Several new nesprin-1 binding partners were identified, of which many are well-characterized RNA-binding proteins involved in various forms of nuclear RNA-processing events. Matrin-3 was one such protein identified, and a new cytoplasmic localization for matrin-3 is shown.
Abstract
Nesprins are highly conserved spectrin repeat–containing scaffold proteins predominantly known to function at the nuclear envelope (NE). However, nesprin isoforms are emerging with localizations and scaffolding functions at sites away from the NE, suggesting their functions are more diverse than originally thought. In this study, we combined nesprin-1 coimmunoprecipitations with mass spectrometry to identify novel nesprin-1 binding partners for isoforms that localize to subcellular compartments beyond the NE. We show that one of these interactors, matrin-3 (matr3), localizes to mRNA processing bodies (PBs), where we have previously shown a nesprin-1 isoform to localize. Furthermore, we show that Matr3 is part of PB mRNP complexes, is a regulator of miRNA-mediated gene silencing, and possibly shuttles to stress granules in stressed cells. More importantly, we identify a new C-terminally truncated Matr3 isoform that is likely to be involved in these functions and PB localization. This study highlights several novel nesprin-1 binding partners and a new function and localization for Matr3 in cytoplasmic RNA granules.
INTRODUCTION
Nesprins are a novel spectrin-repeat (SR) family of cellular scaffolds and linkers, predominantly known to localize to the nuclear envelope (NE; Zhang et al., 2001; Mellad et al., 2011). To date, four nesprin genes have been identified, and their full-length proteins localize to the outer nuclear membrane and provide connections to cytoskeletal filaments. Full-length nesprin-1 and nesprin-2 bind F-actin directly via their N-terminal calponin homology domains (Zhang et al., 2002). The SRs of nesprin-3 and nesprin-4 interact with plectin and Kif5b in a manner that establishes intermediate filament and microtubule connections, respectively (Wilhelmsen et al., 2005; Roux et al., 2009). NE nesprins are one of the major components of the linker of the nucleoskeleton and cytoskeleton (LINC) complex and form connections with inner NE transmembrane SUN domain proteins in the perinuclear space. In turn, the SUN domain proteins interact with the nuclear lamina underlying the inner nuclear membrane to complete the LINC complex (Crisp et al., 2006). This double membrane–spanning complex maintains nuclear integrity, mediates nuclear anchorage and positioning, and plays a definitive role in mechanosensing and cell migration (Razafsky et al., 2014; Chang et al., 2015).
The emergence of KASH-less nesprin isoforms, generated by alternative transcription, has provided evidence for nesprin functionality beyond the NE (Rajgor and Shanahan, 2013). Indeed, all the KASH-less nesprin isoforms examined thus far have localized, often in a cell-specific manner, to an array of different cellular compartments, including focal adhesions, the nucleolus, the Golgi, and promyelocytic leukemia bodies, and diffusively in the cytosol and nucleus (Kobayashi et al., 2006; Warren et al., 2010; Rajgor et al., 2012). Recently we reported the presence of p50Nesp1 in mRNA processing bodies (P-bodies, or PBs). p50Nesp1 is a short variant of nesprin-1 composed entirely of SRs that facilitates PB anchorage to microtubules and plays a role in PB dynamic properties such as localized movement (Rajgor et al., 2014). Data from this and other studies suggest that miniature nesprin-1 isoforms composed of SRs otherwise located in the central rod region of nesprin-1 giant can localize in the nucleus and cytosol (Warren et al., 2010; Rajgor et al., 2012, 2014).
In this study, we identified potential nesprin-1 interacting partners for KASH-less nesprin-1 variants to further characterize the function of nesprins as intracellular scaffolds and linkers. Mass spectrometry identified a host of proteins associated with nesprin-1 involved in RNA-processing events, including matrin-3 (Matr3). Matr3 is a well-established nuclear matrix protein identified two decades ago (Nakayasu and Berezney, 1991). However, its function is only beginning to emerge through the identification of novel binding partners that suggest matr3 functions in a number of key cellular processes, including chromatin remodeling, transcription, RNA splicing, DNA replication and repair, and apoptosis (Zhang and Carmichael, 2001; Zeitz et al., 2009; Salton et al., 2010; Coelho et al., 2015).
Here we report a role for a new potential matr3 isoform in miRNA-mediated translational repression and show for the first time its localization in PBs. By identifying matr3 PB-localizing and PB-binding domains, we suggest cytoplasmic functions for matr3 and speculate that neuronal and muscular degenerative diseases caused by mutations within the matr3 gene could hamper posttranscriptional processes associated with cytoplasmic matr3 RNA-processing events (Senderek et al., 2009; Johnson et al., 2014).
RESULTS
Identification of novel nesprin-1 binding partners
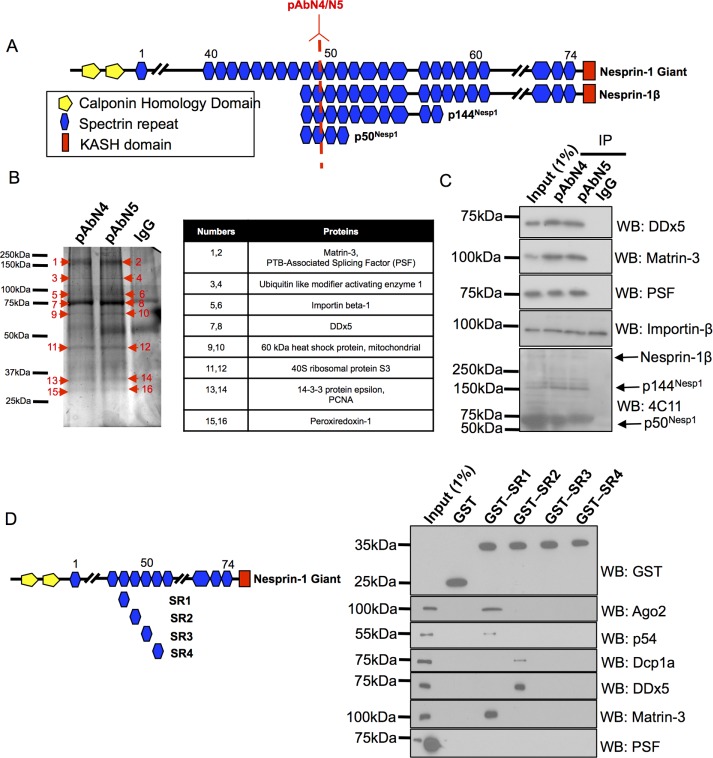
To further characterize the ability of nesprin-1 to function as an intracellular scaffold, we set out to identify novel binding partners for KASH-less nesprin-1 isoforms. Coimmunoprecipitations (co-IPs) with two previously characterized nesprin-1 polyclonal antibodies (pAbN4 and pAbN5) were performed on U2OS whole-cell lysates (Figure 1A). The epitopes for these antibodies are located within SR 49 of the nesprin-1 giant and are separated by several amino acid residues. In U2OS cells, both antibodies only detect PB and nuclear nesprin-1 variants, as previously described (Rajgor et al., 2014).
