Aggregation-prone amyloid β-peptides occurring in Alzheimer’s disease (AD) inhibit the import of nuclear-encoded mitochondrial precursor proteins. The observation of insoluble coaggregates between preproteins and Aβ peptides provides a biochemical explanation for mitochondrial dysfunction typically observed in AD-affected cells.
Abstract
Aβ peptides play a central role in the etiology of Alzheimer disease (AD) by exerting cellular toxicity correlated with aggregate formation. Experimental evidence has shown intraneuronal accumulation of Aβ peptides and interference with mitochondrial functions. Nevertheless, the relevance of intracellular Aβ peptides in the pathophysiology of AD is controversial. Here we found that the two major species of Aβ peptides, in particular Aβ42, exhibited a strong inhibitory effect on the preprotein import reactions essential for mitochondrial biogenesis. However, Aβ peptides interacted only weakly with mitochondria and did not affect the inner membrane potential or the structure of the preprotein translocase complexes. Aβ peptides significantly decreased the import competence of mitochondrial precursor proteins via an extramitochondrial coaggregation mechanism. Coaggregation and import inhibition were significantly stronger for the longer peptide Aβ42, correlating with its importance in AD pathology. Our results demonstrate that direct interference of aggregation-prone Aβ peptides with mitochondrial protein biogenesis represents a crucial aspect of the pathobiochemical mechanisms contributing to cellular damage in AD.
INTRODUCTION
β-Amyloid (Aβ) peptides have been associated with severe human pathological conditions such as Alzheimer disease (AD; Murphy and LeVine, 2010), Down syndrome (Head and Lott, 2004), and cerebral amyloid angiopathy (Weller et al., 2000), all characterized by accumulation and deposition of Aβ peptides in the CNS. Owing to the diversity of pathological aspects connected with a severe neurodegenerative disease like AD, biochemical mechanisms resulting in neuronal cell death and correlation with accumulation of Aβ peptides are not completely clear (Musiek and Holtzman, 2015).
Aβ peptides derive from a proteolytic process mediated by β- and γ-secretases on a type 1 transmembrane precursor called amyloid precursor protein (APP). The most common forms in AD are constituted of 40 (Aβ40) and 42 (Aβ42) amino acids (Zhang et al., 2011). Mutations, environmental factors, and aging can induce changes in the equilibrium between Aβ peptide production and removal (Mawuenyega et al., 2010), as well as imbalance between amyloidogenic and nonamyloidogenic pathways (Agostinho et al., 2015). This causes an increase of Aβ peptide concentration, promoting aggregation and deposition as senile plaques in brain parenchyma. Kinetic and structural studies on Aβ aggregation in vitro reported that unstructured Aβ monomers have an intrinsic tendency to self-assemble spontaneously by a nucleation-polymerization mechanism into higher-order oligomeric, protofibrillar, and fibrillar states (Thal et al., 2015). The aggregation process is enhanced by high peptide concentrations, presence of nucleation seeds, and altered pH, ionic strength, or temperature (Stine et al., 2003). Furthermore, a large variety of posttranslational modifications of the Aβ sequence influence aggregation propensity (Kummer and Heneka, 2014; Thal et al., 2015). Because Aβ42 oligomers represent the most toxic amyloidogenic peptide species, the main component of AD senile plaques, and the first to deposit during senile plaque formation, they play a key pathophysiological role in the development of AD (Haass and Selkoe, 2007). Of interest, although Aβ42 has only small structural differences from the other Aβ peptides, it displays distinct clinical, biological, and biophysical behaviors (Jarrett et al., 1993; Bitan et al., 2003).
