In vivo analysis of the F-BAR–domain protein syndapin indicates that it regulates basolateral recycling of transmembrane cargo in the Caenorhabditis elegans intestine. SDPN-1 may facilitate the fission of membranes from the early endosome that are acquiring recycling endosome characteristics, allowing cargo to exit the early endosome.
Abstract
Syndapin/pascin-family F-BAR domain proteins bind directly to membrane lipids and are associated with actin dynamics at the plasma membrane. Previous reports also implicated mammalian syndapin 2 in endosome function during receptor recycling, but precise analysis of a putative recycling function for syndapin in mammalian systems is difficult because of its effects on the earlier step of endocytic uptake and potential redundancy among the three separate genes that encode mammalian syndapin isoforms. Here we analyze the endocytic transport function of the only Caenorhabditis elegans syndapin, SDPN-1. We find that SDPN-1 is a resident protein of the early and basolateral recycling endosomes in the C. elegans intestinal epithelium, and sdpn-1 deletion mutants display phenotypes indicating a block in basolateral recycling transport. sdpn-1 mutants accumulate abnormal endosomes positive for early endosome and recycling endosome markers that are normally separate, and such endosomes accumulate high levels of basolateral recycling cargo. Furthermore, we observed strong colocalization of endosomal SDPN-1 with the F-actin biosensor Lifeact and found that loss of SDPN-1 greatly reduced Lifeact accumulation on early endosomes. Taken together, our results provide strong evidence for an in vivo function of syndapin in endocytic recycling and suggest that syndapin promotes transport via endosomal fission.
INTRODUCTION
Enclosed by a membrane, cells establish and maintain their architecture in part through a process called endocytosis. Endocytosis is a highly conserved trafficking pathway that begins with the vesicle-mediated internalization of proteins and lipids from the plasma membrane. Cells internalize receptors through clathrin-dependent endocytosis (CDE) and clathrin-independent endocytosis (CIE) pathways (Doherty and McMahon, 2009; Grant and Donaldson, 2009). In CDE, clathrin adaptor proteins can recognize motifs within the cytoplasmic tails of transmembrane proteins. On recognition, these adaptor proteins coassemble with clathrin at the plasma membrane, forming cage-like structures that produce plasma membrane–coated pits. As the process continues, the invaginated clathrin coated pits (CCPs) pinch off into vesicles, uncoat, and fuse with one another and preexisting early endosomes (Doherty and McMahon, 2009). Once in the early endosome, the reduced pH results in the dissociation of many ligand–receptor complexes. As fluid and membrane are added to the early endosome, membrane tubules begin to form and extend. At this point, many internalized receptors are recycled to the plasma membrane either directly (rapid recycling) or indirectly via the endocytic recycling compartment (ERC) (slow recycling) or Golgi (retrograde recycling). Membrane proteins and lumenal content that fail to recycle are transported to the late endosomes and eventually the lysosomes for degradation (Doherty and McMahon, 2009; Grant and Donaldson, 2009).
Endocytic uptake and recycling are tightly regulated and function in a diverse array of biological processes, including cell adhesion and junction formation, cell migration, cytokinesis, cell polarity, and signal transduction (Maxfield and McGraw, 2004; Grant and Donaldson, 2009). In mammalian fibroblasts, the ERC is typically composed of a dense collection of juxtanuclear membrane tubules and vesicles that ultimately carry macromolecules back to the plasma membrane (Maxfield and McGraw, 2004; Grant and Donaldson, 2009). Important aspects of the mechanisms that mediate the return of cargo molecules to the plasma membrane remain to be elucidated.
The Caenorhabditis elegans intestine—a polarized epithelium—is a powerful model system to characterize the molecular components required for endocytic recycling (Chen et al., 2006; Sato et al., 2014). This system allows for genetic manipulation and imaging analysis in the context of an intact living epithelial tube. The worm intestine is surprisingly simple, composed of 20 cells organized into a single layer, forming nine donut-like rings (Leung et al., 1999). The apical domain faces the lumen and is specialized for nutrient uptake, covered in dense microvilli. As in mammalian intestinal epithelia, the C. elegans intestinal lumen is supported by an underlying terminal web and overlying glycocalyx (McGhee, 2007). The basolateral surface faces the pseudocoelom and is responsible for the exchange of molecules between the intestine and the rest of the body (McGhee, 2007).
A number of key conserved recycling regulators were first discovered in C. elegans, including RAB-10 and RME-1 (Grant et al., 2001; Chen et al., 2006). Animals lacking RME-1 accumulate gigantic endosomes that trap basolateral transmembrane recycling cargoes (Grant et al., 2001; Chen et al., 2006). These enlarged organelles are positive for ARF-6 and lack RAB-5, placing RME-1 function at a late step of endocytic recycling (Chen et al., 2006; Shi et al., 2009). Complementary studies in mammalian cell culture demonstrated that loss of mRme-1/EHD1, a mammalian homologue of RME-1, greatly slows the recycling of transferrin and major histocompatibility class I protein (MHCI), trapping recycling cargo in the juxtanuclear ERC (Lin et al., 2001; Caplan et al., 2002). Whereas recycling tubules are concentrated near the nucleus in cultured mammalian cells, in the C. elegans intestine RME-1 labels a network of tubular recycling endosomes located just below the basolateral intestinal plasma membrane (Grant et al., 2001; Chen et al., 2006). Although superficially similar to the intestinal phenotype in rme-1 mutants, animals deficient in RAB-10 display an earlier block in basolateral recycling that is more cargo specific. The grossly enlarged endosomes in rab-10 mutants are positive for RAB-5 and ARF-6 and accumulate the model CIE cargo green fluorescent protein–tagged α-chain of human interleukin-2 receptor (hTAC::GFP) but not the model CDE cargo GFP-tagged human transferrin receptor (hTfR::GFP; Chen et al., 2006; Shi et al., 2010). RAB-10 appears to function just after the early endosome master regulator RAB-5, with feedback from RAB-10 acting to down-regulate RAB-5 as cargo recycles (Liu and Grant, 2015).
Little is known of recycling endosome biogenesis and its relationship to early endosomes. Given the tubular nature of the recycling network, we sought to further analyze the role of BAR domain proteins in the sculpting of membrane tubules, potentially facilitating the biogenesis of recycling cargo carriers. One BAR-domain protein family suggested to work in the endocytic recycling pathway is syndapin/pacsin.
Drosophila syndapin is important for embryonic cellularization, associating with anillin and septin proteins and influencing formin recruitment and actin dynamics on cleavage furrow tips (Takeda et al., 2013; Sherlekar and Rikhy, 2016). Drosophila syndapin has also been shown to associate with postsynaptic membranes at the neuromuscular junction, affecting the formation of a specialized subsynaptic reticulum in muscle cells (Kumar et al., 2009; Koles et al., 2015).
Mammals express three syndapin/pacsin genes encoding F-BAR–domain membrane-bending proteins (Dharmalingam et al., 2009). Syndapin 1 is neuron specific and is required for activity-dependent bulk endocytosis at presynaptic membranes but not direct synaptic vesicle endocytosis via clathrin. Syndapin 2 is ubiquitously expressed and has been variously reported to function in CDE (fibroblasts and apical membrane of epithelia), caveolae dynamics, Golgi traffic, endocytic recycling, actin dynamics, neuronal development, and cell migration (Ritter et al., 1999; Meng et al., 2011; Quan and Robinson, 2013). The first work linking syndapin 2 to endocytic recycling in mammalian cells showed that NPF motifs in syndapin 2 bind to the EH-domain of Ehd1/mRme-1 and that overexpression of the isolated NPF or EH domains interfered with transferrin recycling (Braun et al., 2005). The syndapin 2 SH3 domain interacts with the recycling tubule protein MICAL-L1, further implicating it in recycling regulation (Giridharan et al., 2013). Syndapin 3 expression is up-regulated during adipocyte differentiation, and overexpression of syndapin 3, which lacks NPF sequences, increased the recycling of Arf6-dependent cargo GLUT1 in adipocytes (Roach and Plomann, 2007).
Unlike syndapin 1, for which mouse knockouts were used, many studies on syndapin 2 and syndapin 3 must be interpreted with caution because they mainly tested syndapin function by injecting anti-syndapin antibodies and/or overexpressing the full-length protein or individual syndapin protein domains (Andersson et al., 2008; Clayton et al., 2009). In cases in which syndapin 2 function was studied after knockdown, analysis of its function in recycling was hampered by its requirement in CDE. There is a general lack of knockdown/knockout data analyzing the recycling of cargo. Collectively these results implicate some syndapin family proteins in endocytic recycling but do not definitively establish such a role.