FIGURE 1:
Identification of novel nesprin-1 binding partners. (A) Schematic illustrating the location of pAbN4 and pAbN5 relative to the nesprin-1 giant, nesprin-1β, and smaller KASH-less nesprin-1 isoforms. (B) SDS–PAGE and silver-stained gel of pAbN4 and pAbN5 co-IPs. Table summarizes interacting partners identified by mass spectrometry that were present in both nesprin-1 immune complexes but not the IgG control. (C) Validation of mass spectrometry binding partners by co-IPs and Western blotting (N = 4). (D) GST pull downs using SRs located in the central rod region of nesprin-1 at bait (N = 3).
pAbN4, pAbN5, and control immunoglobulin G (IgG) immune complexes from U2OS cells were separated by SDS–PAGE, and silver-stained bands present in both pAbN4 and pAbN5 lanes, but not the negative control IgG lane, were identified by mass spectrometry (Figure 1B). Only proteins detected by both antibodies were considered as potential novel interactors. These included nuclear components involved in RNA processing such as Matr3, PTB-associated splicing factor (PSF), DDx5, and hnRNP A2/B1. Cytoplasmic proteins were also identified (HSP90, 40S ribosomal protein SA, creatine kinase, cytoskeletal actin) as well as Importin-β, suggesting that these KASH-less nesprin-1 variants may function as intracellular nuclear, cytoplasmic, and nuclear envelope scaffolds consistent with their localizations (Rajgor et al., 2014).
Interestingly, no nesprin-1 was detected by mass spectrometry, which may be due to poor detection of nesprin-1 peptides by mass spectrometry or because the bands corresponding to nesprin-1 were not sufficiently excised or digested from the silver-stained gels. To confirm nesprin-1 was being immunoprecipitated by pAbN4 and pAbN5, we performed Western blotting with a previously described nesprin-1 monoclonal antibody (4C11) to show nesprin-1 enrichment within pAbN4 and pAbN5 immune complexes (Holt et al., 2016). Indeed, nesprin-1 isoforms p50Nesp1, p144Nesp1, and nesprin-1β were detected in pAbN4 and pAbN5 IPs. Further Western blotting was performed to confirm the presence of Matr3, PSF, DDx5, and Importin-β (Figure 1C). Although matr3, PSF, and DDx5 coimmunoprecipitated with nesprin-1, Importin-β was also present within the control IgG immune complexes and therefore not considered as a potential nesprin-1 binding protein.
To confirm the interaction between nesprin-1 with matr3, PSF, and DDx5, we performed glutathione S-transferase (GST) pull-down experiments using previously described GST-nesprin-1 constructs (Figure 1D). GST-SR1, GST-SR2, GST-SR3, and GST-SR4 represent SRs 48–51 of nesprin-1 giant and are also the four SRs that comprise the p50Nesp1 isoform that localizes to PBs. These constructs were selected because they are present in close proximity to the nesprin-1 pAbN4 and pAbN5 epitopes. Interestingly, matr3 and DDx5 associated with SR1 and SR2, respectively, which are the same SRs that we have previously shown to bind miRNA-mediated gene silencing (miRISC) regulators Argonaute-2 (Ago2) and p54/DDx6. PSF did not associate with any of the SRs used in this experiment, suggesting that it likely interacts with another of the many SRs of nesprin-1 (Figure 1D). Together these experiments identify novel nesprin-1 binding partners and potential new functions for nesprin-1.
Matr3 is required for miRISC function
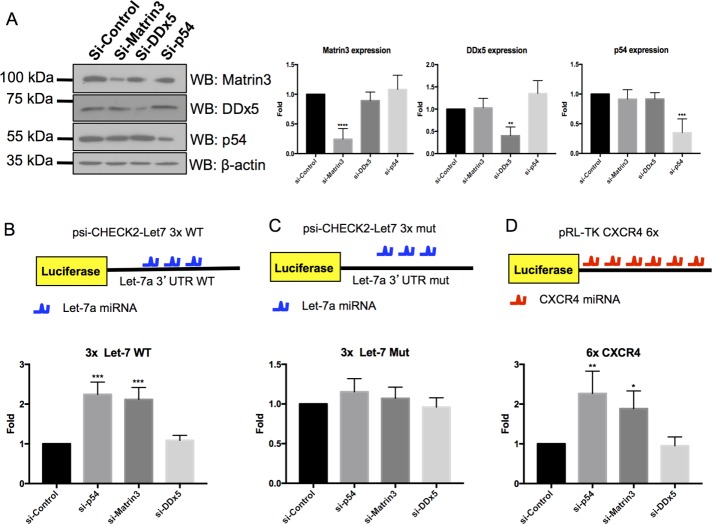
SR48 and SR49 (SR1 and SR2 domains in Figure 1D) of nesprin-1, which Matr3 and DDx5 associate with, have previously been shown to be required for miRISC function (Rajgor et al., 2014). Furthermore, matr3 and DDx5 have also previously been identified in Argonaute protein complexes (Hock et al., 2007; Rajgor et al., 2014). Together these experiments suggest matr3 and DDx5 may be part of the miRISC complex required for miRNA-mediated gene silencing. We therefore decided to perform the Let-7a miRISC reporter assay to determine whether these two proteins are required for miRNA-mediated gene silencing. The psi-CHECK2-let-7a 3× luciferase vector contains 3×Let-7a miRNA-binding sites, and luciferase expression is translationally repressed when transfected into cells expressing endogenous Let-7a and functional miRISC complexes (Lytle et al., 2007; Johnston et al., 2010). Knockdown of core silencing proteins involved in miRISC pathways, for example, Rck/p54, GW182, or p50Nesp1, reduces miRISC activity (Chu and Rana, 2006; Rajgor et al., 2014). To determine whether matr3 or DDx5 is also required for this process, we performed the Let-7a miRISC assay in U2OS cells depleted of either DDx5 or matr3 with small interfering RNAs (siRNAs). Rck/p54 knockdown was also performed and served as a positive control for these assays. Successful knockdown of these proteins was determined by Western blotting, with which we observed a ∼75.6% knockdown of matr3, ∼59.6% knockdown of DDx5, and ∼64.9% knockdown of p54 (Figure 2A). Endogenous Let-7a was unable to attenuate luciferase silencing in DDx5-depleted cells; however, matr3 knockdown enhanced luciferase activity comparable to Rck/p54 depletion, suggesting Matr3 is required for miRNA-mediated gene silencing (Figure 2B). When the Let-7a binding sites of the luciferase reporter were mutated so they could no longer associate with endogenous Let-7a, no change in luciferase activity was observed under any of the knockdown conditions (Figure 2C). These data suggest matr3 induces translational repression in a miRNA-mediated manner.
FIGURE 2:
Matr3 is required for miRISC function. (A) Western blotting demonstrating efficient knockdown of matrin3, DDx5, and p54 using their respective siRNAs (N = 4). (B) Cells depleted of Matr3 have attenuated miRISC function of a Let-7a reporter assay. p54 knockdown cells served as a positive control (N = 4). (C) Endogenous Let-7a does not suppress activity of the reporter under any knockdown condition when the Let-7a binding sites are mutated (N = 4). (C, D) Cells depleted of Matr3 have attenuated miRISC function of CXCR4 reporter assay (N = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001; one-way ANOVA, Dunnett’s post hoc test.