The amyloid cascade hypothesis represents the major theory to explain the etiology and pathology of AD (Hardy and Selkoe, 2002; Musiek and Holtzman, 2015). This hypothesis, strongly supported by genetic studies on familial AD cases (Hardy and Higgins, 1992), proposes that an aggregation of Aβ peptides is responsible for initiation of a multistep pathological cascade eventually resulting in neuronal death. A growing body of evidence also suggests the prominent contribution of intracellular accumulation of Aβ peptides as a trigger of neurodegeneration and AD pathology on the cellular level (Wirths et al., 2004; Wirths and Bayer, 2012; Gouras et al., 2010). Intracellular pools of Aβ peptides may stem from intracellular production, reuptake of secreted peptide molecules, or both. Eventually accumulating also in the cytosol, it is likely that intracellular Aβ peptides interact with membranes or other cellular components and induce structural changes of subcellular compartments (LaFerla et al., 2007).
Mitochondrial dysfunction is now generally accepted as a general pathological feature in AD patients (Mattson et al., 2008; Piaceri et al., 2012; Selfridge et al., 2013). In line with this, a modification of the amyloid cascade hypothesis was postulated that supports the correlation between mitochondrial dysfunction with AD. Named the mitochondrial cascade hypothesis, it considers how individual mitochondrial dysfunctions, accumulating in aging cells, could influence Aβ peptide homeostasis and aggregation and consequently the chronology of AD (Swerdlow et al., 2014). However, it is still disputed whether mitochondrial dysfunctions are early casual events or a consequence of other pathological events in AD patients. Evidence exists for accumulation of Aβ peptides in mitochondria, interactions with protein components of the mitochondrial matrix, and perturbations of mitochondrial functions (Lustbader et al., 2004; Hansson Petersen et al., 2008; Mossmann et al., 2014; Kaminsky et al., 2015). The molecular mechanisms underlying mitochondrial accumulation and the claimed effects of Aβ peptides on mitochondria need critical analysis and clarification. For this reason, we performed a comprehensive biochemical analysis of the interaction between the two Aβ peptides species relevant to AD (Aβ40 and Aβ42) with human mitochondria. One of the major cellular processes responsible for maintaining mitochondrial functions is the import of nuclear-encoded mitochondrial precursor proteins from the cytosol (Chacinska et al., 2009). We used an established import assay based on isolated intact mitochondria (Ryan et al., 2001) to check whether and how Aβ peptides directly interfere with the mitochondrial protein import reaction. Taken together, our results show a strong and direct inhibitory effect of Aβ peptides on mitochondrial protein biogenesis. This inhibition is not caused by a damaging influence of Aβ peptides on mitochondrial functions but is correlated with a coaggregation phenomenon between Aβ peptides and precursor proteins that severely restricts their import competence.
RESULTS
Aβ peptides interfere with the import of mitochondrial precursor proteins
The import of precursor proteins, synthesized at cytosolic ribosomes, represents a crucial process in maintaining mitochondrial function and activity. To test a direct effect of Aβ peptides on mitochondrial protein import, we used an in vitro assay system that measures the uptake of radiolabeled mitochondrial precursor proteins into intact mitochondria isolated from human cell cultures. This assay allows to directly follow the association, uptake, and processing of mitochondrial precursor proteins (Ryan et al., 2001; Chacinska et al., 2009).
As precursor proteins, we used the following 35S-labeled polypeptides: mitochondrial malate dehydrogenase (MDH2), an enzyme of the citric acid cycle; ornithine carbamoyltransferase (OTC), which is involved in the urea cycle, and Su9(86)–dihydrofolate reductase (DHFR) and Su9(70)‑DHFR, both artificial, mitochondrially targeted fusion proteins comprising the presequence of the subunit 9 (Su9) of the F1Fo-ATP synthase (86 and 70 amino acids, respectively) from Neurospora crassa fused to the complete mouse DHFR. All of these precursor proteins contain an N-terminal presequence that is cleaved by the mitochondrial processing peptidase (MPP) after the polypeptide reaches the matrix compartment. Their mitochondrial import depends on the membrane translocase complexes translocase of the outer mitochondrial membrane (TOM) and translocase of the inner mitochondrial membrane with the core component Tim23 (TIM23) and a functional inner membrane potential (Δψmt; Chacinska et al., 2009). In addition, we tested a precursor protein of the metabolite carrier family, the adenine nucleotide translocator 3 (ANT3). This protein is constituted by highly hydrophobic transmembrane subunits and lacks an N-terminal presequence. ANT3 is inserted into the inner mitochondrial membrane, and its import uses a distinct pathway that depends on the TOM and TIM22 complexes (Truscott et al., 2002).
