Abstract
The closely related species Brassica rapa and B. oleracea encompass a wide range of vegetable, fodder and oil crops. The release of their reference genomes has facilitated resequencing collections of B. rapa and B. oleracea aiming to build their variome datasets. These data can be used to investigate the evolutionary relationships between and within the different species and the domestication of the crops, hereafter named morphotypes. These data can also be used in genetic studies aiming at the identification of genes that influence agronomic traits. We selected and resequenced 199 B. rapa and 119 B. oleracea accessions representing 12 and nine morphotypes, respectively. Based on these resequencing data, we obtained 2,249,473 and 3,852,169 high quality SNPs (single-nucleotide polymorphisms), as well as 303,617 and 417,004 InDels for the B. rapa and B. oleracea populations, respectively. The variome datasets of B. rapa and B. oleracea represent valuable resources to researchers working on evolution, domestication or breeding of Brassica vegetable crops.
Subject terms: Plant domestication, Agricultural genetics
Background & Summary
Many important crops have been domesticated and are cultivated from the genus Brassica, including those used as oilseeds, condiments, fodder, or vegetables. The ‘triangle of U’1 describes the relationships between the six economically important Brassica species. Three of the six species are diploids (A genome, Brassica rapa, n=10; B genome, B. nigra, n=8; and C genome, B. oleracea, n=9), while the other three species are allotetraploids resulting from interspecific hybridization between the diploids (AB genomes, B. juncea, n=18; AC genomes, B. napus, n=19; BC genomes, B. carinata, n=17). These different crops are characterized by their specialized development of organs2, and are referred to as morphotypes. Interestingly, similar morphotypes are often selected for in two or more Brassica species, clearly illustrating convergent crop domestication. This includes the leafy heads in Chinese cabbage (B. rapa) and cabbage (B. oleracea) and tubers at stems or hypocotyls/roots in kohlrabi’s (B. oleracea), swede’s (B. napus) and turnips (B. rapa), enlarged stems in marrow stem kale (B. oleracea) and also broccoletto (B. rapa), enlarged inflorescences in cauliflower and broccoli’s (B. oleracea), the many axillary shoots in Brussels sprouts (B. oleracea) and mizuna/mibuna’s (B. rapa) and the increased numbers of enlarged seedpods in oilseed (B. rapa and B. napus).
In recent years, the genomes of B. rapa and B. oleracea have been assembled and released3–5. Brassicaceae comparative genomics provided 24 genomic blocks (GBs) as the framework for genome studies in Brassicas6. With the GBs system, the comparative genomic analysis of B. rapa, B. oleracea, and A. thaliana revealed the whole genome triplication (WGT) event that is shared by all Brassicas7–9, and made it possible to deduce and reconstruct the diploid ancestor for the Brassiceae tribe10. Further studies reported the phenomenon of sub-genome dominance among the three sub-genomes in Brassicas8,9, and found that the biased distribution of small RNA-targeted TEs plays an important role in this phenomenon11,12. Furthermore, the release of the genome sequences has also contributed to the mapping, cloning and functional studies of agronomic genes in Brassica crops, such as two clubroot resistance genes13,14, two blackleg resistance genes15,16, as well as two key genes regulating the time of flowering17–19. The availability of these reference genomes also made it possible to build variome datasets by resequencing populations of B. rapa and B. oleracea, to investigate domestication of the diverse morphotypes.
Here, we selected 199 B. rapa and 119 B. oleracea accessions representing the 12 and nine morphotypes from these two species, respectively (Fig. 1, Supplementary Tables 1 and 2). These accessions originate from a wide geographic range, from North till South Asia and Europe20. Whole genome resequencing data of these 199 B. rapa and 119 B.oleracea accessions was generated using the Illumina HiSeq2000 platform, with paired-end reads from 350 bp insert libraries. After filtering low-quality or duplicated reads, totally, we produced 982.87 Gb resequencing data for the 199 B. rapa accessions and 742.35 Gb for the 119 B. oleracea accessions, which is about on average 10× coverage for accessions of both B. rapa and B. oleracea, ranging from 1.67× till 19.8× coverage. These reads were then mapped to the reference genomes of B. rapa version V1.5 and B. oleracae V1.0, respectively. Finally, we called out 2,249,473 and 3,852,169 reliable SNPs (single-nucleotide polymorphisms), as well as 303,617 and 417,004 InDels for the B. rapa and B. oleracea populations, respectively. With these datasets, we have identified genomic selection signals for traits of leaf-heading and tuberous organs in both B. rapa and B. oleracea, and found that sub-genome parallel selection was associated with morphotype diversification and convergent crop domestication in the two Brassica species21.