Here we analyze the function of the C. elegans syndapin/pacsin-family protein SDPN-1, establishing its role in basolateral endocytic recycling in the context of the C. elegans intestine, a polarized epithelium. We previously found partial colocalization of SDPN-1 with RME-1, and found that loss of RME-1 or ALX-1/ALIX reduced the number and increased the average size of SDPN-1–labeled endosomes in the C. elegans intestine (Shi et al., 2007; Pant et al., 2009). Here we show that SDPN-1 is also enriched on a population of RAB-5–labeled early endosomes located close to the basolateral plasma membrane. We also find that loss of SDPN-1 appears to block basolateral recycling of CIE and CDE cargo and results in accumulation of abnormal endosomes containing both early and recycling endosome markers. Our data suggest that SDPN-1 is required for a step in endosome maturation that allows separation of early and recycling compartments, likely contributing to a fission step in this process.
RESULTS
Full-length recombinant SDPN-1 tubulates acidic membranes in vitro
Like its mammalian homologues, SDPN-1 contains one conserved N-terminal F-BAR domain and a single C-terminal SH3 domain (Figure 1, A and B). Extensive reports show that the isolated F-BAR domain of syndapin 1 is capable of tubulating/vesiculating membranes in vitro (Wang et al., 2009; Rao et al., 2010). To test conservation of function, we expressed and purified full-length recombinant SDPN-1A and examined its function in vitro when reconstituted with negatively charged liposomes. SDPN-1 protein created narrow membrane tubules with an average diameter of ∼40 nm (Figure 1D and E). These results are similar to those reported for the purified F-BAR domain of mammalian syndapin 1, indicating conservation of molecular function (Wang et al., 2009; Rao et al., 2010).
FIGURE 1:
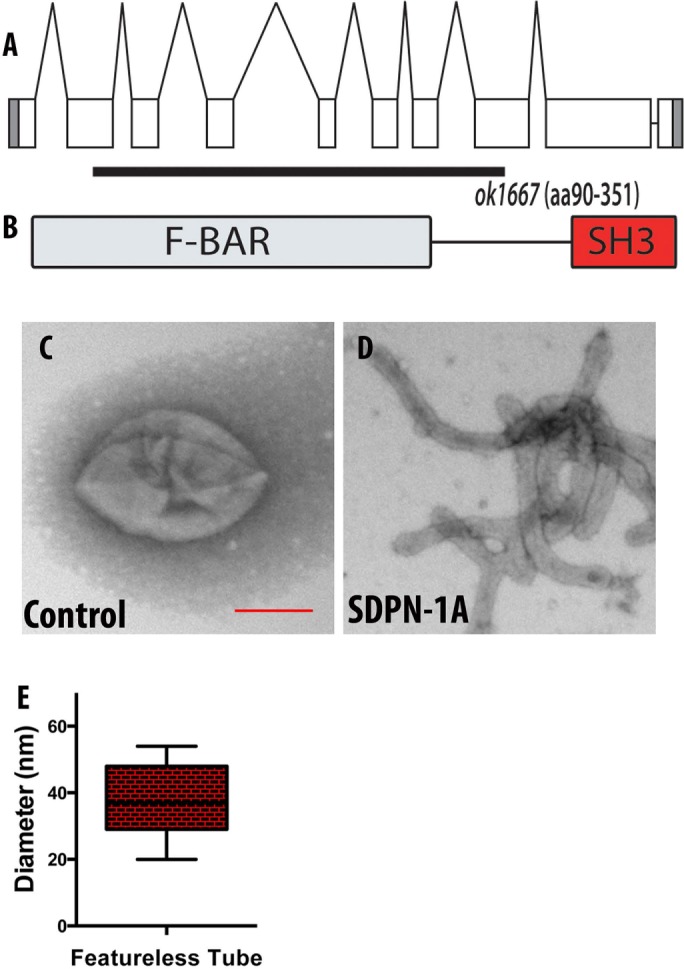
(A) Genomic structure of the sdpn-1 gene and the location of the ok1667 mutant deletion. ok1667 is a 2547–base pair deletion from the second exon to the eighth exon. (B) The ok1667 allele deletes sequences encoding amino acids 90–351, including the majority of the F-BAR domain, and places downstream sequences out of frame. (C–E) In vitro membrane tubulation of PtdSer liposomes by full-length SDPN-1a. Electron micrographs of acidic PtdSer liposomes (0.05 mg/ml, average diameter 400 nm) incubated with 2.5 μM (C) GST or (D) full-length SDPN-1. (E) Statistical analysis. Diameters of membrane tubules shown in D were quantified on at least three independently prepared electron microscopy grids. Scale bar, 200 nm.
SDPN-1 is broadly expressed in multiple tissues
To characterize the expression pattern and subcellular localization of SDPN-1, we created low–copy number integrated transgenic C. elegans lines expressing GFP fused to the C-terminus of SDPN-1 driven by sdpn-1 promoter sequences (Supplemental Figure S1). We observed clear expression in the intestine and pharynx and some additional cells in the head region (Supplemental Figure S1). In the intestine, SDPN-1::GFP localized to distinct puncta near the basolateral cortex, as well as on or near the apical intestinal plasma membrane (Supplemental Figure S1, B and C).
SDPN-1 is required for the transport of CDE and CIE cargo that recycles to the plasma membrane via the recycling endosome
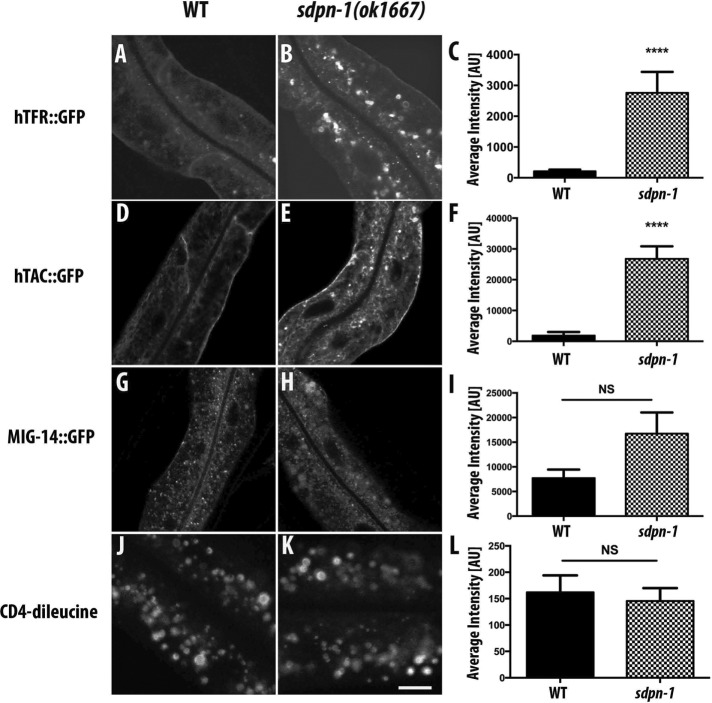
To test the requirements of SDPN-1 in cargo trafficking, we assayed the effect of a sdpn-1 deletion mutant, ok1667, on a diverse panel of basolateral transmembrane cargo with well-characterized postinternalization trafficking routes expressed specifically in the C. elegans intestine (Chen et al., 2006; Shi et al., 2007, 2010; Sun et al., 2012; Sato et al., 2014). The ok1667 allele is missing sequences from within the second to eighth exons, deleting a substantial fraction of the F-BAR–domain coding sequences and producing a predicted frameshift and premature stop codon predicted to remove the remainder of the protein in both predicted splice forms (Figure 1). Thus the ok1667 mutant should not produce any functional SDPN-1 protein. The well-characterized basolateral cargoes that we tested included hTAC, DAF-4 (type II transforming growth factor β [TGF-β] receptor), hTFR (human transferrin receptor), MIG-14/Wntless (a transmembrane chaperone for WNT ligands), SMA-6 (a type I TGF-β receptor), and CD4-dileucine (Chen et al., 2006; Shi et al., 2009; Gu et al., 2013; Gleason et al., 2014). hTAC and DAF-4 recycle via the recycling endosome in an ARF-6-dependent pathway (Shi et al., 2012; Gleason et al., 2014). MIG-14 and SMA-6 recycle via retrograde recycling in a retromer-dependent manner (Shi et al., 2009; Gleason et al., 2014). hTfR, MIG-14, SMA-6, and CD4-dileucine are clathrin dependent in their endocytosis. hTAC is a clathrin-independent cargo. CD4-dileucine does not appear to recycle and appears to enter the degradative pathway after endocytosis (Gu et al., 2013).
hTAC::GFP, hTfR::GFP, and DAF-4::GFP displayed dramatic accumulations at internal sites within the intestinal cells of sdpn-1(ok1667) mutants and did not accumulate on the cell surface (Figure 2, A–F, and Supplemental Figure S2, A–C). The abnormal accumulation of recycling cargo hTFR::GFP was rescued by intestine-specific expression of SDPN-1::tagRFP, further indicating that the cargo transport defect is caused by loss of SDPN-1 (Supplemental Figure S3, A–C). The localization of MIG-14::GFP, SMA-6::GFP, and GFP::CD4-dileucine remained comparable to that of wild type in sdpn-1(ok1667) mutants (Figure 2, G–I, and Supplemental Figure S2, D-F). Taken together, these results indicate that SDPN-1 is not required for uptake from the basolateral plasma membrane or retrograde transport from endosomes to the Golgi. Instead, loss of SDPN-1 appears to specifically affect basolateral cargo proteins that recycle to the plasma membrane via the early endosome–to–recycling endosome route.