Additionally, we monitored the ability of exogenous CXCR4 miRNA to silence a luciferase reporter containing 6×CXCR4 miRNA binding sites (Doench et al., 2003). As with the Let-7a reporter, no change in luciferase activity was seen in DDx5-depleted cells, while matr3- and p54-depleted cells had significantly enhanced luciferase expression (Figure 2D). Together these experiments suggest Matr3 is a novel component of the miRISC complex required for miRNA-mediated gene silencing.
Matr3 localizes to RNA PBs
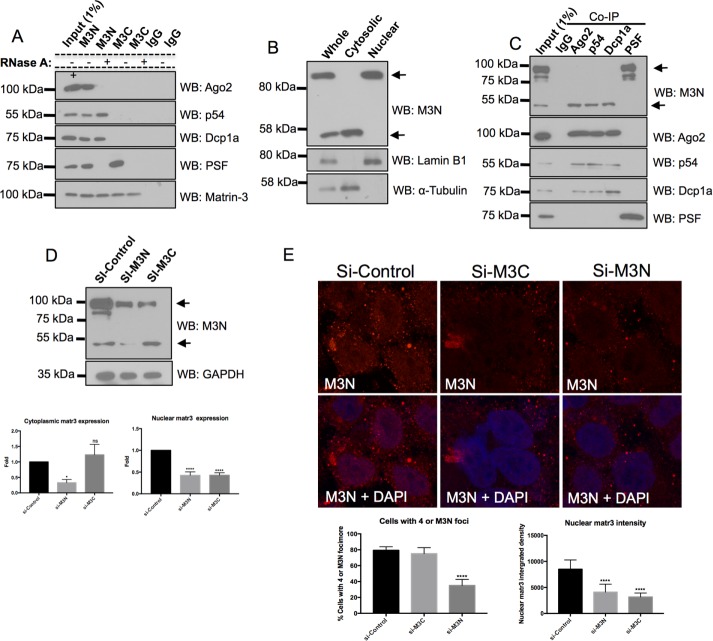
Matr3 is a 95 kDa nuclear matrix protein containing two RNA-recognition motifs (RRMs) flanked by two zinc finger (ZnF) domains (Figure 3A). The functions of these domains remain unclear; however, matr3 has multifunctional properties in many cellular processes (Zhang and Carmichael, 2001; Zeitz et al., 2009; Salton et al., 2010; Coelho et al., 2015). Therefore we examined the cellular localization of matr3 using antibodies targeting the N-terminal (M3N) or C-terminal (M3C) ends of the protein in U2OS cells (Figure 3B). Both M3N and M3C antibodies stained the nuclear matrix; however, M3N additionally detected cytoplasmic foci that colocalized with the PB marker Hedls (Figure 3B). This suggests that matr3 is present in PBs, which is consistent with its miRISC function.
FIGURE 3:
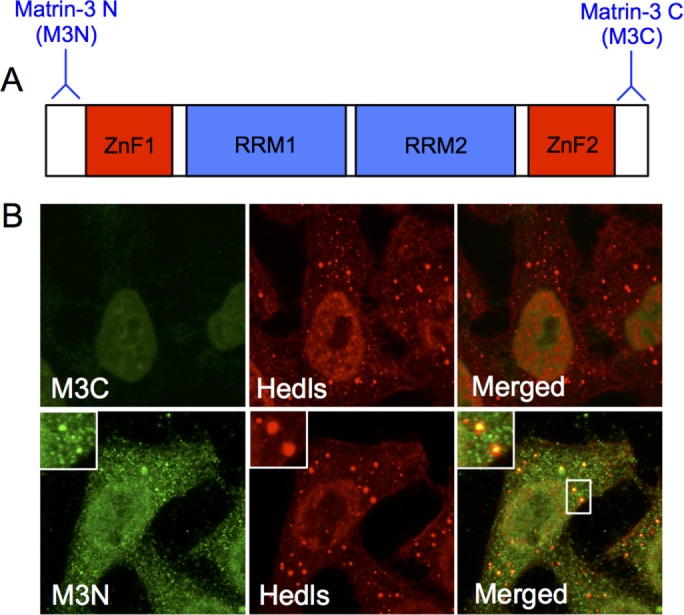
Matr3 localizes to PBs. (A) Schematic representation illustrating the structure of matr3 and the location of M3N and M3C antibodies. (B) M3C detects the nuclear matrix in U2OS cells. M3N detects the nuclear matrix and cytoplasmic foci that colocalize with PB markers Hedls (N = 100–300 over four experimental repeats).
Matr3 interacts with PB proteins
To identify whether PB protein complexes were associated with matr3, we probed M3N and M3C immune complexes for Ago2, p54, and Dcp1a (Figure 4A). M3C immune complexes did not contain any of the PB proteins, while all 3 PB components were present in M3N immune complexes, of which Ago2 association was RNA dependent. Both M3N and M3C contained PSF, a nuclear matr3 binding partner, and served as a positive control for matr3 isolated protein complexes (Figure 4A; Zhang and Carmichael, 2001).
FIGURE 4:
Unique matr3 is present within PBs. (A) Co-IPs with M3N and M3C show that only M3N immune complexes contain PB proteins. PSF served as a positive control (N = 4). (B) M3N detects two Western bands in U2OS whole-cell lysates. Subcellular fractions show the larger ∼95 kDa band is present in the nuclear fraction, and the smaller ∼50 kDa band is present in the cytosol. Lamin B1 and α-tubulin were used to measure purity of nuclear and cytoplasmic fractions respectively (N = 4). (C) PB proteins coimmunoprecipitate with the smaller matr3 variant, while PSF co-IPs with the larger form (N = 4). (D) si-M3N knocks down both M3N bands detected by Western blotting, while M3C only knocks down the larger molecular weight band (N = 5). (E) si-M3N knocks down cytoplasmic foci and nuclear matr3 staining. si-M3C does not affect PB but does eliminate nuclear staining. Cytoplasmic foci knockdown was determined by cells having <4 foci (N = 400 over four experimental repeats). Nuclear matr3 levels were measured by nuclear matr3 fluorescence integrated density (N = 100 over four experiments). *, p < 0.05; ****, p < 0.0001; one-way ANOVA, Dunnett’s post hoc test.
New matr3 isoform localizes to PBs
Next we investigated whether nuclear and cytosolic matr3 represents a common pool of the protein by probing U2OS cytosolic and nuclear extracts with antibodies to M3N, lamin B1, and α-tubulin (Figure 4B). The presence of lamin B1 only in the nuclear extract and α-tubulin only in the cytosolic pool suggested pure fractions of the two compartments. Western blotting with M3N detected the 95 kDa matr3 protein in the nucleus, while a smaller ∼50 kDa band was present in the cytosolic fraction (Figure 4B). Probing for M3C detected only the 95 kDa band in the nuclear fraction (unpublished data). These data indicate that two populations of Matr3 are present: a 95 kDa protein in the nucleus and a smaller ∼50 kDa protein in the cytoplasm.
We performed co-IPs to identify which proteins were associated with the two different matr3 species (Figure 4C). Ago2, Rck/p54, and Dcp1a, but not PSF, immune complexes were enriched with the smaller 50 kDa matr3 variant, while PSF coimmunoprecipitated with the larger 95 kDa nuclear matr3 protein. Western blotting of these co-IPs for the three PB proteins and PSF showed no PSF to be present in PB complexes or visa versa (i.e., no PB components in PSF immune complexes; Figure 4C).