Figure 1. Images of the different morphotypes of Brassica vegetables.
(a) ten images show representative morphotypes of B. rapa; (b) seven images show representative morphotypes of B. oleracea.
The whole genome resequencing data and variome datasets for the B. rapa and B. oleracea populations are valuable resources to researchers studying evolution, domestication or trait discovery, and to breeders of Brassica crops. These data can be exploited to develop genetic markers, and chip arrays for gene mapping and functional studies.
Methods
These methods are expanded versions of descriptions in our related work21.
Sample collection of B. rapa and B. oleracea
Accessions representing the 12 morphotypes of B. rapa and nine morphotypes of B. oleracea were collected (Tables 1 and 2). Almost all morphotypes from the two Brassica species were included in this collection (Supplementary Tables 1 and 2). The 199 B. rapa accessions included 46 Chinese cabbage accessions (B. rapa ssp. pekinensis) (Table 1), 54 turnip accessions (B. rapa ssp. rapa), 25 pak choi accessions (B. rapa ssp. chinensis), 30 caixin accessions (B. rapa ssp. parachinensis), 13 zicaitai accessions (B. rapa ssp. chinensis var. purpurea Bailey), 14 oil seed accessions (B. rapa ssp. oleifera), four taicai accessions (B. rapa ssp. chinensis var. tai-tsai Lin), seven wutacai accessions (B. rapa ssp. narinosa), one edible flower accession (B. rapa ssp. broccoletto) and one yellow sarson (B. rapa ssp. tricolaris), as well as two accessions each of komatsuna (B. rapa ssp. perviridis) and mizuna (B. rapa ssp. nipposinica). This B. rapa population includes 75 DH (doubled haploid) lines, 18 inbred lines, and 106 germplasm lines (Supplementary Table 1). The 119 B. oleracea accesions included 45 cabbage accessions (B. oleracea var. capitata) (Table 2), 19 kohlrabi accessions (B. oleracea var. gongylodes), 20 cauliflower accessions (B. oleracea var. botrytis), 23 broccoli accessions (B. oleracea var. italica), four Chinese kale accessions (B. oleracea var. alboglabra), as well as two accessions each of kale (B. oleracea var. acephala), Brussels sprouts (B. oleracea var. gemmifera), curly kale (B. oleracea var. sabellica), and wild B. oleracea. This B. oleracea population included 69 DH or inbred lines, 44 germplasm lines (inbred lines from genebank accessions), and six genebank accessions (Supplementary Table 2).
Table 1. Summary of the number of accessions per B. rapa morphotype.
| Index | Name | English name morphotype | Number |
|---|---|---|---|
| 1 | ssp. pekinensis | Chinese cabbages | 46 |
| 2 | ssp. rapa | turnip | 54 |
| 3 | ssp. chinensis | pak choi | 25 |
| 4 | ssp. parachinensis | caixin | 30 |
| 5 | ssp. chinensis var. purpurea Bailey | zicaitai | 13 |
| 6 | ssp. oleifera | oil seeds | 14 |
| 7 | ssp. chinensis var. tai-tsai Lin | taicai | 4 |
| 8 | ssp. narinosa | wutacai | 7 |
| 9 | ssp. broccoletto | edible flower | 1 |
| 10 | ssp. tricolaris | yellow sarson | 1 |
| 11 | ssp. perviridis | komatsuna | 2 |
| 12 | ssp. nipposinica | mizuna | 2 |
| Total | / | / | 199 |
Table 2. Overview of the numbers of accessions per B. oleracea morphotype.
| Index | Name | English name morphotype | Number |
|---|---|---|---|
| 1 | var. capitata | cabbage | 45 |
| 2 | var. gongylodes | kohlrabi | 19 |
| 3 | var. botrytis | cauliflower | 20 |
| 4 | var. italica | broccoli | 23 |
| 5 | var. alboglabra | Chinese kale | 4 |
| 6 | var. acephala | kale | 2 |
| 7 | var. gemmifera | brussels sprouts | 2 |
| 8 | var. sabellica | curly kale | 2 |
| 9 | wild type | wild | 2 |
| Total | / | / | 119 |
We have grown all the 199 B. rapa accessions with three replicates in the green house during autumn 2014 to confirm their morphotypes. The phenotypes of these accessions were investigated till their maturity.