FIGURE 2:
sdpn-1 mutants disrupt recycling cargo traveling along the early endosome/recycling endosome/plasma membrane route. Laser scanning confocal micrographs of about one cell length of the C. elegans intestine expressing GFP-tagged transmembrane cargo proteins. In wild-type animals, (A) hTfR::GFP and (D) hTAC::GFP predominantly label the basolateral plasma membrane and sparse intracellular puncta. In sdpn-1(ok1667) mutants, both model cargo proteins accumulate intracellularly: (B) hTfR::GFP and (E) hTAC-GFP. (C, F) Quantification of average intracellular fluorescence intensity. Total intensity was measured for six animals for each genotype sampled in three different regions of each intestine. Localization and signal intensity remained comparable to wild type in sdpn-1(ok1667) mutants expressing MIG-14::GFP (G, H) or the unrecycled model transmembrane cargo GFP::CD4-dileucine (J, K). (I, L) Quantification of MIG-14::GFP and GFP::CD4-dileucine average fluorescence intensity. Total intensity was measured in six animals of each genotype sampled in three different regions of each intestine. Error bars represent SEM. ****p < 0.0001 (Student’s t test). Scale bar, 10 μm.
We also examined the apical intestinal plasma membrane protein PGP-1::GFP in sdpn-1 mutants (Supplemental Figure S4, A and B). PGP-1::GFP in sdpn-1 mutants did not accumulate intracellularly and appeared normally localized to the apical membrane, suggesting that apical secretion/recycling does not depend upon SDPN-1. However, the PGP-1::GFP marker, and electron microcopy of unmarked strains, revealed that the intestinal lumen was expanded in sdpn-1 mutants, with an unusual convoluted structure (Supplemental Figure S4, B and D). Close inspection of electron micrographs suggested that although expanded, the apical membrane itself and the associated microvilli and terminal web appeared normal (Supplemental Figure S4, E and F). These results may indicate a role for SDPN-1 in apical endocytosis, as suggested in other systems (Da Costa et al., 2003).
SDPN-1 affects several endosome types
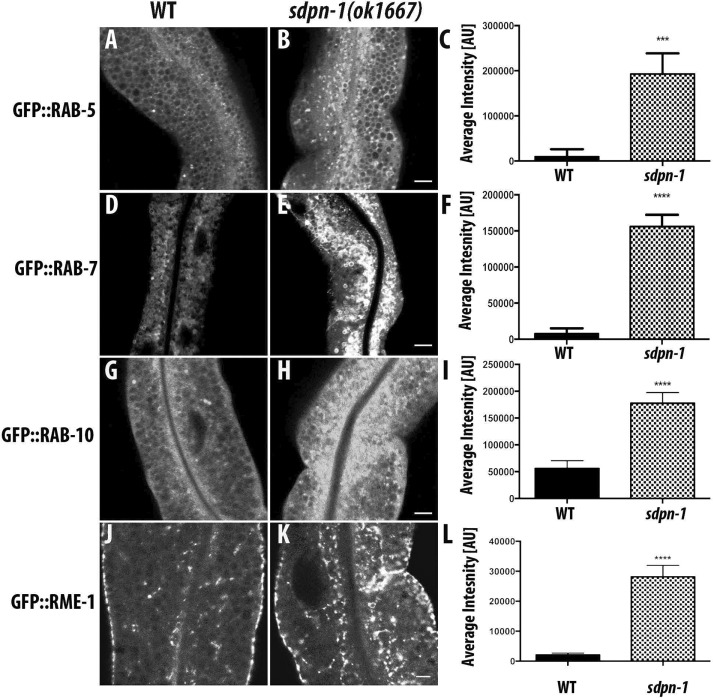
Our transmembrane cargo data indicate a role for SDPN-1 in endocytic recycling. To further investigate how SDPN-1 affects endosomes, we quantified the effects of SDPN-1 loss of function using a group of well-established molecular markers for specific endosome types. Collectively such studies can help to determine the specific transport steps affected. We noted an increase in RAB-5– and RAB-7–labeled endosomal size and fluorescence intensity in sdpn-1 mutants (Figure 3, A–F, and Supplemental Figure 5, A and B), indicating strong effects on early and late endosomes. We also observed an abnormal accumulation of RAB-10-labeled endosomes (Figure 3, G–I, and Supplemental Figure 5C). RAB-10 promotes basolateral recycling and partially overlaps RAB-5 on endosomes (Chen et al. 2006; Liu and Grant, 2015).
FIGURE 3:
sdpn-1 mutants affect early endosomes, basolateral recycling endosomes, and late endosomes. Laser scanning confocal micrographs of the worm intestine expressing GFP-tagged fusion proteins that are resident markers for distinct endosomal compartments. sdpn-1 mutants show abnormal intracellular accumulation of early endosomes labeled with GFP::RAB-5, late endosomes marked with GFP::RAB-7, GFP::RAB-10, which functions at the early endosome/basolateral recycling endosome interface, and basolateral recycling endosomes marked with GFP::RME-1. Control micrographs in the wild type background are shown for (A) GFP::RAB-5, (D) GFP::RAB-7, (G) GFP::RAB-10, and (J) GFP::RME-1. Confocal images in the sdpn-1(ok1667) mutant background are shown for (B) GFP::RAB-5, (E) GFP::RAB-7, (H) GFP::RAB-10, and (K) GFP::RME-1. Quantification of total fluorescence intensity for (C) GFP::RAB-5, (F) GFP::RAB-7, (I) GFP::RAB-10, and (L) GFP::RME-1. Error bars represent SEM: ***p < 0.001, ****p < 0.0001 (Student’s t test). Scale bar, 10 μm (B, E, H, K).
We also examined effects on recycling regulator RME-1 localization. Of importance, sdpn-1 mutants caused striking accumulations of RME-1–positive endosomes in the deep cytoplasm far from the plasma membrane, best viewed in cross section of the intestine, where they are normally very sparse (Figure 3, J–M, and Supplemental Figure S5D). Loss of SDPN-1 function did not alter the distribution of RAB-11, a marker for apical recycling endosomes (Supplementary Figure S6, A–D).
Loss of SDPN-1 traps recycling cargo hTFR in abnormal endosomes
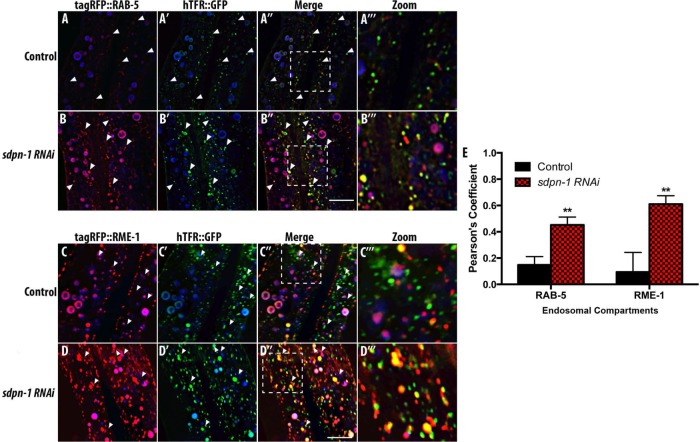
To better understand SDPN-1’s role in cargo recycling, we next analyzed the localization of recycling cargo that accumulated in sdpn-1 mutants, calculating Pearson’s r for accumulated hTfR::GFP and several markers for different steps in endosomal trafficking. In wild-type animals at steady state, we observe a low correlation between intracellular hTFR::GFP puncta positive for early endosome marker tagRFP::RAB-5, suggesting that hTfR traffics through early endosomes quickly (Figure 4, A–A′′′). On depletion of SDPN-1 using RNA interference (RNAi), we observed abnormal accumulation of hTFR, RAB-5, and RME-1, similar to what we observed in sdpn-1(ok1667) mutants, further confirming the phenotypes associated with loss of SDPN-1 (Figure 4, B–B′′′, and Supplemental Figure S7, A–C).
FIGURE 4:
Loss of SDPN-1 traps recycling cargo hTFR::GFP in abnormal endosomes positive for early and recycling endosome markers. All micrographs are from deconvolved three-dimensional (3D) confocal image stacks acquired in intact living animals expressing intestinal-specific GFP- and RFP-tagged proteins. White arrowheads depict positive colocalization. (A–A′′′) Under control conditions, recycling cargo hTFR::GFP- and tagRFP::RAB-5-labeled early endosomes displayed little overlap. (B–B′′′) A strong increase in colocalization of hTFR::GFP and tagRFP::RAB-5 was observed in sdpn-1 RNAi animals. (C–D′′′) hTFR::GFP also displayed an increase in overlap with basolateral recycling endosome marker tagRFP::RME-1 in sdpn-1 RNAi animals. In each image, autofluorescent lysosome-like organelles appear in in all three channels (including blue), whereas GFP appears only in the green channel and RFP appears only in the red channel. Green and red signals that do not overlap with the blue channel represent pure GFP and RFP signals, respectively. (E) Pearson’s r for each endosomal compartment. Six animals. Error bars represent SEM: **p < 0.01. Scale bar, 10 μm.