To identify whether the 50 kDa band was a novel isoform, splice variant, cleavage product, or a nonspecific protein recognized by the M3N antibody, we used siRNAs created against the N-terminal and C-terminal (si-M3N and si-M3C, respectively) open reading frame of matr3. Western blotting demonstrated si-M3N was able to knock down both the 95 and 50 kDa bands, while si-M3C only knocked down the larger 95 kDa band (Figure 4D). Both siRNAs were able to knock down nuclear matr3 levels, as measured by fluorescence intensity within the nucleus (Figure 4E). Previously, U2OS cells have been established to contain several PBs under basal conditions (Rajgor et al., 2014). Therefore, to determine PB knockdown with the siRNAs, we examined 400 U2OS cells and counted the percentage of cells that had between 0 and 4 PBs. si-M3N–transfected cells had a large population of cells containing fewer than 4 PBs (35 ±7.70%, n = 400), while si-M3C–transfected cells had PBs comparable to the control siRNA (si-M3C 75 ±7.62%, and si-control 79.25 ±4.57%, n = 400) (Figure 4E). Together these experiments suggest a novel N-terminal matr3 variant exists in PBs that is different from the nuclear matr3 identified to date.
Matr3 ZnF1 and RRM1 are required for PB and SG localization
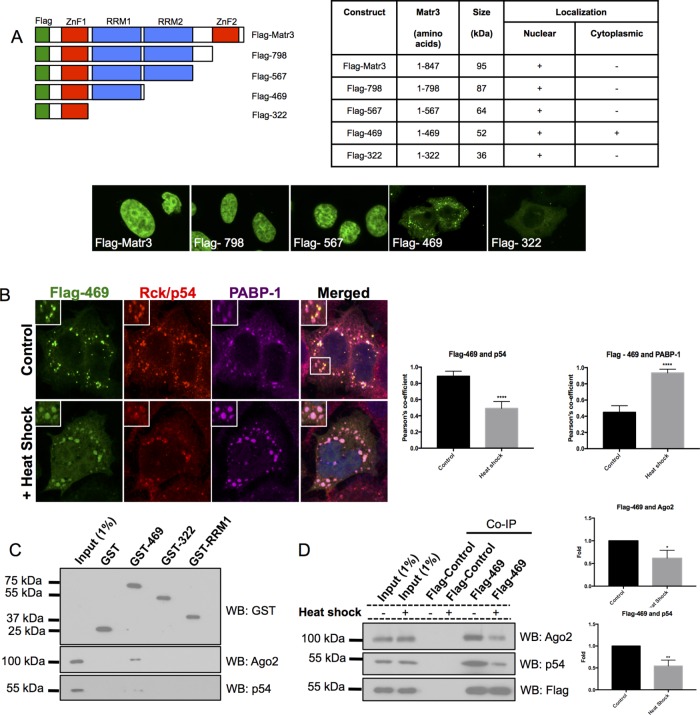
PB detection with the N-terminal antibody and siRNA knockdown of the ∼50 kDa band with si-M3N suggest that PB matr3 is likely to retain its N-terminal domains, assuming no unusual alternative splicing of matr3 occurs. Therefore, to determine which domains of matr3 were required for PB localization, we created Flag-tagged matr3 C-terminal truncation constructs by introducing stop codons after every major domain of matr3 (Figure 5A). Full-length Flag-matr3, Flag-798, and Flag-567 all showed nuclear matrix localization in U2OS cells. However, removal of the second RRM in Flag-469 induced cytoplasmic foci formation, with some transfected cells also displaying weak nuclear localization. Subsequent truncation of RRM1 in Flag-322 resulted in loss of cytoplasmic foci (Figure 5A).
FIGURE 5:
Matr3 ZnF1 and RBD1 are required for PB localization. (A) Flag-tagged C-terminal truncation constructs were created and transfected into U2OS cells. The table indicates which constructs had cytoplasmic and nuclear localizations (N = 30–50 over three experimental repeats). Flag-469 did have weak nuclear staining in a small population of transfected cells (13.33% of cells). (B) Flag-469 foci formed colocalize with PB marker p54 and induce SG formation, as indicated by PABP-1 staining. After heat shock (42°C, 45 min), Flag-469 moves from PBs to SGs. Colocalization of Flag-469 with p54 PBs and PABP-1 SGs before and after SGs were quantified using Pearson’s colocalization coefficient (N = 15 over three experimental repeats). (C) GST-469 pulls down Ago2 and p54 from U2OS cell extract (N = 3). (D) Flag-469 has reduced interactions with Ago2 and p54 after heat shock. *, p < 0.05; **, p < 0.01; ****, p < 0.0001; Student’s t test.
The phenotype of cytoplasmic foci induced by Flag-469 appeared to look like a mixture of PBs and stress granules (SGs) at first glance. As matr3 has previously been detected in SGs by mass spectroscopy and in matr3 overexpression studies (Gallego-Iradi et al., 2015; Jain et al., 2016), we costained Flag-469–transfected cells with the endogenous PB marker p54 and SG marker PABP-1 (Figure 5B). Interestingly, under basal conditions, Flag-469 induced SG formation, even though it colocalized predominantly with p54 PBs. However, when cells were heat shocked at 42°C for 45 min, Flag-469 foci translocated to SGs (Figure 5B). These data suggest that matr3 plays a role in SG induction from PBs and is capable of translocating to SGs during stress.
Next GST pull downs were performed with Matr3 GST-469, GST-322, and GST-RRM1 to determine which portion of the protein contained the relevant binding sites for PB protein complexes (Figure 5C). Matr3 GST-469 pulled down endogenous Ago2 and p54 from U2OS cell extracts, while GST-322 or GST-RRM1 were unable to do so, suggesting that the PB-binding region requires both the ZnF1 and RRM1 domains (Figure 5C).
As Flag-469 translocated from PBs to SGs under thermal shock, we examined whether Flag-469 interactions with PB proteins were disrupted after heat shock. Flag-469 co-IPs had less Ago2 and p54 after stress compared with control cells, suggesting Flag-469 binds to fewer PB proteins after heat shock (Figure 5D). Together these experiments highlight a signicant portion of the matr3 protein that is required for PB and SG localization and interaction with PB proteins.
We then examined whether endogenous PB matr3 was capable of moving from PBs to stress granules. After heat shock, cells were costained with matr3 and Hedls or PABP-1 (Figure 6). Interestingly, we observed a reduction in nuclear matr3 after heat shock. There was also a significant reduction in matr3 foci colocalizing with Hedls PBs, matching the data seen with Flag-469 and PBs following heat shock (Figure 5B). Although matr3 foci did have greater Pearson’s colocalization coefficient with PABP-1 after stress, only very weak colocalization was observed compared with the strong colocalization coefficient observed between Flag-469 and PABP-1 after stress (Figures 6 and 5B). Thus there are slight discrepancies between the behavior of endogenous matr3 and ectopic Flag-469, which are likely due to differing sequences of Flag-469 and endogenous matr3.