Sample preparation and resequencing
Plants of the 199 B. rapa accessions were grown in a greenhouse, each accession with five replicates. At the six-leaf stage, the two youngest leaves were collected from one of the five plants and DNA was extracted from these leaves. For B. oleracea, the DNA of the 44 germplasm lines and six genebank accessions was extracted as described for B. rapa. However for the DH and inbred lines, 50–100 seedlings per genotype were grown on moist filter paper. Cotyledons and hypocotyls were harvested after 12 days and DNA was then extracted.
DNA libraries with approximately 350 bp insert sizes were constructed following the manufacturer’s instructions (Illumina GAII) and paired-end resequencing reads were generated by commercial Illumina HiSeq 2000 service provided by Biomarker Technologies Corporation, Beijing, and BGI, Shenzhen.
Raw data were filtered before alignment
We filtered the raw reads before alignment to the corresponding reference genomes of B. rapa3 or B. oleracea5. First, low quality reads were filtered based on the following three rules. If one end of a paired-end read had >5% ‘N’ bases, then the paired-end read was removed. Secondly, for each paired-end read pair, if one of two reads had an average base-quality less than 20 (Phred-like score), then both reads were removed. Thirdly, for each read, we trimmed its 3’ bases if the quality scores are below 13. The trimming was stopped at the base with quality score ≥13. After trimming, if the remaining read was less than 40, the paired-end reads were removed. Moreover, considering that the duplicates of paired-end reads were generated from a single amplicon, we further removed redundant duplicated reads and only kept a single pair. We completed this raw read filtering process using an in-house made Perl script.
Alignment and variants calling
Filtered reads of each re-sequenced sample were mapped onto the corresponding reference genomes using the software Burrows-Wheeler Aligner (BWA version 0.7.5a-r405)22. We used the B. rapa genome version 1.5 and B. oleracae V1.0 as the references for the two species4. The reference genomes were first indexed by using commands as ‘bwa index reference.fa’ and ‘samtools faidx reference.fa’. The clean reads of each accession were then mapped to the indexed reference genomes one by one with the algorithm ‘mem’ of BWA. The command line was ‘bwa mem reference.fa sample_1.fq sample_2.fq>sample.sam’. It generated a sam file ‘sample.sam’ as the mapping output for each accession. This sam file can be handled by Samtools23 to call variants.
Samtools was used to call SNPs and InDels for each resequenced Brassica accession from the mapping results reported by BWA.
Samtools (version 0.1.19–44428 cd)23 was first used to transfer the sam file to bam file by command ‘samtools view -bS sample.sam sample.bam’. The bam file was then sorted by ‘samtools sort sample.bam sample.sorted.bam’. After index the sorted bam file with ‘samtools index sample.sorted.bam’, candidate genomic variants were called out by using the algorithm ‘mpileup’ of Samtools. The full command was ‘samtools mpileup -q 20 -Q 30 -ugf reference.index sample.sorted.bam | bcftools view -p 0.9 -cg ->candidateVariants.list’. We set ‘-q 20’ to use nucleotides with Phred-like quality scores higher than 20 as reliable nucleotides of a read to report variants. ‘-Q 30’ denotes that mapping quality of reads should be higher than 30 to be considered as a reliable mapping. Bcftools was used to transfer the vcf file generated by ‘mpileup’ and reported candidate variants in an output file23. We used ‘-p 0.9’ to ask Bcftools to report variants at a locus with more than 10% reads showing a different genotype to that of the reference.