Of importance, quantification of the degree of colocalization between hTFR::GFP and tagRFP::RAB-5 in sdpn-1(RNAi) animals revealed a significant increase in colocalization, indicating that loss of SDPN-1 traps recycling cargo in RAB-5–positive endosomes (Figure 4E). We detected only minor overlap between hTFR::GFP and basolateral recycling endosome marker tagRFP::RME-1 in wild-type animals (Figure 4, C–C′′′) but a significant increase in colocalization between hTFR::GFP and tagRFP::RME-1 after sdpn-1 RNAi (Figure 4, D–D′′′ and E). Of importance, the site of hTFR accumulation coincided with redistribution of RME-1 to endosomes that appear in the medial plane of the intestine, where RME-1 is normally very sparse. By contrast, the degree of overlap of hTfR::GFP and tagRFP::RAB-7 or tagRFP::RAB-10 after sdpn-1 RNAi was similar to that of wild type (Supplemental Figure S8, A–D′′′).
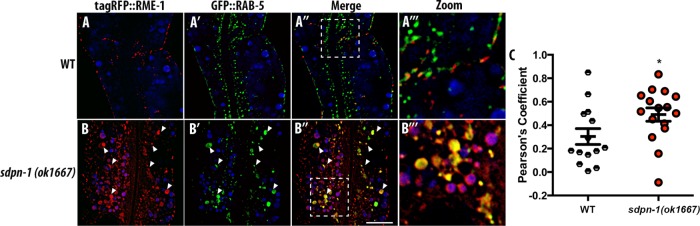
Given the accumulation of recycling cargo hTfR::GFP in the central cytoplasm with endosome markers that typically represent two separate organelles along the recycling pathway, and particularly the accumulation of RME-1 in this region of the cell where it is normally rare, we decided to test for changes in the colocalization of RAB-5 and RME-1. As expected from our previous work, control animals displayed very low correlation between GFP::RAB-5 and tagRFP::RME-1 signals (Figure 5, A–A′′′). However, upon sdpn-1 knockdown, we observed a significant increase in the correlation of GFP::RAB-5 and tagRFP::RME-1 signals (Figure 5, B and C), suggesting that without SDPN-1, the cells accumulate abnormal endosomes that display mixed characteristics of early and recycling endosomes and such endosomes are defective in cargo recycling.
FIGURE 5:
Loss of SDPN-1 results in the accumulation of abnormal endosomes containing markers of both early endosomes and basolateral recycling endosomes. All micrographs are from deconvolved 3D confocal image stacks acquired in intact living animals expressing intestinal-specific GFP- and RFP-tagged proteins. White arrowheads depict positive colocalization. (A–A′′′) In control animals, basolateral recycling endosome marker tagRFP::RME-1 (A) displays little overlap with early endosome marker GFP::RAB-5 (A′). (A″, A′′′) Merged image and enlarged merge. (B–B′′′) In sdpn-1(ok1667) mutant animals, enlarged pleiomorphic endosomes, many of which are positive for both markers, accumulate. (C) Pearson’s r for GFP and RFP signals under wild-type and mutant conditions. In each image autofluorescent lysosome-like organelles appear in in all three channels (including blue), whereas GFP appears only in the green channel and RFP appears only in the red channel. Green and red signals that do not overlap with the blue channel represent pure GFP and RFP signals, respectively. Error bars represent SEM: *p < 0.05. Scale bar, 10 μm.
SDPN-1 predominately resides on early and recycling endosomes
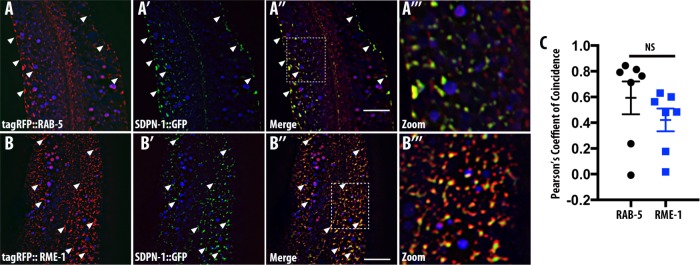
If SDPN-1 functions along the early endosome–to–recycling endosome pathway, we would expect to find SDPN-1 protein enriched on early endosomes, recycling endosomes, or both. Intestine-specific expression of GFP-tagged SDPN-1 labels abundant cytoplasmic puncta near the basolateral surface. Previous work indicated that SDPN-1::GFP overlaps with RFP::RME-1–labeled recycling endosomes (Shi et al., 2007; Pant et al., 2009). To address this question in more depth, we used confocal microscopy to compare the localization of SDPN-1::GFP with additional organelle markers. We found that SDPN-1::GFP also colocalizes well with a subset of RAB-5–positive endosomes that reside close to the basolateral plasma membrane at the periphery of the adult intestinal cells (Figure 6, A–A′′′) as judged by Pearson’s r (Figure 6C). We also confirmed colocalization of SDPN-1::GFP with tagRFP::RME-1 (Figure 6, B–B′′′). Our results indicate that the F-BAR protein SDPN-1 is a resident component of early and recycling endosomes, where it could function at the interface of early and recycling endosome separation.
FIGURE 6:
SDPN-1 resides on early and basolateral recycling endosomes. All micrographs are from deconvolved 3D confocal image stacks acquired in intact living animals expressing intestinal-specific GFP- and RFP-tagged proteins. (A–A′′′) SDPN-1::GFP colocalizes with RAB-5–labeled early endosomes. White arrowheads indicate endosomes labeled by both SDPN-1::GFP and tagRFP-RAB-5. (A′′′) Magnified image designated by the rectangular outline in A″. (B–B′′′) SDPN-1::GFP is also enriched on tagRFP-RME-1–labeled basolateral recycling endosomes. White arrowheads indicate positive colocalization between SDPN-1::GFP and tagRFP::RME-1. (B′′′) Magnified image designated by the rectangular outline in B″. (C) Pearson’s r for colocalization of SDPN-1::GFP with tagRFP-RAB-5 and tagRFP-RME-1. Six animals. Error bars represent SEM.
We also observed sparse colocalization of SDPN-1::GFP with late endosomal marker RAB-7, and this was mainly restricted to puncta rather than large RAB-7 rings, suggesting residence of some SDPN-1 on early endosomes beginning to acquire RAB-7 but not mature late endosomes (Supplemental Figure S9, A–A′′). Additional analysis indicated that SDPN-1::GFP failed to colocalize with the Golgi marker AMAN-2::mCherry (Supplemental Figure S9, B–B′′).
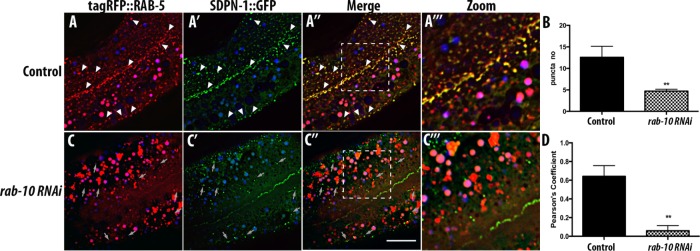
RAB-10 is important for SDPN-1 endosomal recruitment
Because the sdpn-1 phenotype indicates a likely function at the intersection of early endosomes and recycling endosomes, a step in transport that we previously showed involves RAB-10, we sought to determine whether SDPN-1 localization depends on RAB-10 function. On depletion of RAB-10 protein, SDPN-1::GFP appeared more diffusive, with a reduction in the number of remaining SDPN-1–labeled structures (Figure 7, B and C). This is similar to the diffusive GFP::RME-1 labeling in rab-10 mutants, which is associated with concomitant loss of the tubulovesicular meshwork found near the basolateral plasma membrane (Chen et al., 2006; Shi et al., 2010). As noted earlier, wild-type animals displayed extensive colocalization between SDPN-1 and RAB-5 on cortical endosomes (Figure 7, A–A′′′). On loss of RAB-10, RAB-5 positive-endosomes are more numerous and enlarged (Chen et al., 2006; Shi et al., 2010; Liu and Grant, 2015) but fail to colocalize with the remaining SDPN-1 puncta (Figure 7, C–C′′′), as determined by Pearson’s r (Figure 7D). These results suggest that RAB-10 functions upstream or at the same step as SDPN-1 to coordinate recycling events.