FIGURE 6:
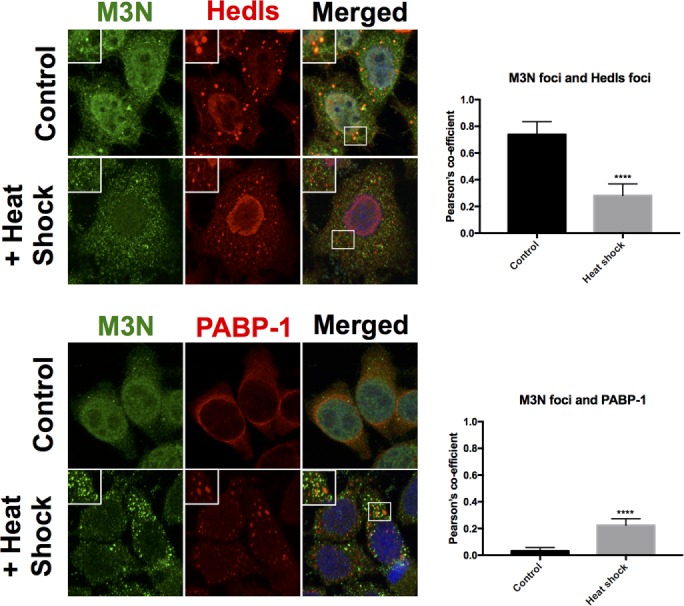
After heat shock, endogenous matr3 does not colocalize with SGs; endogenous matr3 foci have reduced colocalization with Hedls PBs; and Matr3 foci have enhanced colocalization with PABP-1 after heat shock, even though the degree of colocalization remains weak. Furthermore, nuclear matr3 levels were reduced after heat shock. Colocalizations quantified using Pearson’s colocalization coefficient (N = 15 over three experimental repeats). ****, p < 0.0001; Student’s t test.
Next we attempted to identify the origin of the truncated Matr3 PB variant by performing 5′ and 3′ rapid amplification of cDNA ends (RACE) on a HeLa cell cDNA library. However, no new sequences were amplified (unpublished data). Additionally, publicly available databases were screened for clones that could represent this isoform. However, this also failed to identify novel variants, suggesting that further investigation of matr3 sequences is required to identify this variant.
DISCUSSION
Nesprins are a diverse family of intracellular scaffolds and linkers required for maintaining cellular organization and architecture through the generation of multiple isoforms. Here we identified several new nesprin-1 binding partners that provide further evidence that nesprin-1 is a versatile intracellular scaffold. Nesprin-1 scaffolding has already been shown to have an important role in PB function, and in this study, we identify matr3 as a new binding partner and define a previously unrecognized function for a well-studied nuclear protein in PB function.
Matr3 as a novel miRISC regulator, PB, and SG protein
We identified matr3 as both a nesprin-1 binding partner and as a novel candidate for miRISC function, as demonstrated using the Let-7a and CXCR4 miRISC reporter constructs. Using an antibody raised against the N-terminus of matr3, we detected PB localization, which is consistent with the PB localization of nesprin-1 and its ability to interact with other PB proteins (Hock et al., 2007; Rajgor et al., 2014). Matr3 and DDx5 associated with the nesprin-1 SRs, which have previously been shown to interact with Ago2 and p54 and to be required for miRISC function. Although DDx5-depleted cells did not change Let-7a or CXCR4 miRSC activity, matr3 knockdown significantly attenuated miRISC function. The role matr3 plays in miRISC function has yet to be determined. Matr3 has previously been shown to bind small RNA, suggesting that it may be able to bind and sequester miRNAs from binding to mRNA targets (Salton et al., 2011). However, as matr3’s interaction with Ago2 appears to be RNA dependent, it is likely to have a more direct role with the RISC complex.
As an antibody targeting the C-terminal end of Matr3 was unable to detect matr3 localization in PBs or coimmunoprecipitate with PB proteins, either a novel matr3 isoform or a cleavage product must reside within PBs. Indeed, by comparing nuclear and cytoplasmic fractions, we identified an ∼50 kDa cytosolic matr3 variant, and using co-IPs, we showed this variant interacts with PB proteins. Because matr3-containing PBs were depleted along with the 50 kDa Western band using siRNAs targeting the N-terminal region of the protein, but not the C-terminal region, at least two variants of matr3 exist, one that localizes to the nuclear matrix and another that localizes to PBs. As the C-terminal siRNA was unable to knockdown the 50 kDa Western band or PBs, the possibility of matr3 localizing to PBs as a result of a cleavage product of the larger nuclear protein is unlikely.
To identify the major domains required for matr3 PB localization, we created various C-terminal truncation constructs. Matr3-469, which terminates after the first RRM, encodes a 52 kDa construct that we show to localize within PBs. The degree of homology that exists between this construct and the actual ∼50 kDa matr3 isoform expressed in cells has yet to be determined. Interestingly, PB localization through truncation constructs suggests that RRM1 is necessary for localization, as its removal abolished cytoplasmic foci. Furthermore, the PB protein-binding sites on matr3 are likely to be located on both RRM1 and ZnF1, as the individual domains do not associate with PB proteins. Identifying which mRNA or miRNA transcripts bind with RRM1, along with any other protein-interacting partners that associate with the N-terminal portion of the protein, may help determine the role of matr3 in PBs.
The cytoplasmic foci formed by Flag-469 appeared to be a mixture of PBs and SGs at first glance. However, staining for endogenous PBs and SGs showed ectopic matr3-469 to predominantly localize in PBs, even though it was able to induce SG formation. These data indicate matr3 is important for SG formation from localizing to PBs, even though the mechanism of how it does this requires further work. Heat shock resulted in a stronger localization of matr3-469 in SGs, indicating it is also an SG protein, which is consistent with previous mass spectrometry data of SGs and overexpression studies showing matr3 in SGs (Gallego-Iradi et al., 2015; Jain et al., 2016). Furthermore, matr3 interactions with Ago2 and p54 were disrupted after heat shock, suggesting PB functions may be hampered after stress. Whether Flag-469 is able to translocate bound RNA species with it from PBs to SGs warrants further investigation. When we examined whether endogenous matr3 displayed similar effects after heat shock, we did see a reduction of matr3 with PBs and only a weak colocalization with SGs. The discrepancies between matr3-469 and endogenous matr3 are likely to be due to differences in sequences between the two forms. Interestingly, we did observe a significant reduction in endogenous nuclear matr3 levels after heat shock. Whether heat shock induces nuclear matr3 degradation or cytosolic translocation warrants further investigation. Also, whether a similar distribution is seen by Flag-469 and endogenous matr3 after other forms of stress are applied (e.g. oxidative stress by arsenite) is worth exploring.
Using 3′ RACE we attempted to identify a novel 3′ mRNA end but were unsuccessful, possibly due to several limitations associated with this approach, which include difficulty in amplifying rare or potentially long transcripts. Alternate matr3 variants were also not detected on exploration of publicly available databases, making the elucidation of the origin of this 50 kDa variant a focus of future studies.
The localization of our matr3 constructs did provide additional interesting findings about matr3. The nuclear localization signal (NLS) of matr3 has previously been shown to be located between residues 596 and 602, which makes the nuclear localization of Flag-567 interesting. Furthermore, a small population of Flag-469–transfected cells had weak nuclear localization, whereas Flag-322 was entirely cytoplasmic. These experiments suggest that there are at least two additional NLSs located within matr3; one between residues 470 and 567 and a weak NLS between residues 333 and 469.