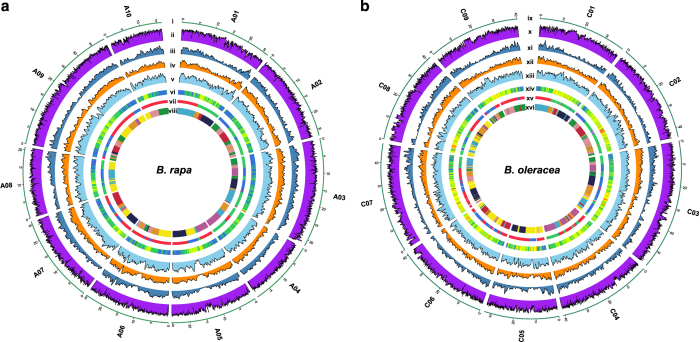
We further filtered the candidate variant list for reliable variants. For each accession, we screened its variations one by one. Here, we called the genotype that is the same as the reference ‘reference allele’, while the one that is different to the reference as ‘alternative allele’. For each variant, only alleles that were covered by sufficient reads (≥3 reads) were considered as confident alleles, and the variant was kept as a reliable variant. We performed this process for all Brassica accessions. This removed potentially false variants produced by sequencing errors. Since most of the Brassica samples (except six genebank accessions of B. oleracea) are from homozygous or almost homozygous accessions, we filtered our data further by retaining the variant calls that were homozygous in individual homozygous accessions (loci were considered as heterozygous if 0.2>DR/(DA+DR)>0.8, where DR denotes the number of reads with the reference allele, and DA denotes the number of reads with the alternative allele). We developed in-house Perl scripts to complete the candidate variants filtering processes. In order to remove rare SNPs and use common SNPs to scan for selection signals, we filtered out SNPs that had a MAF (minor allele frequency)<0.05 in both B. rapa and B. oleracea. Totally, we obtained 2,249,473 SNPs and 303,617 InDels for the B. rapa population, as well as 3,852,169 SNPs and 417,004 InDels for the B. oleracea population. For the two accessions with lowest re-sequencing depth in B. rapa and B. oleracea, we still obtained genotypes of 1,577,624 and 2,690,136 SNPs, respectively. We plotted genomic heterozygosity, density of SNPs and InDels along with the gene density to show the distribution features of these variants in the two Brassica genomes (Fig. 2).
Figure 2. The distribution of genomic variations on chromosomes of the resequenced populations of 199 B. rapa and 119 B. oleracea accessions.
(a) The genomic information of the B. rapa population; i: the ten chromosomes of B. rapa, the physical positions are indicated in units of million bases; ii: the genomic heterozygosity of the B. rapa population. Area charts quantify iii: functional SNPs/InDels, iv: InDels, v: SNPs. vi: heat map for gene density. vii: subgenome partition of the B. rapa genome, red, green, and blue corresponding to subgenomes LF, MF1, and MF2, respectively10. viii: The triplicated 24 genomic blocks in B. rapa10. (b) The genomic information of the B. oleracea population; ix: the nine chromosomes of B. oleracea; x: the genomic heterozygosity of the B. oleracea population. Area charts quantify xi: functional SNPs/InDels, xii: InDels, xiii: SNPs. xiv: heat map for gene density. xv: subgenome partition in B. oleracea genome, red, green, and blue corresponding to subgenomes LF, MF1, and MF2, respectively5. xvi: The triplicated 24 genomic blocks in B. oleracea5. LF denotes the least fractionated subgenome, MF1 and MF2 denote for more fractionated sub-genomes 1 and 2 (ref. 3).
