Abstract
Software to manage automated laboratories interfaces with hardware instruments, gives users a way to specify experimental protocols, and schedules activities to avoid hardware conflicts. In addition to these basics, modern laboratories need software that can run multiple different protocols in parallel and that can be easily extended to interface with a constantly growing diversity of techniques and instruments.
We present Clarity: a laboratory automation manager that is hardware agnostic, portable, extensible and open source. Clarity provides critical features including remote monitoring, robust error reporting by phone or email, and full state recovery in the event of a system crash. We discuss the basic organization of Clarity; demonstrate an example of its implementation for the automated analysis of bacterial growth; and describe how the program can be extended to manage new hardware.
Clarity is mature; well documented; actively developed; written in C# for the Common Language Infrastructure; and is free and open source software. These advantages set Clarity apart from currently available laboratory automation programs.
Introduction
Robotic automation is revolutionizing research in fields including clinical science (1), genomics (2), and systems biology (3, 4). Automated laboratories can produce better, more consistent data; can have lower operating costs; and can be scaled up easily. As more laboratories begin to embrace the benefits of automation, the programs that are used to manage laboratory instruments will have to confront the needs of a new and more diverse group of users.
Software to manage automated laboratories has to interface with hardware instruments, give users a way to describe the activities that make up experimental protocols, and schedule these activities in a way that avoids hardware conflicts. In addition to these basic requirements, modern laboratories need software that can run multiple different protocols in parallel. In a laboratory with multiple independent investigators who share common equipment, software has to be able to schedule parallel protocols on demand without interrupting running protocols. It is also essential for software to be easily extensible so that it can adapt to a constantly growing diversity of techniques and instruments.
Here we present Clarity, a laboratory automation manager designed to meet the challenges of modern laboratory automation. Clarity is hardware agnostic, portable, extensible and open source. Furthermore, it provides critical features that include remote monitoring, robust error reporting by phone or email, and full state recovery in the event of a system crash. We present the basic organization of Clarity; an example of its implementation for the automated analysis of bacterial growth; and a description of how the program can be extended with new instrument interfaces and graphical user interfaces.
General attributes of the organization of Clarity
Hardware and Task Management
The automation of even the most rudimentary laboratory procedures often requires the orchestration of multiple specialized instruments. Instruments usually serve different functions and communicate with the computer in different ways. However, instruments are unified by their purpose: to perform meaningful activities during an experiment.
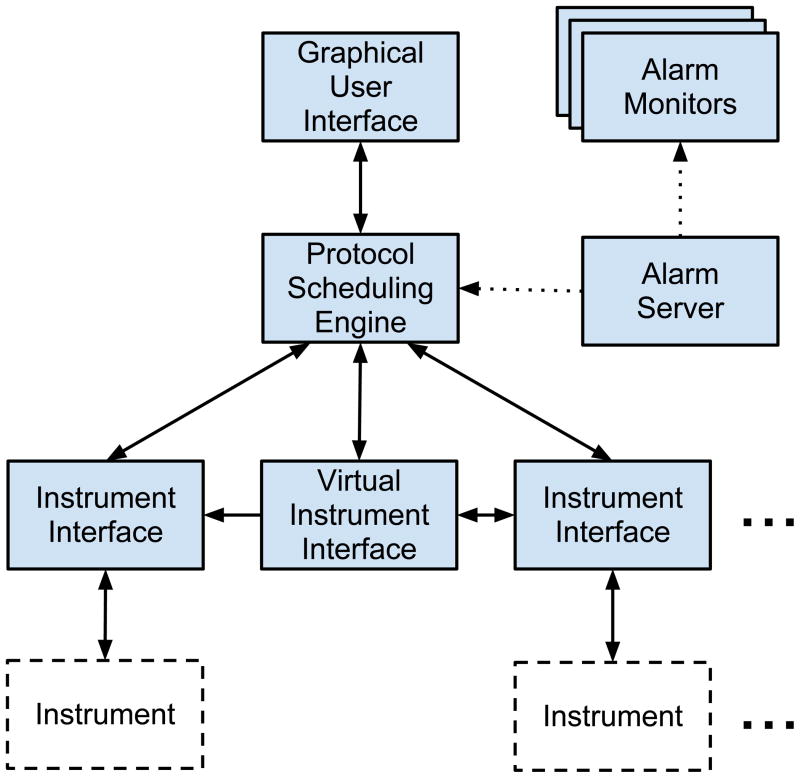
In Clarity, each instrument is associated with a dedicated instrument interface (Figure 1). The instrument interface handles the low-level communication between software and hardware and defines a set of meaningful activities that the instrument can perform during the course of an experiment. The interface to a robotic arm, for example, might define an activity to move the arm to a specific location or lift a microtiter plate. This activity could then appear as a step in the protocol for an experiment. The instrument interface would be responsible for loading the positions of the incubator and spectrometer from a configuration file, for instructing the robotic arm to power the right motors in the right sequence, and for reporting back to Clarity in the event of any hardware errors.