FIGURE 7:
rab-10(RNAi) impairs SDPN-1 recruitment to endosomes and segregates RAB-5 from the remaining SDPN-1–labeled structures. All micrographs are from deconvolved 3D confocal image stacks acquired in intact living animals expressing intestinal-specific GFP- and RFP-tagged proteins. Under control conditions (A–A′′′) SDPN-1::GFP and tagRFP::RAB-5 colocalize extensively on endosomes. White arrowheads indicate positive overlap. (C–C′′′) rab-10(RNAi) animals displayed more-diffusive SDPN-1::GFP signal and a decrease in overlap between SDPN-1::GFP and RAB-5::GFP. Gray arrows indicate residual SDPN-1::GFP-labeled structures lacking RAB-5. In each image, autofluorescent lysosome-like organelles appear in in all three channels (including blue), whereas GFP appears only in the green channel and RFP appears only in the red channel. Green and red signals that do not overlap with the blue channel represent pure GFP and RFP signals, respectively. (B) Quantification of SDPN-1::GFP puncta number. (D) Pearson’s r for SDPN-1::GFP and tagRFP::RAB-5. Six animals. Error bars represent SEM: **p < 0.01. Scale bar, 10 μm.
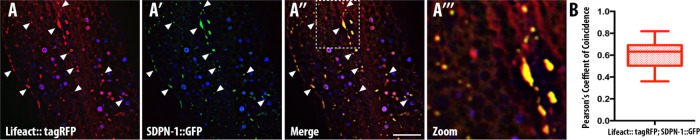
Filamentous actin localizes to SDPN-1–positive endosomes
Mammalian studies of syndapin function at the plasma membrane suggest that syndapin promotes actin polymerization (Qualmann et al., 1999; Qualmann and Kelly, 2000). The syndapin 2 F-BAR domain has also been reported to bind directly to actin filaments (Kostan et al., 2014). Because actin polymerization on endosomes could function to promote endosomal fission, we sought to determine whether SDPN-1–positive endosomes display enrichment of F-actin (Kessels and Qualmann, 2006; Puthenveedu et al., 2010; Giridharan et al., 2013; Granger et al., 2014). To test this, we quantified the localization of F-actin, as reported by tagRFP::LifeAct, an F-actin biosensor, on SDPN-1::GFP–positive endosomes in the intestine of living intact animals (Figure 8, A–A′′′). Under steady-state conditions, confocal micrographs revealed considerable colocalization of LifeAct and SDPN-1, with Pearson’s r = 0.6 (Figure 8B). These data indicate that SDPN-1–positive endosome structures are enriched in filamentous actin.
FIGURE 8:
Filamentous actin localizes to SDPN-1 positive endosomes. All micrographs are from deconvolved 3D confocal image stacks acquired in intact living animals expressing intestinal-specific GFP- and RFP-tagged proteins. LifeAct-tagRFP, an F-actin biosensor containing 17 amino acids of the yeast actin-binding protein ABP-140, was used to report filamentous actin in intestinal cells. White arrowheads depict positive overlap. (A–A′′′) SDPN-1::GFP-positive endosomes are enriched in filamentous actin. Arrowheads correspond to positive colocalization between SDPN-1::GFP and Life Act::tagRFP. (A′′′) Magnified image designated by the region in rectangular outline in A″. In each image, autofluorescent lysosome-like organelles appear in in all three channels (including blue), whereas GFP appears only in the green channel and RFP appears only in the red channel. Green and red signals that do not overlap with the blue channel represent pure GFP and RFP signals, respectively. (B) Pearson’s r for colocalization of SDPN-1::GFP with LifeAct-tagRFP. Six animals. Error bars represent SEM. Scale bar, 10 μm.
sdpn-1 mutant early endosomes are depleted in filamentous actin
If SDPN-1–mediated actin polymerization is necessary for efficient transport along the recycling pathway, we might expect to observe reduced F-actin levels on early endosomes upon loss of SDPN-1. To test this, we analyzed colocalization of GFP-LifeAct and early endosome marker RFP::RAB-5 in the intestinal epithelium. As with SDPN-1, we found that LifeAct is enriched on early endosomes positive for RAB-5 (Figure 9, A–A′′′). After sdpn-1 RNAi, RAB-5–labeled endosomes were visibly enlarged and mainly depleted of LifeAct (Figure 9, B and C). RAB-5–labeled endosomes in rab-10(RNAi) animals were also depleted of the LifeAct reporter, consistent with a function for RAB-10 upstream or at the same step as SDPN-1 (Supplemental Figure S11, A–E). We were unable to detect enrichment of LifeAct on RAB-7–labeled or RME-1–labeled endosomes in wild-type animals (Supplemental Figure 10, A–B′′).
FIGURE 9:
Loss of SDPN-1 disrupts association of filamentous actin with early endosomes. All micrographs are from deconvolved 3D confocal image stacks acquired in intact living animals expressing intestinal-specific LifeAct-GFP and tagRFP-RAB-5 in control RNAi (A–A′′′) and sdpn-1(RNAi) (B–B′′′). In control animals, many tagRFP::RAB-5-positive endosomes are positive for LifeAct::GFP. White arrowheads indicate positive colocalization. (B–B′′′) A striking decrease in localization of LifeAct::GFP and tagRFP::RAB-5 was seen in sdpn-1 RNAi animals. Gray arrows indicate early endosomes depleted of filamentous actin. In each image, autofluorescent lysosome-like organelles appear in in all three channels (including blue), whereas GFP appears only in the green channel and RFP appears only in the red channel. Green and red signals that do not overlap with the blue channel represent pure GFP and RFP signals, respectively. (C) Pearson’s r for colocalization of LifeAct::GFP with tagRFP::RAB-5. Six animals. Error bars represent SEM.***p < 0.001 (Student’s t test) Scale bar, 10 μm.
DISCUSSION
In mammalian cell culture, the syndapin 2 protein has been implicated in endocytic recycling, primarily because it binds to recycling regulators EHD1/mRme-1 and MICAL-L1 (Braun et al., 2005; Giridharan et al., 2013). However, analysis of the requirement of syndapin in endocytic recycling in mammalian cells has been hampered by a requirement for syndapin in clathrin-mediated uptake from the plasma membrane and potential redundancy among the three separate genes encoding syndapins in mammals. To analyze more clearly the requirements for syndapin in recycling, we took advantage of an uncharacterized deletion mutant in the single syndapin/pacsin family gene encoded in the C. elegans genome. Although the sequence reported to link syndapin 2 to EHD proteins is not evolutionary conserved in worm SDPN-1, and C. elegans lacks a MICAL-L1 homologue, our analysis further indicates that SDPN-1 indeed functions in endocytic cargo recycling.
We defined the requirements for SDPN-1 in recycling by analyzing a variety of recycling cargo proteins in an sdpn-1 deletion mutant. Loss of SDPN-1 did not appear to trap any of these cargo proteins at the plasma membrane, allowing a clearer analysis of syndapin function in endocytic recycling possible in mammalian cells. A specific group of cargo proteins that recycle along the early endosome/recycling endosome/plasma membrane route (hTFR, hTAC, and DAF-4) accumulated intracellularly in sdpn-1 mutants, with trapped recycling cargo clearly accumulating in organelles positive for RAB-5 and RME-1. These results suggested that SDPN-1 functions at the interface of early endosomes and recycling endosomes, an interpretation supported by our finding that tagged forms of SDPN-1 are clearly enriched on endosomes that contain early and basolateral recycling endosome markers. Of interest, the cargo proteins trapped in sdpn-1 mutants include examples internalized by clathrin-dependent and clathrin-independent mechanisms, supporting the idea that these cargo types converge in their endocytic transport at the level of the early endosome (Naslavsky et al., 2003). Of importance, loss of SDPN-1 did not perturb the localization of retrograde recycling cargo, suggesting that SDPN-1 functions in the exit of specific recycling cargo from the endosome. sdpn-1 mutants also did not perturb the trafficking of the model degradative cargo CD4-dileucine, suggesting that SDPN-1 is not required for the degradative function of late endosomes. Instead, we propose that the observed abnormal morphology of late endosomes in sdpn-1 mutants is a results of defects in maturation of endosomes and suggest that such late endosomes are still capable of membrane protein degradation and retromer-based recycling to the Golgi.
We consider it likely that the observed accumulation of recycling cargo is a consequence of a defect in the separation of early endosomes and recycling endosomes in sdpn-1 mutants. This is supported by our finding that sdpn-1 mutants accumulate a class of abnormal endosomes positive for early endosome and recycling endosome markers that are normally separate. SDPN-1 may support fission of membranes of the early endosome that are acquiring recycling endosome characteristics, trapping recycling cargo when such fission fails.