Functional annotation of genomic variants
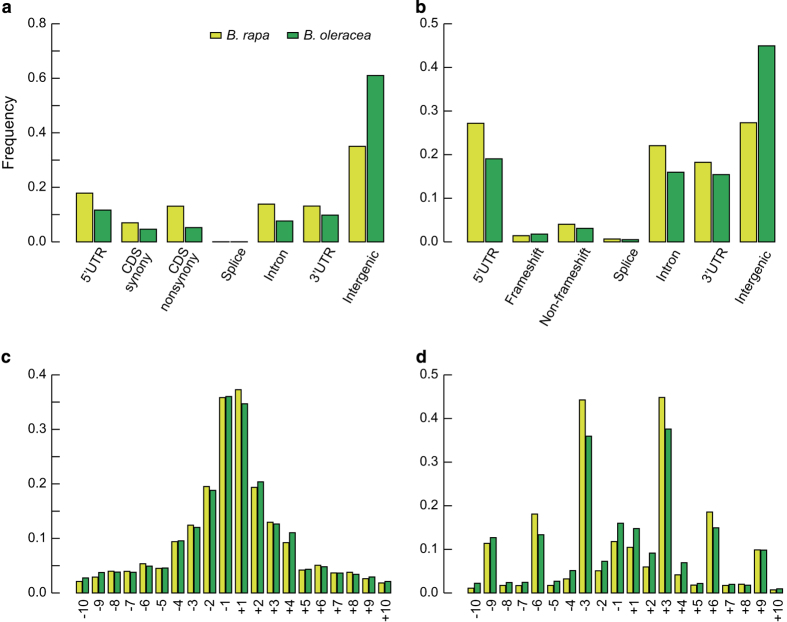
The variants identified in the two Brassica genomes were further annotated into different groups (Table 3 and Table 4). According to the genomic positions of SNPs and InDels relevant to predicted gene models, we first separated them into 1) variants located at genic regions and 2) at inter-genic regions. Variants located at genic regions were further separated into three subgroups: a) variants in coding sequences (CDs), b) in introns and c) in untranslated regions (UTRs). The SNPs and InDels located at CDs (1a) were classified into two subgroups: the subgroup causing changes to the coding amino acids, including non-synonymous SNPs and frame shift InDels, and the subgroup causing no changes to the amino acids, including synonymous SNPs and InDels that do not cause frame-shifts. Intronic variants (1b) were also separated into two subgroups: causing (8 bp to the splice site) or not causing splice site mutations; UTR variants (1c) are divided into two subgroups: variants in 5′ or 3′UTR regions (Fig. 3a,b). The results show that among these 2,249,473 SNPs and 303,617 InDels in B. rapa, 161,319 SNPs (nonsynonymous and splice site) and 16,905 InDels (CDS and splice site) respectively, introduced changes to the protein sequences (Table 3); for the 3,852,169 SNPs and 417,004 InDels in B. oleracea, 154,863 SNPs and 16,687 InDels respectively introduced changes to the protein sequences (Table 4). Additionally, we analyzed the length distribution of InDels, and found that 1 bp deletions or insertions are the dominant InDels (Fig. 3c) in both Brassica populations. The length distribution of InDels located in coding regions of genes was also investigated, and it was found that InDels of three or fold changes of three nucleotides are clearly the dominant types (Fig. 3d). These InDels will not introduce frame-shift mutations to genes, thus are under less stringent selection compared to InDels that don’t correspond to fold changes of three.
Table 3. The distribution of SNPs in the 199 B. rapa and 119 B. oleracea accessions.
|
Genic |
Intergenic | Total | ||||||
|---|---|---|---|---|---|---|---|---|
|
CDS |
Intron |
UTR |
||||||
| nonSyn | Syn | splice | intron | 5UTR | 3UTR | |||
| Abbreviations: CDS for coding sequences; nonSyn for non-synonymous; Syn for synonymous; UTR for untranslated region. | ||||||||
| B. rapa | 160,639 | 298,671 | 680 | 320,898 | 402,207 | 297,313 | 769,065 | 2,249,473 |
| B. oleracea | 154,038 | 176,338 | 825 | 270,474 | 474,953 | 373,303 | 2,402,238 | 3,852,169 |
Table 4. The distribution of InDels in the 199 B. rapa and 119 B. oleracea accessions.
|
Genic |
Intergenic | Total | ||||||
|---|---|---|---|---|---|---|---|---|
|
CDS |
Intron |
UTR |
||||||
| Frameshift | Nonframeshift | splice | intron | 5UTR | 3UTR | |||
| Note: frameshift means an InDel variation that causes a frameshift mutation; abbreviations are the same as in Table 3. | ||||||||
| B. rapa | 4,467 | 10,728 | 1,710 | 68,514 | 81,779 | 56,341 | 82,441 | 303,617 |
| B. oleracea | 5,724 | 9,516 | 1,447 | 57,427 | 82,447 | 59,386 | 203,441 | 417,004 |
Figure 3. The distribution of genomic variations (SNPs and InDels) determined from the resequenced accessions of B. rapa and B. oleracea.