To assess AD-related pathological effects in the import assay, we used the most relevant Aβ peptides found in AD cases, constituted by 40 (Aβ40) and 42 (Aβ42) amino acids. The Aβ peptides and the radiolabeled precursor protein were incubated together with energized human mitochondria isolated from cultured HeLa cells. After the import incubation, samples were treated with proteases to digest residual nonimported polypeptides represented by the precursor form (p) and leaving the completely imported and processed mature form (m). Import reactions were analyzed by tricine-SDS–PAGE and Western blot followed by autoradiography to detect the 35S-labeled imported polypeptides, and the presence of Aβ peptides was detected by immunodecoration with a specific antibody against Aβ. Because ANT3 does not contain an N-cleavable presequence and is not processed in the matrix, complete import was analyzed by blue-native gel electrophoresis (BN–PAGE), indicating the Δψmt-dependent formation of a dimeric complex after insertion into the inner membrane.
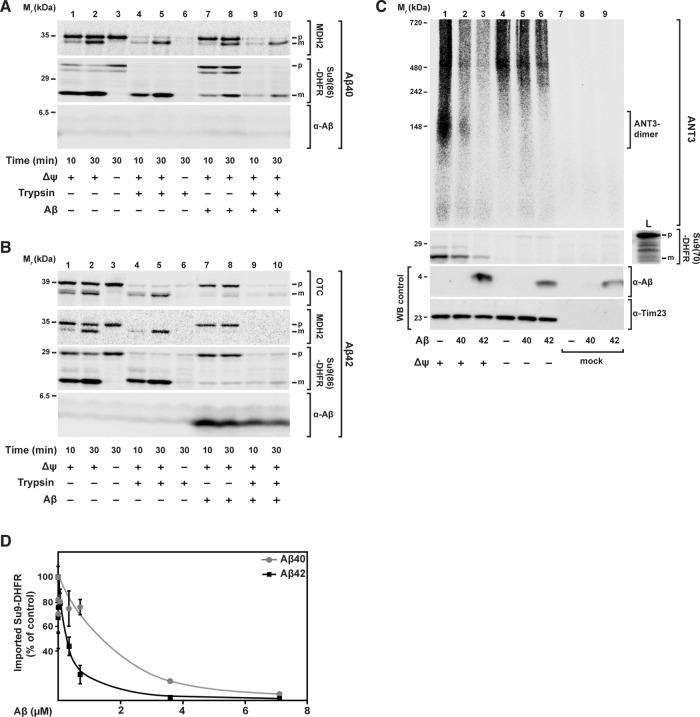
We found that Aβ peptides strongly interfered with the mitochondrial import of all precursor proteins analyzed (Figure 1). The two Aβ peptides showed a different degree of inhibitory effect. Using the same concentration, Aβ40 partially inhibited the import reaction (Figure 1A), whereas Aβ42 showed complete inhibition (Figure 1B), as indicated by the absence of the mature (m) form of a fully imported and processed precursor protein. ANT3 import was analyzed by BN–PAGE to visualize the formation of the complex around 150 kDa (Figure 1C, lane 1). Also in this case, Aβ peptides were able to inhibit the import reaction. Again, Aβ42 was more effective in inhibiting the import reaction than Aβ40. The inhibitory effect, in particular of Aβ42, resulted in a full elimination of the generation of mature forms (m), as well as a complete protease sensitivity of the precursor protein (p) in the import reaction. Taken together, these two criteria indicate a full block of the mitochondrial translocation process and a general phenomenon affecting different import pathways.