Figure 1.
A diagram of Clarity’s important components and their organization. Solid arrows indicate direct communication (e. g. the protocol scheduling engine calls methods in the instrument interfaces that define activities); dashed arrows indicate connections over internet protocols.
Clarity also supports virtual instruments. Virtual instruments do not correspond to hardware; instead, they are meant to perform completely computational activities during experiments. They can be used to write log or data files, to organize hardware instrument interfaces for multi-instrument activities, or to monitor data output to make decisions about the course of an experiment.
Protocol execution
Protocols define the activities that make up an experiment (5). A simple protocol can consist of a list of activities and the times at which they should be performed. More complex protocols can incorporate control flow elements like loops and conditional statements. Conditions are evaluated by virtual instruments and can therefore depend on anything that the program has access to, including data files, protocol details, and program state. Protocols can be written using a simple protocol description language based on XML, the eXtensible Markup Language (6). Figure 2 contains a simplified snippet from a typical protocol file. Alternatively, Clarity is equipped with a graphical protocol editor. Using the protocol editor, the user simply chooses activities from an interactive list and arranges them to specify her protocol. After providing an email address and phone number, for error reporting, the user can save the protocol to an XML file and use Clarity to execute it.
Figure 2.
A simplified example of an XML protocol file.
Clarity’s scheduling engine keeps track of running protocols and uses the instrument interfaces to call the right activities at the right times. Scheduling is complicated by conflicts, which can arise when multiple users run protocols at the same time and on the same instruments. To resolve conflicts, the scheduler runs a simple and flexible algorithm: 1) When an activity finishes it activates the scheduler. 2) The scheduler inspects the uncompleted activities of the remaining protocols, and identifies the activity with the earliest prescribed time. 3) If that time is in the future, Clarity waits, otherwise, it executes the activity immediately. This algorithm is not ideal for procedures that are extremely time sensitive, but it is easy to run dynamically, meaning that new protocols can be added at any time without stopping the execution of running protocols.
Clarity’s basic scheduling algorithm is designed to be easy to understand and to simply avoid any resource conflicts between different protocols. It runs multiple protocols in parallel by alternating which protocol is running serially at any moment, giving exclusive control of the entire system to one executing protocol, and passing control of the system to another protocol (or context switching) only after the currently executing protocol has stopped using the system resources and returned them to a “ready” state. Clarity however, being open source, can also implement more complex scheduling schemes and run protocols in a truly concurrent manner that uses multiple instruments simultaneously for different tasks. Such parallel execution can however create resource conflicts, race conditions, deadlocks and other problems. This is particularly difficult in the laboratory automation context, because which state a protocol is suspended in can be very important. For instance, we might not want to remove an item from an incubator and place it in a liquid handler if it will be some time before the liquid handler finishes its current task and is available to do the next step. For this reason, specifying a framework that optimally handles all possible concurrency issues, ensures that all the available instrument interfaces can provide enough information for the framework to appropriately make decisions (such as the time required to execute instructions) and does not introduce too much complexity to new users is difficult.
Instead of providing a general solution, Clarity’s design assumes that the parallel execution problem for any specific usage scenario will be easier to solve by writing code for an idiosyncratic implementation than specifying that problem for a general framework will be. Clarity provides the tools for a user to relatively easily code a more truly concurrent scheduler. Clarity allows users to implement concurrent operations through the use of virtual instrument classes. Instrument interfaces and virtual instrument objects can have direct access to the scheduler, the instruments and all the loaded protocols. A user can simply write a virtual instrument that examines the entire system state, and adjust the protocols and their execution order accordingly. Clarity also allows virtual instruments to handle events generated by its engine based on instrument processes, allowing them to respond to the actions taken by different protocols. A walk-through tutorial showing how to create concurrent or dynamic protocols and allow users to write more sophisticated scheduling algorithms to replace Clarity’s simple scheduler is part of Clarity’s online documentation. Clarity’s modular organization, open source license, thorough documentation, and community support, make it especially amenable to this kind of customization. However, we emphasize that it will be the responsibility of anyone implementing such custom scheduling operations to ensure that the problems that can arise in parallel computing, such as deadlocks and race conditions, do not occur.