The distribution of 2,148,937 and 3,471,818 SNPs (a), as well as 312,618 and 276,587 InDels (b) in genomes of B. rapa and B. oleracea, respectively. The length distributions of total InDels (c), as well as 17,018 and 13,502 InDels located at coding regions (d) in genomes of B. rapa and B. oleracea, respectively. On the X-axis of (c,d), ‘−’ denotes deletion variants in numbers of bases, while ‘+’ denotes insertion variants in numbers of bases.
We further investigated the genetic diversity within each morphotype group in both species with the annotated genomic variations. We performed the analysis by counting the number of polymorphic variants in each morphotype group. The results showed that groups of turnip (54) and Chinese cabbages (46) in B. rapa had most polymorphic loci based on both total SNPs as well as functional SNPs (non-synonymous SNPs or SNPs located at splicing sites) (Supplementary Table 3). Interestingly, the groups of turnip and pak choi had the most polymorphic loci based on total InDels and functional InDels (InDels located at coding sequences or splicing sites). For B. oleracea, groups of cabbage (45) and kohlrabi (19) had both most polymorphic loci of SNPs and InDels (Supplementary Table 4). The number of polymorphic variants reflects the genetic diversity in each group. However, the number of variants is also impacted by the numbers of samples studied.
Code availability
Custom perl scripts used to filter raw reads and candidate genomic variants can be downloaded through http://brassicadb.org/brad/datasets/pub/ReseqPars/codes/ in Brassica database BRAD, or are freely available when requested by email to the authors. Other tools used in this work including version details are BWA (version 0.7.5a-r405) and SAMtools (version 0.1.19–44428 cd). Command lines with details of parameters when using these tools are described in the method section.
Data Records
All the raw reads of 199 B. rapa and 119 B. oleracea accessions used in this work have been deposited in the NCBI Sequence Read Archive with the accession ID SRP071086 (Data Citation 1). The SNP and InDel datasets identified on the resequencing data and related vcf files are public available in Brassica Database BRAD (Data Citation 2).
Technical Validation
In order to measure the quality of variants called out from the resequencing data, we selected five SNPs (Table 5), and genotyped 95 out of the 199 B. rapa accessions for these SNPs using the method of KASP (kompetitive allele specific PCR). With the same method, we also test the polymorphic level of five SNPs determined from 119 B. oleracea accessions in another group of 281 B. oleracea accessions. KASP is a homogeneous, fluorescence-based endpoint SNP genotyping platform (LGC Genomics LLC, Beverly, MA, USA). The five selected SNPs satisfied the following criteria of KASP experiments: 1) No other genomic matches were found for the 50 bp sequences flanking the two sides of each candidate SNP; 2) There are no other SNPs or InDels located at the 50 bp flanking regions of the candidate SNP.
Table 5. Genotype verification of five SNP loci in 95 B. rapa accessions.
| Loci | Chromosome | Position | #Samples | Ratio of correction |
|---|---|---|---|---|
| 1 | A01 | 8278402 | 95 | 100% |
| 2 | A05 | 5655627 | 95 | 100% |
| 3 | A08 | 8149675 | 95 | 100% |
| 4 | A08 | 8028973 | 95 | 100% |
| 5 | A10 | 1610572 | 95 | 100% |
| Total | / | / | 475 | 100% |
For each SNP, two allele-specific primers A1 and A2 and one common reverse primer C were designed by LGC Genomics. The PCR reaction mixture had a total volume of 5 μl and included 2.5 μl DNA, 2.5 μl 2×master mix with 0.07 μl primer mix according to the manufacturer’s guidelines. Three no-template controls were included for each SNP locus. The amplification process was ran in Gene Amp PCR System 9700 (Applied Biosystems) using the following program: 94 °C for 15 min followed by 10 touchdown cycles of 94 °C for 20 s, 61–55 °C for 60 s (decreasing by 0.6 °C per cycle), followed by 26 cycles of 94 °C for 20 s, 55 °C for 60 s. Fluorescence detection was then performed in a 7900 HT Fast Real-Time PCR System (Applied Biosystems), and the results were analysed using SDS2.3 Software (supplied by Applied Biosystems).
We further compared the genotypes of the five SNPs in the 95 samples that were called out by resequencing data, with the genotypes of these loci that were reported by the KASP experiments. Results show that all the 475 loci analyzed have consistent genotypes between the two methods of resequencing and the KASP experiment (Table 5), supporting the fact that the genomic variants are of high quality. The polymorphism level of the five SNPs in B. oleracea is shown in Supplementary Table 5.