Although RAB-10 and SDPN-1 both seem to affect the interface of early endosomes and recycling endosomes, their relationship is unclear. RAB-10 primarily affects recycling of CIE cargo, with little effect on CDE cargo, whereas SDPN-1 strongly affects both types of recycling cargo. In addition, loss of SDPN-1 causes intracellular accumulation of RME-1–labeled structures, whereas loss of RAB-10 causes a general loss of RME-1–labeled structures. RAB-10 affects levels of phosphatidylinositol-(4,5)-bisphosphate on endosomes, a phosphoinositide largely generated from phosphatidylinositol-4-phosphate (PI(4)P) (Shi et al., 2012). One possibility is that RAB-10 affects the amount of PI(4)P on early endosomes acquiring recycling endosome characteristics, and such phosphoinositide levels may affect recruitment or activity of SDPN-1 and RME-1 on endosomal membranes. PI(4)P generation has been implicated in the exit of recycling cargo from early endosomes in other systems (Jovic et al., 2009; Ketel et al., 2016)
Local actin polymerization is also closely associated with membrane budding and fission (Merrifield, 2004; Itoh et al., 2005; Romer et al., 2010). Actin polymerization on vesicles has been proposed to provide mechanical tension needed to drive vesiculation from donor membrane compartments (Merrifield, 2004; Itoh et al., 2005; Romer et al., 2010). In fact, syndapin-dependent actin polymerization has been proposed to promote many actin-driven cellular processes, including release of clathrin-coated vesicles from the plasma membrane and their movement into the cytoplasm (Qualmann and Kelly, 2000). The SH3 domain of mammalian syndapin has been shown to bind to actin nucleation–promoting factors (NPFs), including N-WASP and Cordon-Bleu (Cobl), which in turn activate ARP2/3 to polymerize actin, although it is not clear whether the these known syndapin-interacting NPFs are found on endosomes (Qualmann et al., 1999; Qualmann and Kelly, 2000; Modregger et al., 2000; Ahuja et al., 2007). Another role proposed for membrane-associated actin is in the stabilization of tubular microdomains on endosomes, as demonstrated for the β2-adrenergic receptor, although syndapin is not known to participate in this process (Puthenveedu et al., 2010). Studies in Drosophila report that syndapin localizes to the cleavage furrow and recruits actin regulators to mediate cytoskeletal remodeling during cytokinesis (Takeda et al., 2013; Sherlekar and Rikhy, 2016).
Because of the importance of actin-based mechanisms in various membrane-sculpting events and the association of syndapin with actin regulation, we considered it likely that early endosomal SDPN-1 promotes endosome fission via actin polymerization or recruitment, concomitant with F-BAR–mediated membrane bending. Indeed, we observed a clear enrichment of filamentous actin on SDPN-1–positive endosomes and found that loss of SDPN-1 or RAB-10 disrupts the normal enrichment of F-actin on early endosomes. Thus, although further analysis is required to determine the precise function of such syndapin-regulated endosomal actin, it is likely that syndapin-mediated endosomal cargo recycling depends on local F-actin accumulation.
MATERIALS AND METHODS
General methods and strains
All C. elegans strains used in this study were derived from the wild-type Bristol strain N2. Genetic crosses and other C. elegans husbandry were performed according to standard protocols (Brenner, 1974). Strains expressing transgenes were grown at 20°C. Supplemental Table S1 gives a complete list of the strains used in this study.
RNAi studies were performed using the feeding method (Timmons and Fire, 1998). Feeding constructs were obtained from the Ahringer Library and sequence verified. For all experiments, L4-stage animals were treated for 30 h, and F1 progeny were scored as adults.
The extent of the sdpn-1(ok1667) deletion mutation was determined by PCR and DNA sequencing. For routine crosses, sdpn-1(ok1667) was either balanced or kept homozygous/hemizygous, with final confirmation of genotype using either of two three-primer combinations: tacaccatcccaacgaatca sdpn1 extF, ttatgcacttcgatgcctca sdpn1extR, and gtgtctccagatgcggcaacca sdpn1 intR; or gggctactctttccgacatc ok1667_OF, cgttgatcgaacggtatctg ok1667_OR, and tgtatacatggaggcgatgg ok1667_IR. We found that sdpn-1 animals displayed reduced fecundity, producing 141 ± 22.4 eggs, compared with wild-type controls producing 187 ± 39.2. The U-shaped gonad arms of sdpn-1 mutants remained comparable to wild type (n = 30), suggesting that this defect was not caused by mismigration of gonad arms.
Plasmid and transgenic strains
For intestinal-specific expression, GFP or red fluorescent protein (tag RFP)/mCherry fusion transgenes were cloned into a previously described vha-6 promoter-driven vector modified with a Gateway cassette inserted at the Asp-718I site just upstream of the GFP and RFP coding region (Chen et al., 2006). All PCR products of the genes of interest were first cloned into the Gateway entry vector pDONR221 by BP reaction (Invitrogen, Carlsbad, CA). Isolation of pDONR221 plasmids carrying genes of interest was transferred into the intestinal expression vectors by Gateway recombination cloning in the LR clonase II (Invitrogen) reaction to generate N-terminal/C-terminal fusions. Low-copy integrated transgenic animals expressing all of these plasmids were obtained by microparticle bombardment (Praitis et al., 2001; Schweinsberg and Grant, 2013).
To construct the GFP-tagged sdpn-1 transgene driven by its own promoter, sdpn-1 genomic and promoter sequences were PCR amplified from the C. elegans genomic DNA. Amplified PCR products were then cloned into the entry vector pDONR221 and then transferred into the C. elegans pPD117.01 vector containing the Gateway cassette (Invitrogen), followed by GFP coding sequences, let-858 3′ untranslated region sequences, and the unc-119 gene of Caenorhabditis briggsae. The GFP-tagged construct was bombarded into unc-119(ed3) mutant animals to establish low-copy integrated transgenic lines by particle bombardment (Praitis and Maduro, 2011).
Protein expression
A PreScission site was added to the full-length SDPN-1A cDNA by standard PCR and cloned into a pGEX2T expression plasmid (Amersham Pharmacia, Piscataway, NJ) to yield an N-terminally glutathione S-transferase (GST)–PreScission fusion protein.
Control GST (Zwaagstra et al., 2011) and GST-SDPN-1A fusion proteins were expressed in Escherichia coli Artic Express cells (Stratagene, La Jolla, CA). Bacterial cultures grown in Luria–Bertani broth were induced at an OD600 of 0.8 with 0.1 mM isopropyl-β-d-thiogalactoside and grown overnight at 12°C. Bacterial cells were lysed in 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; pH 7.5), 400 mM NaCl, 1 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich, St. Louis, MO) using an Avestin Emulsiflex-C3 (Avestin, Ontario, Canada) high-pressure homogenizer. Lysed cells were centrifuged at 10,000 × g for 30 min in a Sorvall SS-34 rotor (Sorvall, Geulph, ON, Canada). The soluble supernatant fraction was centrifuged for another 40 min at 4°C at 100,000 × g in a Beckman Ti-70 ultracentrifuge rotor (Beckman, Fullerton, CA). Equilibrated with lysis buffer, the supernatant was applied to the glutathione–Sepharose 4B column (Amersham). The column with bound protein was washed thoroughly with wash buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM DTT). The GST moiety was cleaved using 225-μg PreScission protease (Amersham). To check the purity of tagless protein, cleaved SDPN-1A was analyzed by SDS–PAGE followed by Coomassie staining. Cleaved protein was eluted with several volumes of wash buffer, snap frozen in liquid nitrogen, and stored at −80°C. For proteoliposome assays, the frozen aliquots were rapidly thawed and then spun at 20,000 × g at 4°C to remove any aggregated protein. Protein concentration was determined under denaturing conditions by absorbance at 280 nm.
Liposome preparation
Phosphatidylserine (PtdSer) lipids were purchased from Avanti Polar Lipids (Alabaster, AL). For liposome preparation, chloroform was evaporated under a continuous stream of argon gas and subjected to vacuum desiccation overnight. PtdSer lipids were then resuspended at 1 mg/ml concentration in argon-purged liposome buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM MgCl2). Liposomes of 0.4-μm average diameter were formed by using 0.4-μm polycarbonate track-etched membrane filters (Whatman) with an Avanti Mini Extruder as per manufacturer’s instructions (Polar Lipids).
Liposome tubulation assays
For proteoliposome tubulation assays, 2.5 μM SDPN-1 was incubated with 0.5 mg/ml final concentration of 100% PtdSer liposomes in liposome buffer. All samples were incubated on ice for 4 min, and 12 min after the start of protein addition, the samples were spotted on 300 mesh carbon-Formvar–coated copper grids. Grids were negatively stained with 1% uranyl acetate at 25°C for 1.5 min. To examine membrane morphologies, electron microscopy was performed using a JEOL 1200 EX or JEOL 100 CX transmission electron microscope at 80 kV. Images were obtained at 50,000× magnification as indicated. Quantification of liposome tubule diameter was performed using Fiji (National Institutes of Health [NIH], Bethesda, MD).
Measurement function
Mean values were calculated and plotted on graphs. SEM was calculated and used as Y-error bars on graphs.