Matr3 mutations have been identified in amyotrophic lateral sclerosis (ALS), and these mutations also map to the N-terminal region of matr3 that localizes to PBs and SGs. Although nuclear matr3 functions such as transcription and splicing could be disrupted as a result of ALS-causing mutations, there is a possibility that specific functions of PB matr3, such as miRISC function or SG-assembly properties, are also hampered. This is further supported by other RNA granule proteins, such as TDP-43 and FUS, also being mutated in ALS (Li et al., 2013). Therefore further understanding of the role of matr3 in PB biology may provide useful insight into understanding the onset of ALS.
New novel functions for Nesprin-1
Recently matr3 has been shown to interact with the C-terminal tail of lamin A/C, and this interaction is important in maintaining a correct interface between the lamina and nuclear matrix (Depreux et al., 2015). Smaller KASH domain–containing isoforms of nesprin-1 also reside at the inner nuclear membrane and bind to lamin A/C. As nesprin-1 SRs share a high degree of homology, it is possible that SRs other than SR48 (SR1 in Figure 1D) bind matr3. Thus it is likely that nesprin variants present within the LINC complex also form a scaffold with nuclear matr3, further suggesting that nuclear envelopathies due to nesprin and lamin mutations may also exhibit impaired connections with matr3. It is also plausible that many of the matr3 functions identified, including alternative splicing, DNA damage response, chromatin organization, and retention of hyperedited RNA, could be regulated by mechanical signals via the nesprins and the LINC complex (Ma et al., 1999; Zhang and Carmichael, 2001; Salton et al., 2010, 2011; Coelho et al., 2015). This may be particularly important in the central nervous system and muscle, where nesprin (Emery-Dreifuss muscular dystrophy and autosomal-recessive cerebellar ataxia]) and matr3 (ALS) mutations have been shown to induce disease (Zhang et al., 2007; Johnson et al., 2014).
We also identified other nesprin-1 binding partners involved in a variety of cellular activities, including transcription (DDx5, PSF), alternative splicing (PSF, DDx5, hnRNP A2/B1, Matr3), and DNA replication/damage repair (PCNA, Matr3) in this study (Emili et al., 2002; Fuller-Pace and Ali, 2008; Chen and Manley, 2009; Coelho et al., 2015). These findings illustrate the potential multifunctional nature of nesprin-1, wherein different isoforms generated through alternative transcription are likely to be involved in distinct biological roles. No nesprin-1 was detected by mass spectrometry in our screen, which initially suggests that these may not be novel nesprin-1 binding partners. However, we were able to show by Western blotting that nesprin-1 was indeed present within pAbN4 and pAbN5 immune complexes. There are several reasons for the absence of nesprin-1 as seen by mass spectrometry. One possibility is that the bands corresponding to nesprin-1 were not sufficiently excised or digested from the silver-stained gels. Another more likely scenario is that the binding partners associate with more than one SR within nesprin-1 or that multiple nesprin-1 isoforms have the same binding partners. As the central rod domain of nesprin-1 is present in a large number of isoforms, and as many SRs share a high homology, either case is possible and would result in a greater enrichment of binding partners relative to nesprin-1 proteins. Furthermore, we were able to map matr3 and DDx5 to specific SRs of nesprin-1 using GST pull downs, which strongly suggests that our screen did identify novel nesprin-1 binding partners. Identifying which proteins partner with the various nesprin-1 isoforms and the subcellular compartment in which these complexes localize will provide further insights into the importance of nesprin-1 variants as intracellular scaffolds and possible clues to how mutations in nesprin-1 may affect diverse signaling pathways in disease.
MATERIALS AND METHODS
Plasmids
Matr3 C-terminal truncations mutants were created by introducing termination codons into pCI-Flag-Matrin3DNA3.1, which was kindly provided by Christopher Smith (University of Cambridge), using the QuikChange II site-directed mutagenesis kit (Agilent, Santa Clara, CA). Primers used for creating the truncations were as follows: Flag-798 (forward: 5′GACTATGTGATACCTAATAAAGGGTTTTACTGTAAGCTG3′; reverse: 5′CAGCTTACAGTAAAACCCTTTATTAGGTATCACATAGTC3′), Flag-567 (forward: 5′GAGATGTGTGAAGGTTGACTAGTCTGAGAAATATAAAAAACTG3′; reverse: 5′CAGTTTTTTATATTTCTCAGACTAGTCAACCTTCACACATCTC3′), Flag-469 (forward: 5′GCCAGTGAGAGTTCATTAATCCCAGAAGTATAAAA-GA3′; reverse: 5′ TCTTTTATACTTCTGGGATTAATGAACTCTCACTGGC3′), Flag-322 (forward: 5′CGTCGATGCCAGCTTCTTTAAGAAATCTACCCAGAATGG3′; reverse: 5′CCATTCTGGGTAGATTTCTTAAAGAAGCTGGCATCGACG3′). The psi-CHECK2-let-7 ×3 and psi-CHECK2-let-7 3× mut vectors were kindly provided by G. Hutvagner (University of Dundee, Scotland, UK; Johnston et al., 2010). pRL-TK CXCR4 6× vector was kindly provided by P. Sharp (MIT; Doench et al., 2003) via Addgene.
Tissue culture
U2OS cells were passaged when reaching ∼70% confluency and maintained in DMEM complete (Sigma, St. Louis, MO) supplemented with 10 U/ml penicillin, 10 mg/ml streptomycin, 10 mg/ml glutamine, and 10% fetal bovine serum. When appropriate, cells were treated with sodium arsenite (0.5 mM), nocodazole (10 μg/ml), or cycloheximide (10 μg/ml) or in fresh culture media for 1 h.
Co-IPs
Cells were washed with phosphate-buffered saline (PBS), and extracts were prepared by scraping cells in ice-cold co-IP buffer (10 mM Tris, pH 7.4, 150 mM KCl, 1 mM ethylenediaminetetraacetic acid [EDTA], 1% Triton, and protease inhibitor cocktail). For identifying novel nesprin-1 binding partners using mass spectrometry, 250 mM KCl was used to increase stringency. Lysed cells were centrifuged at 13,050 × g for 20 min at 4°C and were precleared with protein A– or protein G–Sepharose beads (Sigma) for 1 h at 4°C with rotation. Protein complexes were obtained by incubating 2 μg of relevant antibody with 200 μg of cell lysate overnight at 4°C with gentle rotation. Immune complexes were purified using Sepharose beads, washed three times with co-IP buffer, and boiled in loading buffer, and then the entire co-IP was analyzed by Western blotting or silver staining.
The co-IPs for the RNase treatment were performed under identical conditions. However, washed beads were resuspended into co-IP buffer and divided into two equal volumes. RNase mixture (10 μg/ml; DNase free; Roche, Basel, Switzerland) was added to one, and both were incubated at room temperature (RT) for 20 min. After two washes in cold co-IP buffer, proteins were eluted in loading buffer.