Usage Notes
All the resequencing datasets of this work are deposited in the NCBI database with the accession number SRP066057 (alias: PRJNA301642). Access to the plant materials (or DNA samples for materials provided from companies) described in this paper is possible for research purposes via a material transfer agreement with the material owners. Please contact Dr Xiaowu Wang (wangxiaowu@caas.cn) or Guusje Bonnema (guusje.bonnema@wur.nl).
Additional Information
How to cite this article: Cheng, F. et al. Genome resequencing and comparative variome analysis in a Brassica rapa and Brassica oleracea collection. Sci. Data 3:160119 doi: 10.1038/sdata.2016.119 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This study was funded by the National Program on Key Research Project (2016YFD0100307), the 973 Program (2013CB127000), National Science and Technology Ministry (2014BAD01B09), the National Natural Science Foundation of China (NSFC grants 31630068, 31301771, 31272179, and 31301784), and a grant from the Technological Top Institute (TTI) Green genetics, Rijk Zwaan, and Bejo zaden. Research was supported by the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, P.R. China, and the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences.
Footnotes
The authors declare no competing financial interests.
Data Citations
- 2016. NCBI Sequence Read Archive. SRP071086
- 2016. BRAD Brassica database. http://brassicadb.org/brad/datasets/pub/ReseqPars/genotypes/
References
- Nagaharu U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japan. J. Bot 7, 389–452 (1935). [Google Scholar]
- Boswell V. R. Our Vegetable Travelers. National Geographic Magazine 96, 145–217 (1949). [Google Scholar]
- Wang X. et al. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43, 1035–1039 (2011). [DOI] [PubMed] [Google Scholar]
- Cheng F. et al. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol 11, 136 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun 5, 3930 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz M. E., Lysak M. A. & Mitchell-Olds T. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci 11, 535–542 (2006). [DOI] [PubMed] [Google Scholar]
- Cao J. et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43, 956–963 (2011). [DOI] [PubMed] [Google Scholar]
- Cheng F. et al. Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS One 7, e36442 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. et al. Altered patterns of fractionation and exon deletions in Brassica rapa support a two-step model of paleohexaploidy. Genetics 190, 1563–1574 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F. et al. Deciphering the diploid ancestral genome of the Mesohexaploid Brassica rapa. Plant Cell 25, 1541–1554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F. et al. Epigenetic regulation of subgenome dominance following whole genome triplication in Brassica rapa. New Phytol 211, 288–299 (2016). [DOI] [PubMed] [Google Scholar]
- Woodhouse M. R. et al. Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc Natl Acad Sci USA 111, 5283–5288 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama K. et al. Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L. PLoS One 8, e54745 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Hatakeyama K., Fukino N. & Matsumoto S. Fine mapping of the clubroot resistance gene CRb and development of a useful selectable marker in Brassica rapa. Breed Sci 63, 116–124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkan N. J. et al. The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol 197, 595–605 (2012). [DOI] [PubMed] [Google Scholar]
- Tollenaere R. et al. Identification and characterization of candidate Rlm4 blackleg resistance genes in Brassica napus using next-generation sequencing. Plant Biotechnol J 10, 709–715 (2012). [DOI] [PubMed] [Google Scholar]
- Xiao D. et al. The Brassica rapa FLC homologue FLC2 is a key regulator of flowering time, identified through transcriptional co-expression networks. J Exp Bot 64, 4503–4516 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. et al. A naturally occurring InDel variation in BraA.FLC.b (BrFLC2) associated with flowering time variation in Brassica rapa. BMC Plant Biol 12, 151 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnema G. in The Brassica rapa Genome (ed.Wang X. 131–144 (Springer, 2015). [Google Scholar]
- Ignatov A. N., Artemyeva A. M. & Hida K. Orign and Expansion of Cultivated Brassica rapa in EURASIA: Linguistic Facts. Acta Horticulturae 867, 81–88 (2008). [Google Scholar]
- Cheng F. et al. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat Genet 48, 1218–1224 (2016). [DOI] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2016. NCBI Sequence Read Archive. SRP071086
- 2016. BRAD Brassica database. http://brassicadb.org/brad/datasets/pub/ReseqPars/genotypes/