Error reporting and recovery
An unavoidable aspect of laboratory automation is that instruments can malfunction in the course of protocol execution. Some instrument interfaces can recognize and recover from common errors without user intervention. When an instrument interface encounters an error that it cannot handle, Clarity logs the error and immediately stops protocol execution. At this point, Clarity tries to alert the owners of the affected protocols about the error. Based on the user’s preference, Clarity can send emails or text messages with detailed information about the probable causes of the error. Clarity can also call users’ phone numbers to alert them at all hours of the day and night.
Error reporting is handled jointly by Clarity and a remote alarm server. The alarm server runs on a separate computer and exchanges information with Clarity over the internet. This ensures that users continue to get error reports in the event that one of the computers malfunctions. The alarm server also lets users monitor running protocols. Users can install a monitoring program on a home computer and use it to connect to the alarm server over the internet. The monitor displays upcoming activities, protocol information, and video from user-installed cameras.
Once notified, a user can often resolve problems remotely. Each instrument interface defines methods to re-initialize its associated instrument. The user can use Clarity’s logs or the video cameras to determine whether an instrument needs to be re-initialized. If so, the user can activate the right recovery method from Clarity’s graphical user interface.
Clarity always maintains a backup of the program’s state: the list of running protocols and the list of activities that have yet to be performed. Before and after executing any activity, Clarity updates this backup. This ensures that when errors occur, there is a record of the program’s state that can be used to rescue experiments. Once the problem is fixed, the backup can loaded into a new instance of Clarity to continue running experiments as before.
Graphical interface
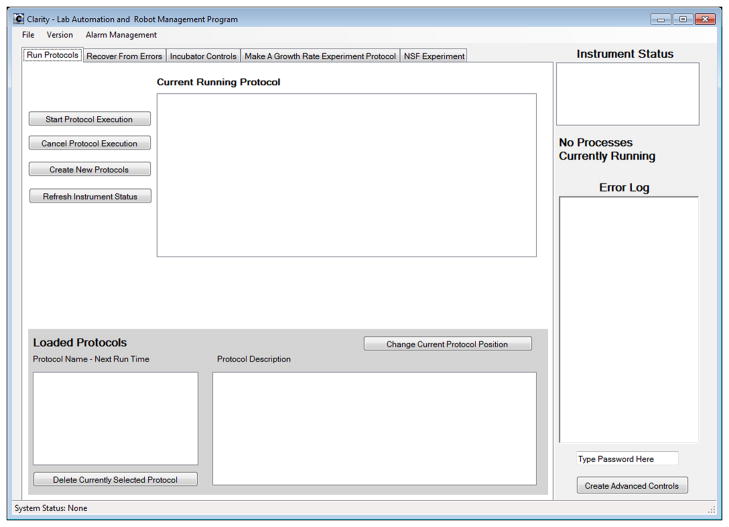
Users interact with Clarity’s components—hardware and virtual instrument interfaces, the protocol scheduling engine, and the remote alarm server—through a graphical interface (Figure 3). The main menu allows users to load and save program states, load protocols, and manage the remote alarm server. The body of the interface is organized into tabs, making it easy for users to add location specific features. The main tab displays running protocols, instrument statuses, a log of errors, and controls to start and stop protocol execution. The error recovery tab provides methods to recover and reinitialize connected instruments. Additional tabs can be implemented to control specific instruments, or to carryout location specific tasks.
Figure 3.
The main tab of Clarity’s graphical use interface. This is where users can control protocol execution. The main tab also displays information about the currently running protocols, all scheduled activities, the statuses of hardware instruments, and an error log.
Clarity’s graphical interface updates itself automatically to accommodate new instrument interfaces. For example, the graphical protocol editor automatically includes activities from new or custom instruments. When it starts, the protocol editor generates its list of activities dynamically by inspecting all available instrument interfaces. The error recovery tab is also generated at run-time. Clarity’s self updating user interface means that users can create custom instrument interfaces without worrying about integrating them with the rest of the program.