Transmission electron microscopy
Young adult wild-type animals were prepared for electron microscopy by a standard immersion fixation protocol (Hall, 1995). Well-fed animals were moved from the culture plate into buffered aldehyde fixative and immediately cut open with a razor blade to allow access to the fixative past the cuticle. After fixation in aldehydes, worms were rinsed in buffer and refixed in buffered osmium tetroxide, en bloc stained with uranyl acetate, and then dehydrated and embedded into Epon for thin sectioning. Mutant animals (ok1667) prepared by high-pressure freezing and freeze substitution (HPF/FS) following a standard protocol (Hall et al., 2012). Briefly, animals were moved from the culture plate into an HPF sample carrier in a slurry of E. coli, and the sample carrier was closed and fast frozen in a Baltec HM 010 high-pressure freezer. Frozen samples were freeze substituted into 2% osmium tetroxide and 2% water in acetone over 5 d and then rinsed in cold acetone and embedded in plastic. For all samples, thin sections were collected on a diamond knife, mounted on Formvar-coated slot grids, poststained with uranyl acetate, and examined in a Philips CM10 electron microscope fitted with an Olympus Morada digital camera. For best views of the intestinal lumen, we chose animals that had been sectioned lengthwise, and we viewed the anterior lumen at the level of INT 1 and INT2, before the intestine is squeezed by the gonad, and behind the swollen lumen of the anteriormost portion of INT1.
Microscopy and image analysis
Live worms were mounted on 10% agarose pads with 10 mM levamisole as described previously (Sato et al., 2005). Multiwavelength fluorescence colocalization images were obtained using the Axio Imager Z1 (Carl Zeiss MicroImaging, Oberkochen, Germany) equipped with a Yokagawa CSU-X1 spinning disk and a Photometrics Evolve 512 electron-multiplying charge-coupled device camera and captured using MetaMorph software (Universal Imaging, West Chester, PA). Out-of-focus light from captured images was removed with a constrained iterative deconvolution algorithm using AutoQuant X3 (AutoQuant Imaging, Watervliet, NY). Images taken in the 4′,6-diamidino-2-phenylindole channel were used to identify broad-spectrum intestinal autofluorescences caused by lipofuscin-positive, lysosome-like organelles (Clokey and Jacobson, 1986; Hermann et al., 2005). Quantification of colocalization images was done using open source ImageJ (Fiji) software (Schindelin et al., 2012). To obtain intestinal images of GFP fluorescence without interference from autofluorescence, we used argon 488-nm excitation and the spectral fingerprinting function of the Zeiss LSM510/710 Meta confocal microscope system (Carl Zeiss Micro Imaging) as described previously (Chen et al., 2006). Quantification of images was performed with MetaMorph version 6.3r2 (Universal Imaging).
Supplementary Material
Acknowledgments
We thank David Lo, Tanvi Goyal, and Peter Schweinsberg for technical assistance in making plasmid clones and biolistic transgenic lines and Michael Pierce and Joseph Kramer for help with confocal microscopy. This work was supported by NIH Grants GM067237 and GM103995 (to B.D.G.) and 6R24OD010943 (to D.H.H.) and an Anne B. and James B. Leathem Fellowship (to A.M.G.).
Abbreviations used:
- AMAN-2
alpha mannosidase
- ARF-6
ADP ribosylation factor-6
- CCPs
clathrin coated pits
- CDE
clathrin-dependent endocytosis
- CIE
clathrin-independent endocytosis
- DAF-4
dauer formation-defective-4
- ERC
endocytic recycling compartment
- GLUT1
glucose-transporter 1
- hTAC
human interleukin-2 receptor
- hTfr
human transferrin receptor
- MICAL-L1
molecules interacting with CAsL-like1
- MIG-14
abnormal cell migration-14
- PGP-1
P-glycoprotein-1
- RME-1
receptor-mediated endocytosis-1
- SDPN-1
syndapin/pascin-family F-BAR domain protien
- SMA-6
small-6.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-02-0116) on September 14, 2016.
REFERENCES
- Ahuja R, Pinyol R, Reichenbach N, Custer L, Klingensmith J, Kessels MM, Qualmann B. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell. 2007;131:337–350. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson F, Jakobsson J, Low P, Shupliakov O, Brodin L. Perturbation of syndapin/PACSIN impairs synaptic vesicle recycling evoked by intense stimulation. J Neurosci. 2008;28:3925–3933. doi: 10.1523/JNEUROSCI.1754-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun A, Pinyol R, Dahlhaus R, Koch D, Fonarev P, Grant BD, Kessels MM, Qualmann B. EHD proteins associate with syndapin I and II and such interactions play a crucial role in endosomal recycling. Mol Biol Cell. 2005;16:3642–3658. doi: 10.1091/mbc.E05-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan S, Naslavsky N, Hartnell LM, Lodge R, Polishchuk RS, Donaldson JG, Bonifacino JS. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 2002;21:2557–2567. doi: 10.1093/emboj/21.11.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Schweinsberg PJ, Vashist S, Mareiniss DP, Lambie EJ, Grant BD. RAB-10 is required for endocytic recycling in the Caenorhabditis elegans intestine. Mol Biol Cell. 2006;17:1286–1297. doi: 10.1091/mbc.E05-08-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton EL, Anggono V, Smillie KJ, Chau N, Robinson PJ, Cousin MA. The phospho-dependent dynamin-syndapin interaction triggers activity-dependent bulk endocytosis of synaptic vesicles. J Neurosci. 2009;29:7706–7717. doi: 10.1523/JNEUROSCI.1976-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clokey GV, Jacobson LA. The autofluorescent “lipofuscin granules” in the intestinal cells of Caenorhabditis elegans are secondary lysosomes. Mech Ageing Dev. 1986;35:79–94. doi: 10.1016/0047-6374(86)90068-0. [DOI] [PubMed] [Google Scholar]
- Da Costa SR, Sou E, Xie J, Yarber FA, Okamoto CT, Pidgeon M, Kessels MM, Mircheff AK, Schechter JE, Qualmann B, Hamm-Alvarez SF. Impairing actin filament or syndapin functions promotes accumulation of clathrin-coated vesicles at the apical plasma membrane of acinar epithelial cells. Mol Biol Cell. 2003;14:4397–4413. doi: 10.1091/mbc.E03-05-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmalingam E, Haeckel A, Pinyol R, Schwintzer L, Koch D, Kessels MM, Qualmann B. F-BAR proteins of the syndapin family shape the plasma membrane and are crucial for neuromorphogenesis. J Neurosci. 2009;29:13315–13327. doi: 10.1523/JNEUROSCI.3973-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty G, McMahon H. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- Giridharan SS, Cai B, Vitale N, Naslavsky N, Caplan S. Cooperation of MICAL-L1, syndapin2, and phosphatidic acid in tubular recycling endosome biogenesis. Mol Biol Cell. 2013;24:1776–1790. doi: 10.1091/mbc.E13-01-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason RJ, Akintobi AM, Grant BD, Padgett RW. BMP signaling requires retromer-dependent recycling of the type I receptor. Proc Natl Acad Sci USA. 2014;111:2578–2583. doi: 10.1073/pnas.1319947111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger E, McNee G, Allan V, Woodman P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin Cell Dev Biol. 2014;31:20–29. doi: 10.1016/j.semcdb.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B, Zhang Y, Paupard MC, Lin SX, Hall DH, Hirsh D. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat Cell Biol. 2001;3:573–579. doi: 10.1038/35078549. [DOI] [PubMed] [Google Scholar]
- Gu M, Liu Q, Watanabe S, Sun L, Hollopeter G, Grant BD, Jorgensen EM. AP2 hemicomplexes contribute independently to synaptic vesicle endocytosis. Elife. 2013;2:e00190. doi: 10.7554/eLife.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DH. Electron microscopy and three-dimensional image reconstruction. Methods Cell Biol. 1995;48:395–436. doi: 10.1016/s0091-679x(08)61397-7. [DOI] [PubMed] [Google Scholar]
- Hall DH, Hartwieg E, Nguyen KC. Modern electron microscopy methods for C. elegans. Methods Cell Biol. 2012;107:93–149. doi: 10.1016/B978-0-12-394620-1.00004-7. [DOI] [PubMed] [Google Scholar]
- Hermann GJ, Schroeder LK, Hieb CA, Kershner AM, Rabbitts BM, Fonarev P, Grant BD, Priess JR. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol Biol Cell. 2005;16:3273–3288. doi: 10.1091/mbc.E05-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Jovic M, Kieken F, Naslavsky N, Sorgen PL, Caplan S. Eps15 homology domain 1-associated tubules contain phosphatidylinositol-4-phosphate and phosphatidylinositol-(4,5)-bisphosphate and are required for efficient recycling. Mol Biol Cell. 2009;20:2731–2743. doi: 10.1091/mbc.E08-11-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessels MM, Qualmann B. Syndapin oligomers interconnect the machineries for endocytic vesicle formation and actin polymerization. J Biol Chem. 2006;281:13285–13299. doi: 10.1074/jbc.M510226200. [DOI] [PubMed] [Google Scholar]
