnest A, Paton NI. Prevalence of and risk factors for nasal colonization with Staphylococcus aureus among human immunodeficiency virus-positive outpatients in Singapore. Infect Control Hosp Epidemiol 2004; 25:438–40. [DOI] [PubMed] [Google Scholar]