Clarity in action: implementation for automated analysis of bacterial growth
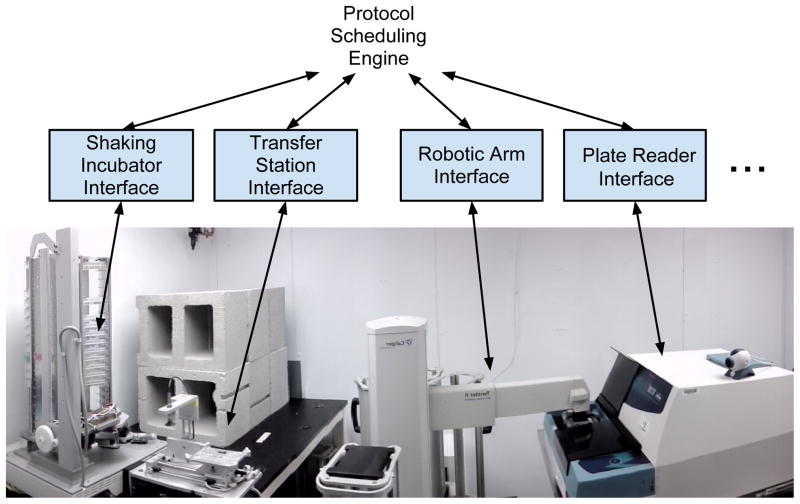
To demonstrate a typical use case, we describe our laboratory’s use of Clarity to manage a series of instruments for automated monitoring of bacterial growth (Figure 4). A major effort in our group is to evolve replicate populations of one or more bacterial species in the laboratory as a means to study the physiological basis of adaptation (7). Given that single experiments can involve hundreds of replicate populations, we maintain populations in 48-well microtiter plates that are stacked on an arbitrary-access, shaking tower that holds up to 38 plates. In order to maintain optimal growth of our study organism, Methylobacterium extorquens, we house the shaking tower, as well as the rest of the system, in a temperature-controlled environmental room at 30 °C, and use a commercial humidifier to augment the humidity to ~75% relative humidity to minimize evaporation. Under these conditions, the primary component of fitness is the exponential growth rate of the culture (8). By using a multi-well plate reader to take optical density readings over multiple days, we can assay the fitness of nearly 2,000 strains concurrently.
Figure 4.
Our hardware configuration for tracing bacterial growth curves in an automated manner.
Users load a 48-well plate—already containing the needed media and cultures—onto the shaking tower. Using Clarity’s graphical interface, the user specifies parameters of the growth curve protocol (e.g. number of measurements); selects the correct position on the tower; enters a file name for the data; and provides email addresses and phone numbers for error reporting. At this point the user can also chose to apply the protocol to multiple plates, specifying their positions on the shaking tower. Then, the user initiates the protocol. A video demonstrating an automated optical density measurement is available in the supplemental materials. Briefly, the protocol proceeds as follows: 1) The robotic spatula removes the microtiter plate from the specified position and places it on the transfer station. 2) A vacuum suction cup raises the plate’s lid and the plate base is moved toward the robotic arm. 3) The arm grasps the plate base, swings into position above the multi-well plate reader, and lowers the plate onto that instrument’s loading platform. 5) The plate is lowered into the plate reader and optical density readings are taken. A virtual instrument records the reading and all the relevant metadata to a spreadsheet. 6) Activities 1–4 are repeated in reverse to return the plate to the transfer station, replace the lid, and load the plate back into the tower. Each of the above activities is specified by the protocol, initiated by the scheduler, mediated through an instrument interface, and carried out by a particular instrument.
The data produced by Clarity are not only high-throughput, but also high quality. Because the 48-well plates allow for effective mixing and aeration—and due to our efforts to optimize the growth medium for Methylobacterium (9)—we routinely observe per capita growth that is incredibly stable throughout exponential phase (Figure 5A). Using a software package we’ve developed (10) we can reliably measure small differences in growth rates (Figure 5B). Since these small differences can have dramatic evolutionary consequences, it is crucial that our data be as high quality as possible. The required precision, scale, and extended timeline of our work preclude performing these experiments in the absence of automation.
Figure 5.