FIGURE 1:
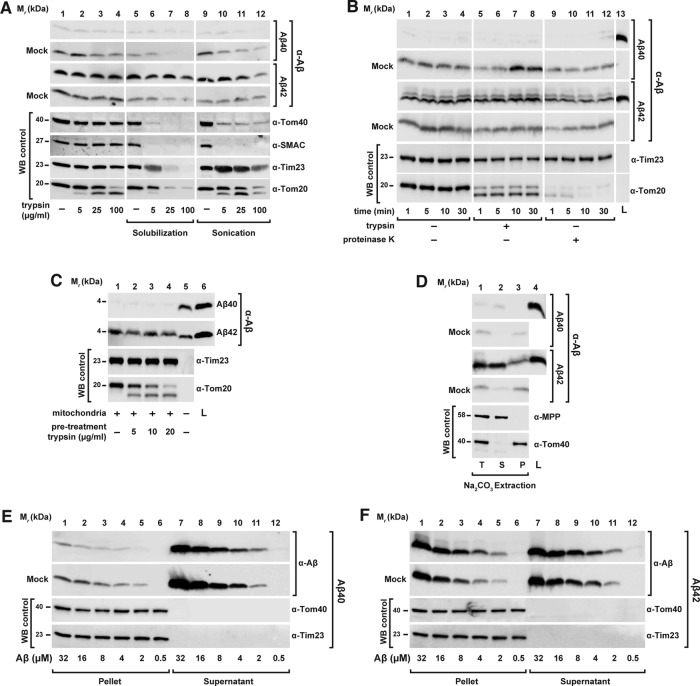
Effect of Aβ peptides on mitochondrial import of nuclear-encoded precursor proteins. 35S‑labeled radioactive precursor proteins were incubated with energized and isolated mitochondria from HeLa cell cultures in the presence of same amounts (3.5 μM) of Aβ40 and Aβ42 peptides. (A, B) Import of the precursor proteins mitochondrial MDH2, the artificial reporter construct Su9(86)-DHFR, and OTC for the indicated incubation times. After the import reaction, half of the samples (lanes 4–6, 9, and 10) were treated with trypsin (100 μg/ml) to remove nonimported preproteins. Imported proteins were analyzed by tricine-SDS–PAGE, followed by Western blot, digital autoradiography, and immunodecoration against Aβ peptides. (C) Import of ANT3 in comparison with Su9(70)-DHFR. After import, all samples were treated with PK (50 μg/ml) and analyzed by BN- (ANT3) or SDS–PAGE (Su9(70)-DHFR), Western blot, and digital autoradiography. As control, immunodecoration against Tim23 was carried out. (D) Quantification of import-inhibitory effect of Aβ peptides. Import experiments with the precursor protein [35S]Su9(86)DHFR and different amounts of Aβ peptides (0.007–7.0 μM) were performed as described. The signals of processed and protease-resistant preprotein bands (m form) were quantified using ImageJ. The amount of imported protein in the absence of Aβ peptide was set to 100%. Mean values and SD were determined for three independent experiments. L, loading control; m, mature processed form; p, precursor protein; WB, Western blot.
To investigate the concentration dependence of the inhibitory effect of Aβ peptides on mitochondrial import, we performed a titration of Aβ peptide amounts during the [35S]Su9(86)-DHFR import assay (Figure 1D and Supplemental Figure S1, A and B). After import, samples were digested by trypsin and analyzed by tricine-SDS–PAGE, autoradiography, and Western blot. We quantified the protease-resistant mature form (m) of the imported [35S]Su9(86)-DHFR. We found that the inhibitory effect of Aβ42 was ∼10-fold stronger than that of Aβ40 (Figure 1D). Inhibition of import by Aβ42 started at a concentration of ∼0.1 μM, whereas for Aβ40, a concentration of >1 μM was required. Note that only at the highest concentration was the Aβ40 band detectable also in the mitochondrial fraction (Supplemental Figure S1B).