GST pull down
GST and GST fusion proteins were induced from 100 ml of bacterial culture for 2 h using 0.2 mM isopropyl β-d-1-thiogalactopyranoside at 30°C. Ten to twenty microliters of culture was used for purification (the rest was pelleted and frozen) according to the GE Healthcare protocol using glutathione–Sepharose 4B beads (GE Healthcare, Little Chalfont, United Kingdom). Pull downs were performed from U2OS whole-cell lysates prepared as described above. Four hundred micrograms of protein lysates was incubated with 30 μl beads overnight at 4°C with rotation. Bound proteins were eluted into protein loading buffer, and the entire pull down was analyzed by Western blotting.
Mass spectrometry
Bands from silver-stained gels were excised and cut into small pieces of ∼2 mm using a sterile scalpel. In-gel digestion was performed with trypsin on a robotic digestion system (Investigator ProGest; GENOMIC SOLUTIONS, Ann Arbor, MI). Peptides were separated by a nanoflow liquid-chromatography (LC) system on a reverse-phase column (C18 PepMap100, 3 μm, 100 Å, 25 cm; Dionex) and subjected to tandem mass spectrometry (MS/MS) analysis (LTQ-Orbitrap XL, Thermo Fisher Scientific). The spectra were matched against the human database (UniProtKB/Swiss-Prot Release 14.6, 20,333 protein entries). Protein hits with identification probability > 99.0% and at least two unique peptides with identification probability > 95% were selected using Scaffold software (version 2.0; Proteome Software, Portland, OR) and considered for further work. Mass spectrometry was performed by Xiaoke Yin (King’s College London).
Cytoplasmic and nuclear fractionations
Plated cells were washed twice with PBS and harvested by scraping cells in an appropriate volume of nuclear fractionation buffer (250 mM sucrose, 20 mM HEPES, pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, and protease inhibitor cocktail). The cells were collected and incubated on ice for 10 min; this was followed by a centrifugation at 845 × g for 10 min in a 4°C centrifuge to pellet nuclei. The supernatant containing the cytoplasmic fraction was transferred to a fresh Eppendorf. The pelleted nuclei were resuspended in an appropriate volume of RIPA buffer (150 mM KCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, and protease inhibitor cocktail), vortexed, incubated on ice for 10 min, and centrifuged at 13,050 × g for 10 min in a 4°C centrifuge. The supernatant was collected as the nuclear fraction, and 10–20 μg of protein per fraction was analyzed by Western blotting.
Western blotting
Cell extracts were resolved on a 10% SDS–PAGE gel and transferred to polyvinylidene fluoride membrane (Millipore, San Diego, CA) using a semidry apparatus. The membranes were blotted with the antibodies indicated in each figure, and bands were visualized using the ECL Western blotting substrate (Thermo Fisher Scientific, Waltham, MA). Membranes were incubated with the following primary antibodies: anti-Rck/p54 (ab54611; Abcam, Cambridge, United Kingdom), anti-Ago2 (ab57113; Abcam), anti-Dcp1a (ab57654; Abcam), anti–β-actin (A2228; Sigma), anti-Matr3C (ab84422; Abcam), anti-Matr3N (ab51081; Abcam), PSF (sc-101137; Santa Cruz, Santa Cruz, CA), DDx5 (ab21696; Abcam), Importin-β (ab2811; Abcam) mouse anti–α-tubulin (T5168; Sigma-Aldrich), rabbit-LaminB1 (sc-20682; Santa Cruz). Secondary antibodies conjugated to horseradish peroxidase (GE Healthcare) were used at 1:10,000 dilutions and developed using ECL reagent and X-ray film. Densitometry measurements were performed using ImageJ.
Let-7 miRISC luciferase assay
Twelve thousand U2OS cells were plated into each well of a 24-well plate and cultured under standard growth conditions overnight. The following day, three wells per experimental condition were cotransfected with 1 μl Matr3, DDx5, Rck/p54, or scrambled siRNA oligos (all at 20-μM stock solutions) and 1 μg of psi-CHECK2-let-7 ×3 or psi-CHECK2-let-7 ×3 m using Hiperfect transfection reagent (Qiagen). siMatr3 and si-DDx5 si-p54 sequences used were 5′GUCAUUCCAGCAGUCAUCUUU3′ and 5′AAGAGGUGGAAACAUACAGAA3′, respectively. Si-p54 was a Smart pool obtained from Dharmacon (L-006371-00-000). Luciferase readings were taken 72 h after transfection using the Dual-Luciferase Reporter assay (Promega, Madison, WI). Renilla luciferase activity was normalized to firefly luciferase activity to control for transfection efficiency (both transcribed off the psi-CHECK2 plasmids).
CXCR4 miRISC luciferase assay
Twelve thousand U2OS cells were plated into each well of a 24-well plate and cultured under standard growth conditions overnight. The following day, three wells per experimental condition were transfected with Hiperfect siRNA-plasmid complexes containing 1 μg pRL-TK CXCR4 6× Renilla luciferase plasmids, 50 ng pGL3.4 firefly luciferase plasmid, 1 μl CXCR4 siRNA oligo (20 μM stock), and 1 μl of siRNA oligo (20 μM stock) targeting proteins to be knocked down. Luciferase readings were taken 72 h after transfection using the Dual-Luciferase Reporter assay (Promega). Renilla luciferase activity was normalized to firefly luciferase activity to control for transfection efficiency.
siRNA-mediated gene silencing
Sixty thousand U2OS cells were plated into each well of a six-well plate and cultured under standard growth conditions overnight. The following day, cells were transfected with 1 μl siM3N, siM3C or scrambled siRNA oligos (all at 20-μM stock solutions) using Hiperfect transfection reagent (Qiagen, Hilden, Germany). siM3N and siM3C were 5′GUCAUUCCAGCAGUCAUCUUU3′ and 5′GCUCCUCCAAGUAGCAAUAUU3′, respectively. Knockdown efficiency was analyzed using 10–20 μg of cell extract by Western blotting.
Immunofluorescence microscopy
Cells were fixed for 5 min in 3.7% paraformaldehyde (PFA) (Sigma-Aldrich) followed by 2 min permeabilization with 0.5% NP-40 (EMD Millipore). The coverslips were incubated with blocking solution (1% bovine serum albumin) for 1 h at RT. Primary antibodies used for immune staining were diluted in blocking solution and applied to the coverslips for 1 h at RT; this was followed by three short rinses in PBS. Primary antibodies were as follows: anti-Matr3C (ab84422; Abcam), anti-Matr3N (ab51081; Abcam), and mouse anti-S6K used to detected Hedls (sc-8418; Santa Cruz) at 1:200; rabbit anti-Rck/p54 (A300-461A; Bethyl Laboratories, Montgomery, TX) and mouse anti-PABP-1 (10E10, sc-32318; Santa Cruz) at 1:200. Next the coverslips were incubated with secondary antibodies, diluted 1:1000 in blocking buffer, and conjugated to Alexa Fluor 405, 488, or 546 (Invitrogen, Carlsbad, CA) for 1 h at RT. The coverslips were next stained with 4′,6-diamidino-2-phenylindole and rinsed with PBS before being mounted onto slides using Mowiol mounting media. Images were acquired at RT using a using an Olympus IX81 wide-field microscope with a 40× air objective lens attached to a Hamamatsu Orca-R2 cooled CCD camera. Pearson’s colocalization coefficients were determined using the Coloc2 plug-in in imageJ.