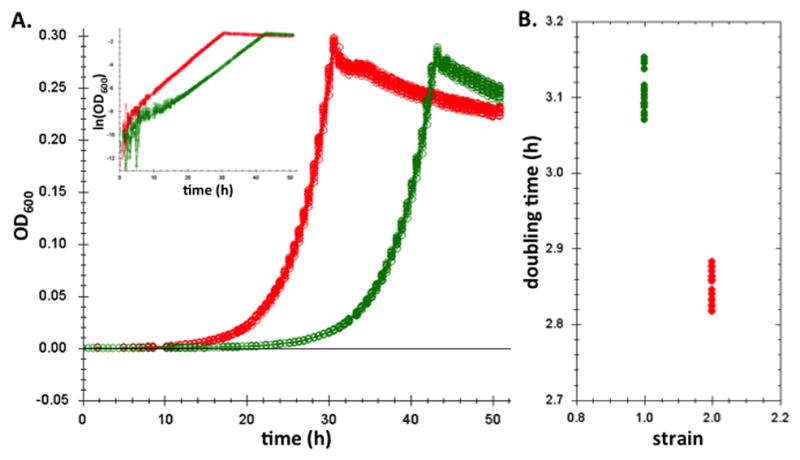
An example set of bacterial growth curves measured by Clarity. A.) The replicates represent a comparison of the optical density measured for over fifty hours for two isolates (red and green; 18 replicates of each) of a strain of Methylobacterium extorquens AM1 evolved during an evolution experiment from the same ancestor. Note in the inset the log-linear increase in density indicates the remarkable constancy of per-capita growth throughout these conditions. B.) Analysis of the doubling times of the two strains indicates the precision by which we can estimate this exponential rate.
Customizing Clarity
Clarity is easy to extend or customize with new instrument interfaces. Custom instrument interfaces can be written to control new hardware instruments, or to carry out computational activities by implementing virtual instruments. Instrument interfaces can be written in any language that conforms to the Common Language Infrastructure (11). Since most modern programming languages have a Common Language Infrastructure implementation, almost anyone with some programming experience can write a custom instrument. Furthermore, instrument interfaces are implemented as independent libraries that are loaded dynamically; this means that they can be included in the program without recompiling Clarity.
Clarity’s online documentation (12) includes a tutorial on implementing a new instrument interface. There we demonstrate how to implement a virtual instrument to send email updates on the progress of running protocols. Every instrument interface will be different in the way that it handles communication with a hardware instrument—or email server—but all conform to a standard way of communicating with Clarity. Basically, every instrument interface needs to inherit from Clarity’s BaseInstrumentClass class. This ensures that all interfaces define an instrument status flag, a recovery method, and a method to release system resources. In addition, BaseInstrumentClass, implements a method to initialize location specific variables (e.g., network details) using XML configuration files. The class membership also serves to identify instrument interface classes. When the program starts, it looks for members of BaseInstrumentClass to include in the list of available instruments.
It is also possible to customize Clarity’s graphical user interface. For example, we make extensive use of a tab that facilitates the design of growth curve protocols. The online documentation includes a template graphical user interface that can be tweaked and customized easily. Alternatively, since protocols reside in XML files, one can write a standalone protocol-generating application that would not need to interface with Clarity directly. Instructions for customizing the user interface and sample interface templates are available in Clarity’s documentation. Clarity is currently designed to manage lab instruments and execute instructions with them; however, it does not include a specific framework to manage the data generated by these experiments. A recent open-source database schema was published, AutoLabDB, (13) that would be useful for this endeavor, and future development will likely focus on implementing database interactions through virtual instruments in Clarity.
Conclusions
Clarity is mature, well documented, and actively developed software for managing laboratory automation. We manage two automated laboratories with Clarity and plan on continuing to release bug fixes and new features. Because the success of a software project depends on the availability of quality support for new users, we maintain up-to-date documentation for Clarity online, and are available to answer questions on a dedicated email list (12). Clarity is written in the C# language and runs on the Common Language Infrastructure (11). This means that the program can run on most software and hardware platforms (including Windows, OS X, and GNU/Linux operating systems). It also means that Clarity can be developed in almost any programming language.
Although Clarity currently has support for only a few instrument types, if a common protocol existed for interfacing with devices, it would be possible to have Clarity generically interact with any device implementing that protocol. Recently, the Standardization in Lab Automation (SiLA) consortium, have created standards for device control interfaces that expose devices as web services and communicates with them using the Simple Object Access Protocol (SOAP) (14). Devices using such a protocol could readily be made available for use in Clarity, and this is an active goal of the development team, but we are only hindered because we do not have access to any devices implementing the standardized protocols.