Aβ peptides do not interfere with general mitochondrial functions
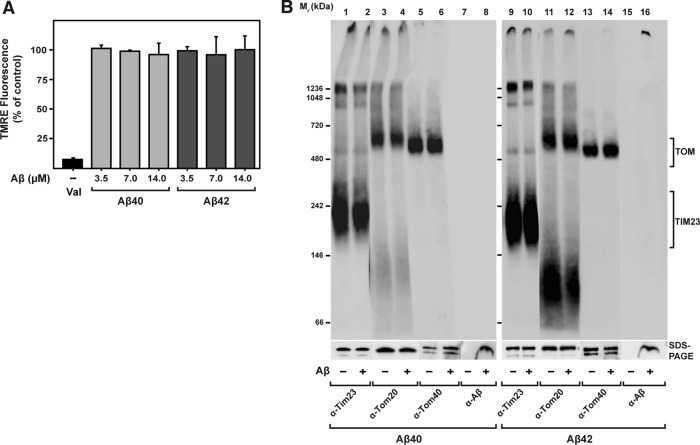
Because it was previously reported that Aβ peptides exert direct damage on mitochondria in vitro (Lustbader et al., 2004; Hansson Petersen et al., 2008; Mossmann et al., 2014), we assayed the state of specific import-related mitochondrial functions in our experimental setup. An electric potential across the mitochondrial inner membrane (Δψmt) is indispensable for import of precursor proteins into the matrix, as well as insertion into the inner membrane (Ryan et al., 2001). We measured Δψmt in our model by the potential-dependent accumulation of the fluorescent dye tetramethylrhodamine ethyl ester (TMRE) after incubation of isolated and energized mitochondria with increasing amounts of Aβ peptides (Figure 2A). Both Aβ40 and Aβ42 did not exhibit any effect on Δψmt, even at high concentrations. As negative control, we incubated the mitochondria with 0.5 μM valinomycin, which causes complete dissipation of the membrane potential and concomitant strong reduction of the fluorescence signal. Using BN–PAGE, we inspected the structure and composition of translocase complexes responsible for the import reaction under native conditions. In the BN–PAGE, the translocase complexes of both the outer membrane (TOM) and the inner membrane (TIM23) migrate as distinct high–molecular weight bands. Incubations with both Aβ peptides did not have any visible effect on the running behavior of the translocase complexes, indicating no significant change in structure and composition (Figure 2B). Furthermore, the absence of effects in the native PAGE indicated that there is no significant stable interaction between the mitochondrial import complexes and Aβ peptides themselves. Note that the detection of Aβ peptides in BN gels (Figure 2B) revealed a signal for Aβ42 localized in the upper part of the stacking gel, consistent with formation of high–molecular weight aggregates. In addition, we also checked the running behavior of the five respiratory chain complexes of the inner membrane in native PAGE and again found no significant differences caused by the presence of Aβ peptides (Supplemental Figure S2). These results demonstrated that Aβ peptides did not negatively affect mitochondrial activities that are directly relevant for the import reaction. In line with this, resistance of mitochondrial control proteins against proteinase K (PK) treatment after import also suggests that mitochondrial membranes remained largely intact after Aβ treatment.
FIGURE 2:
Effect of Aβ peptides on import-related mitochondrial functions. (A) The Δψmt was evaluated after treatment of energized mitochondria with increasing amount of Aβ peptides as indicated, followed by incubation with the potential-dependent fluorescent dye TMRE. After removal of excess TMRE, fluorescence was determined. Mean values and SD were determined from three independent experiments. (B) After treatment of isolated and energized mitochondria with Aβ peptides (3.5 μM), structure and composition of import translocase complexes were analyzed by BN–PAGE, SDS–PAGE, and Western blotting. Before loading, mitochondria were solubilized in a buffer containing 1% digitonin. Immunodecorations were performed against components of the translocase complexes TOM and TIM23—responsible for the import of presequence-containing preproteins through the mitochondrial membranes— Tom20, Tom40, and Tim23 (lanes 1–6 and 9–14), and Aβ peptides (lanes 7, 8, 15, and 16).