- Ketel K, Krauss M, Nicot AS, Puchkov D, Wieffer M, Muller R, Subramanian D, Schultz C, Laporte J, Haucke V. A phosphoinositide conversion mechanism for exit from endosomes. Nature. 2016;529:408–412. doi: 10.1038/nature16516. [DOI] [PubMed] [Google Scholar]
- Koles K, Messelaar EM, Feiger Z, Yu CJ, Frank CA, Rodal AA. The EHD protein Past1 controls postsynaptic membrane elaboration and synaptic function. Mol Biol Cell. 2015;26:3275–3288. doi: 10.1091/mbc.E15-02-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostan J, Salzer U, Orlova A, Toro I, Hodnik V, Senju Y, Zou J, Schreiner C, Steiner J, Merilainen J, et al. Direct interaction of actin filaments with F-BAR protein pacsin2. EMBO Rep. 2014;15:1154–1162. doi: 10.15252/embr.201439267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Fricke R, Bhar D, Reddy-Alla S, Krishnan KS, Bogdan S, Ramaswami M. Syndapin promotes formation of a postsynaptic membrane system in Drosophila. Mol Biol Cell. 2009;20:2254–2264. doi: 10.1091/mbc.E08-10-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung B, Hermann G, Priess J. Organogenesis of the Caenorhabditis elegans intestine. Dev Biol. 1999;216:114–134. doi: 10.1006/dbio.1999.9471. [DOI] [PubMed] [Google Scholar]
- Lin S, Grant B, Hirsh D, Maxfield F. Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat Cell Biol. 2001;3:567–572. doi: 10.1038/35078543. [DOI] [PubMed] [Google Scholar]
- Liu O, Grant BD. Basolateral endocytic recycling requires RAB-10 and AMPH-1 mediated recruitment of RAB-5 GAP TBC-2 to endosomes. PLoS Genet. 2015;11:e1005514. doi: 10.1371/journal.pgen.1005514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield F, McGraw T. Endocytic recycling. Nat Rev Mol Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- McGhee JD. The C. elegans intestine. WormBook; 2007. 2007 (Mar 27), 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Tian L, Zhou J, Li Z, Jiao X, Li WW, Plomann M, Xu Z, Lisanti MP, Wang C, Pestell RG. PACSIN 2 represses cellular migration through direct association with cyclin D1 but not its alternate splice form cyclin D1b. Cell Cycle. 2011;10:73–81. doi: 10.4161/cc.10.1.14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield CJ. Seeing is believing: imaging actin dynamics at single sites of endocytosis. Trends Cell Biol. 2004;14:352–358. doi: 10.1016/j.tcb.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Modregger J, Ritter B, Witter B, Paulsson M, Plomann M. All three PACSIN isoforms bind to endocytic proteins and inhibit endocytosis. J Cell Sci. 2000;113:4511–4521. doi: 10.1242/jcs.113.24.4511. [DOI] [PubMed] [Google Scholar]
- Naslavsky N, Weigert R, Donaldson JG. Convergence of non-clathrin- and clathrin-derived endosomes involves Arf6 inactivation and changes in phosphoinositides. Mol Biol Cell. 2003;14:417–431. doi: 10.1091/mbc.02-04-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant S, Sharma M, Patel K, Caplan S, Carr CM, Grant BD. AMPH-1/Amphiphysin/Bin1 functions with RME-1/Ehd1 in endocytic recycling. Nat Cell Biol. 2009;11:1399–1410. doi: 10.1038/ncb1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V, Maduro MF. Transgenesis in C. elegans. Methods Cell Biol. 2011;106:161–185. doi: 10.1016/B978-0-12-544172-8.00006-2. [DOI] [PubMed] [Google Scholar]
- Puthenveedu MA, Lauffer B, Temkin P, Vistein R, Carlton P, Thorn K, Taunton J, Weiner OD, Parton RG, von Zastrow M. Sequence-dependent sorting of recycling proteins by actin-stabilized endosomal microdomains. Cell. 2010;143:761–773. doi: 10.1016/j.cell.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B, Kelly RB. Syndapin isoforms participate in receptor-mediated endocytosis and actin organization. J Cell Biol. 2000;148:1047–1062. doi: 10.1083/jcb.148.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B, Roos J, DiGregorio PJ, Kelly RB. Syndapin I, a synaptic dynamin-binding protein that associates with the neural Wiskott-Aldrich syndrome protein. Mol Biol Cell. 1999;10:501–513. doi: 10.1091/mbc.10.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan A, Robinson PJ. Syndapin–a membrane remodelling and endocytic F-BAR protein. FEBS J. 2013;280:5198–5212. doi: 10.1111/febs.12343. [DOI] [PubMed] [Google Scholar]
- Rao Y, Ma Q, Vahedi-Faridi A, Sundborger A, Pechstein A, Puchkov D, Luo L, Shupliakov O, Saenger W, Haucke V. Molecular basis for SH3 domain regulation of F-BAR-mediated membrane deformation. Proc Natl Acad Sci USA. 2010;107:8213–8218. doi: 10.1073/pnas.1003478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter B, Modregger J, Paulsson M, Plomann M. PACSIN 2, a novel member of the PACSIN family of cytoplasmic adapter proteins. FEBS Lett. 1999;454:356–362. doi: 10.1016/s0014-5793(99)00830-3. [DOI] [PubMed] [Google Scholar]
- Roach W, Plomann M. PACSIN3 overexpression increases adipocyte glucose transport through GLUT1. Biochem Biophys Res Commun. 2007;355:745–750. doi: 10.1016/j.bbrc.2007.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer W, Pontani LL, Sorre B, Rentero C, Berland L, Chambon V, Lamaze C, Bassereau P, Sykes C, Gaus K, Johannes L. Actin dynamics drive membrane reorganization and scission in clathrin-independent endocytosis. Cell. 2010;140:540–553. doi: 10.1016/j.cell.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Sato K, Norris A, Sato M, Grant BD. C. elegans as a model for membrane traffic. WormBook; 2014. 2014 (Apr 25), 1–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Sato K, Fonarev P, Huang CJ, Liou W, Grant BD. Caenorhabditis elegans RME-6 is a novel regulator of RAB-5 at the clathrin-coated pit. Nat Cell Biol. 2005;7:559–569. doi: 10.1038/ncb1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg PJ, Grant BD. C. elegans gene transformation by microparticle bombardment. WormBook; 2013. 2013 (Dec 30), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlekar A, Rikhy R. Syndapin promotes pseudocleavage furrow formation by actin organization in the syncytial Drosophila embryo. Mol Biol Cell. 2016;27:2064–2079. doi: 10.1091/mbc.E15-09-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A, Chen CC, Banerjee R, Glodowski D, Audhya A, Rongo C, Grant BD. EHBP-1 functions with RAB-10 during endocytic recycling in Caenorhabditis elegans. Mol Biol Cell. 2010;21:2930–2943. doi: 10.1091/mbc.E10-02-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A, Liu O, Koenig S, Banerjee R, Chen CC, Eimer S, Grant BD. RAB-10-GTPase-mediated regulation of endosomal phosphatidylinositol-4,5-bisphosphate. Proc Natl Acad Sci USA. 2012;109:E2306–2315. doi: 10.1073/pnas.1205278109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A, Pant S, Balklava Z, Chen CC, Figueroa V, Grant BD. A novel requirement for C. elegans Alix/ALX-1 in RME-1-mediated membrane transport. Curr Biol. 2007;17:1913–1924. doi: 10.1016/j.cub.2007.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A, Sun L, Banerjee R, Tobin M, Zhang Y, Grant BD. Regulation of endosomal clathrin and retromer-mediated endosome to Golgi retrograde transport by the J-domain protein RME-8. EMBO J. 2009;28:3290–3302. doi: 10.1038/emboj.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Liu O, Desai J, Karbassi F, Sylvain MA, Shi A, Zhou Z, Rocheleau CE, Grant BD. CED-10/Rac1 regulates endocytic recycling through the RAB-5 GAP TBC-2. PLoS Genet. 2012;8:e1002785. doi: 10.1371/journal.pgen.1002785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Robinson IM, Savoian MM, Griffiths JR, Whetton AD, McMahon HT, Glover DM. Drosophila F-BAR protein Syndapin contributes to coupling the plasma membrane and contractile ring in cytokinesis. Open Biol. 2013;3:130081. doi: 10.1098/rsob.130081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Wang Q, Navarro MV, Peng G, Molinelli E, Goh SL, Judson BL, Rajashankar KR, Sondermann H. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc Natl Acad Sci USA. 2009;106:12700–12705. doi: 10.1073/pnas.0902974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaagstra JC, El-Alfy M, O’Connor-McCourt MD. Transforming growth factor (TGF)-beta 1 internalization: modulation by ligand interaction with TGF-beta receptors types I and II and a mechanism that is distinct from clathrin-mediated endocytosis. J Biol Chem. 2001;276:27237–27245. doi: 10.1074/jbc.M100033200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.