Clarity is open source under a free software license. This ensures that experiments performed by Clarity are as reproducible as possible because anyone can inspect the source code to determine how the program works, and because experiments are specified in full detail by sharable XML protocols. It also ensures that Clarity remains flexible because anyone can modify the source code to fit particular needs. Most importantly, the open source paradigm means that every contribution to Clarity benefits the whole community of users. These advantages set Clarity apart from the other currently available laboratory automation programs (15, 16, 17).
Acknowledgments
We thank D. G. Robinson for his useful comments on this manuscript. This work was supported by grants from the NIH (GM078209) and NSF (DEB-0845893).
Footnotes
The source code and documentation for Clarity is available at: http://code.google.com/p/osla/
References
- 1.Boyd J. Robotic Laboratory Automation. Science. 2002;295:517–8. doi: 10.1126/science.295.5554.517. [DOI] [PubMed] [Google Scholar]
- 2.Mardis ER. A decade’s perspective on DNA sequencing technology. Nature. 2011;470:198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- 3.King RD, Rowland J, Oliver SG, Young M, Aubrey W, Byrne E, Liakata M, Markham M, Pir P, Soldatova LN, Sparkes A, Whelan KE, Clare A. The Automation of Science. Science. 2009;324:85–9. doi: 10.1126/science.1165620. [DOI] [PubMed] [Google Scholar]
- 4.Zimmermann HF, Degussa JR. A Fully Automated Robotic System for High Throughput Fermentation. Journal of Laboratory Automation. 2006;11:134–7. doi: 10.1016/j.cll.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Schäfer R. Concepts for Dynamic Scheduling in the Laboratory. Journal of Laboratory Automation. 2004;9:382–97. [Google Scholar]
- 6.Bray T, Paoli J, Sperberg-McQueen CM, Maler E, Yergeau F. Extensible markup language (XML) 2008 [Google Scholar]
- 7.Lee M-C, Chou H-H, Marx CJ. Asymmetric, bimodal trade-offs during adaptation of Methylobacterium to distinct growth substrates. Evolution. 2009;63:2816–30. doi: 10.1111/j.1558-5646.2009.00757.x. [DOI] [PubMed] [Google Scholar]
- 8.Chou H-H, Chiu H-C, Delaney NF, Segrè D, Marx CJ. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science. 2011;332:1190–2. doi: 10.1126/science.1203799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delaney NF, Kaczmarek ME, Ward LM, Lee M-C, Marx CJ. Development of an optimized medium, strain and high-throughput culturing methods for Methylobacterium extorquens. PLoS One. 2012 doi: 10.1371/journal.pone.0062957. In preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delaney NF, Kaczmarek ME, Marx CJ. Minimizing sources of variance in microbial growth curves and development of an open-source growth curve fitter. PLoS One. 2012 In preparation. [Google Scholar]
- 11.ECMA (European Association for Standardizing Information and Communication Systems) Geneva, Switzerland: 2002. Standard ECMA-335: Common language infrastructure (CLI) [Google Scholar]
- 12. [accessed June 2012];Clarity’s web page. http://code.google.com/p/osla/
- 13.Sparkes A, Clare A. AutoLabDB: a substantial open source database schema to support a high-throughput automated laboratory. Bioinformatics. 2012;28:1390–7. doi: 10.1093/bioinformatics/bts140. [DOI] [PubMed] [Google Scholar]
- 14.Bär H, Hochstrasser R, Papenfulß B. SiLA: Basic Standards for Rapid Integration in Laboratory Automation. Journal of Laboratory Automation. 2012;17(2):86–95. doi: 10.1177/2211068211424550. [DOI] [PubMed] [Google Scholar]
- 15. [accessed June 2012];Overlord3, real-time static dynamic scheduling laboratory automation software. http://www.paa.co.uk/labauto/products/software/p-overlord3.asp.
- 16.Gentsch J. Flexible laboratory automation to meet the challenge of the ‘90s. Chemometrics and Intelligent Laboratory Systems. 1993;21:229–33. [Google Scholar]
- 17.Benn ND, Liscouski J. Discussion of Open-Source Methodologies in Laboratory Automation. Journal of Laboratory Automation. 2009;14:82–9. [Google Scholar]