Acknowledgments
We thank the British Heart Foundation and Medical Research Council for funding this work.
Abbreviations used:
- Ago2
Argonaute-2
- ALS
amyotrophic lateral sclerosis
- co-IP
coimmunoprecipitation
- GST
glutathione S-transferase
- IgG
immunoglobulin G
- LINC
linker of the nucleoskeleton and cytoskeleton
- Matr3
matrin-3
- miRISC
miRNA-mediated gene silencing
- NE
nuclear envelope
- NLS
nuclear localization signal
- PB
processing body
- PBS
phosphate-buffered saline
- PSF
PTB-associated splicing factor
- RRM
RNA-recognition motif
- RT
room temperature
- SG
stress granule
- siRNA
small interfering RNA
- SR
spectrin repeat
- ZnF
zinc finger.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-06-0346) on October 12, 2016.
REFERENCES
- Chang W, Worman HJ, Gundersen GG. Accessorizing and anchoring the LINC complex for multifunctionality. J Cell Biol. 2015;208:11–22. doi: 10.1083/jcb.201409047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho MB, Attig J, Bellora N, Konig J, Hallegger M, Kayikci M, Eyras E, Ule J, Smith CW. Nuclear matrix protein Matrin3 regulates alternative splicing and forms overlapping regulatory networks with PTB. EMBO J. 2015;34:653–668. doi: 10.15252/embj.201489852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depreux FF, Puckelwartz MJ, Augustynowicz A, Wolfgeher D, Labno CM, Pierre-Louis D, Cicka D, Kron SJ, Holaska J, McNally EM. Disruption of the lamin A and matrin-3 interaction by myopathic LMNA mutations. Hum Mol Genet. 2015;24:4284–4295. doi: 10.1093/hmg/ddv160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A, Shales M, McCracken S, Xie W, Tucker PW, Kobayashi R, Blencowe BJ, Ingles CJ. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA. 2002;8:1102–1111. doi: 10.1017/s1355838202025037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace FV, Ali S. The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem Soc Trans. 2008;36:609–612. doi: 10.1042/BST0360609. [DOI] [PubMed] [Google Scholar]
- Gallego-Iradi MC, Clare AM, Brown HH, Janus C, Lewis J, Borchelt DR. Subcellular localization of Matrin 3 containing mutations associated with ALS and distal myopathy. PLoS One. 2015;10:e0142144. doi: 10.1371/journal.pone.0142144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt I, Duong NT, Zhang Q, Lam LT, Sewry CA, Mamchaoui K, Shanahan CM, Morris GE. Specific localization of nesprin-1-alpha2, the short isoform of nesprin-1 with a KASH domain, in developing, fetal and regenerating muscle, using a new monoclonal antibody. BMC Cell Biol. 2016;17:26. doi: 10.1186/s12860-016-0105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Pioro EP, Boehringer A, Chia R, Feit H, Renton AE, Pliner HA, Abramzon Y, Marangi G, Winborn BJ, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci. 2014;17:664–666. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Geoffroy MC, Sobala A, Hay R, Hutvagner G. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Mol Biol Cell. 2010;21:1462–1469. doi: 10.1091/mbc.E09-10-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Katanosaka Y, Iwata Y, Matsuoka M, Shigekawa M, Wakabayashi S. Identification and characterization of GSRP-56, a novel Golgi-localized spectrin repeat-containing protein. Exp Cell Res. 2006;312:3152–3164. doi: 10.1016/j.yexcr.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Siegel AJ, Berezney R. Association of chromosome territories with the nuclear matrix. Disruption of human chromosome territories correlates with the release of a subset of nuclear matrix proteins. J Cell Biol. 1999;146:531–542. doi: 10.1083/jcb.146.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellad JA, Warren DT, Shanahan CM. Nesprins LINC the nucleus and cytoskeleton. Curr Opin Cell Biol. 2011;23:47–54. doi: 10.1016/j.ceb.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Nakayasu H, Berezney R. Nuclear matrins: identification of the major nuclear matrix proteins. Proc Natl Acad Sci USA. 1991;88:10312–10316. doi: 10.1073/pnas.88.22.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajgor D, Mellad JA, Autore F, Zhang Q, Shanahan CM. Multiple novel nesprin-1 and nesprin-2 variants act as versatile tissue-specific intracellular scaffolds. PLoS One. 2012;7:e40098. doi: 10.1371/journal.pone.0040098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajgor D, Mellad JA, Soong D, Rattner JB, Fritzler MJ, Shanahan CM. Mammalian microtubule P-body dynamics are mediated by nesprin-1. J Cell Biol. 2014;205:457–475. doi: 10.1083/jcb.201306076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajgor D, Shanahan CM. Nesprins: from the nuclear envelope and beyond. Expert Rev Mol Med. 2013;15:e5. doi: 10.1017/erm.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razafsky D, Wirtz D, Hodzic D. Nuclear envelope in nuclear positioning and cell migration. Adv Exp Med Biol. 2014;773:471–490. doi: 10.1007/978-1-4899-8032-8_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, Burke B. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci USA. 2009;106:2194–2199. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salton M, Elkon R, Borodina T, Davydov A, Yaspo ML, Halperin E, Shiloh Y. Matrin 3 binds and stabilizes mRNA. PLoS One. 2011;6:e23882. doi: 10.1371/journal.pone.0023882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salton M, Lerenthal Y, Wang SY, Chen DJ, Shiloh Y. Involvement of matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle. 2010;9:1568–1576. doi: 10.4161/cc.9.8.11298. [DOI] [PubMed] [Google Scholar]
- Senderek J, Garvey SM, Krieger M, Guergueltcheva V, Urtizberea A, Roos A, Elbracht M, Stendel C, Tournev I, Mihailova V, et al. Autosomal-dominant distal myopathy associated with a recurrent missense mutation in the gene encoding the nuclear matrix protein, matrin 3. Am J Hum Genet. 2009;84:511–518. doi: 10.1016/j.ajhg.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren DT, Tajsic T, Mellad JA, Searles R, Zhang Q, Shanahan CM. Novel nuclear nesprin-2 variants tether active extracellular signal-regulated MAPK1 and MAPK2 at promyelocytic leukemia protein nuclear bodies and act to regulate smooth muscle cell proliferation. J Biol Chem. 2010;285:1311–1320. doi: 10.1074/jbc.M109.032557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, van den Bout I, Raymond K, Sonnenberg A. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitz MJ, Malyavantham KS, Seifert B, Berezney R. Matrin 3: chromosomal distribution and protein interactions. J Cell Biochem. 2009;108:125–133. doi: 10.1002/jcb.22234. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Bethmann C, Worth NF, Davies JD, Wasner C, Feuer A, Ragnauth CD, Yi Q, Mellad JA, Warren DT, et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet. 2007;16:2816–2833. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Ragnauth C, Greener MJ, Shanahan CM, Roberts RG. The nesprins are giant actin-binding proteins, orthologous to Drosophila melanogaster muscle protein MSP-300. Genomics. 2002;80:473–481. [PubMed] [Google Scholar]
- Zhang Q, Skepper JN, Yang F, Davies JD, Hegyi L, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J Cell Sci. 2001;114:4485–4498. doi: 10.1242/jcs.114.24.4485. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]