Aβ peptides affect the initial steps of the mitochondrial import reaction
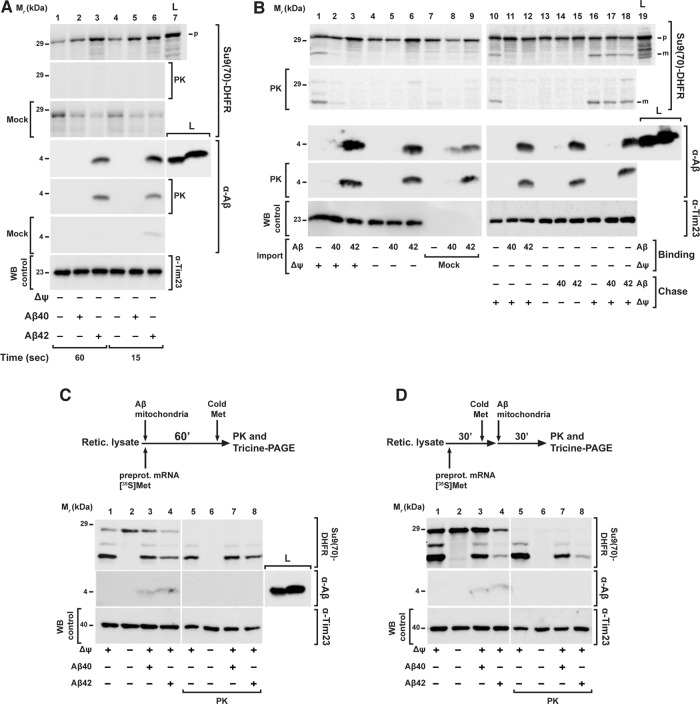
On the basis of the observation of a significant inhibition of the overall import process, we set out to identify the particular step of the import reaction that was affected by Aβ peptides. Most cases of precursor protein import can be generally distinguished into three steps: 1) binding to the receptors of the import machinery of the outer mitochondrial membrane (OMM); 2) Δψmt-dependent transport through the membranes via the translocase complexes; and 3) processing of the precursor to the mature form. To investigate the effect of Aβ peptides on the initial step of the import reaction, we dissipated Δψmt as an import driving force, allowing only binding of precursor proteins to OMM import receptors and/or insertion into the TOM translocase channel. Because the OMM binding reaction is very quick, we incubated the isolated mitochondria with the radioactive precursor protein for short times (range of seconds) in the presence of Aβ peptides and tested for a cofractionation of the precursor polypeptides with the mitochondria. Neither Aβ peptide negatively affected the binding of the precursor protein [35S]Su9(86)‑DHFR (Figure 3A), indicating that the interaction with the mitochondrial surface receptors was not influenced. On the other hand, in particular with Aβ42, we consistently observed elevated amounts of precursor protein associated with mitochondria that were proportional to the amount of peptide used (Supplemental Figure S1C). Because nonspecific radioactive protein bands generated during in vitro translation in addition to the genuine precursor band were also found in association with the mitochondrial pellet after centrifugation, the increase in signal intensity of the precursor protein is probably due to an aggregation phenomenon (see later discussion).
FIGURE 3:
Mitochondrial import steps affected by Aβ peptides. (A) Binding of the precursor protein to the OMM import machinery receptors. After removing the Δψmt, mitochondria were incubated for short times (range of seconds) with Aβ peptides (3.5 μM) and precursor protein [35S]Su9(70)DHFR. Half of the samples were incubated with PK (50 μg/ml) to digest nonimported precursor protein. (B) Separation of preprotein binding (Binding) to OMM from inner membrane translocation and processing steps (Chase). For precursor binding and insertion into the OMM, Δψmt was dissipated by CCCP (1 μM) during incubation with [35S]Su9(70)DHFR in the presence (lanes 11 and 12) and absence (lanes 10 and 13–18) of Aβ peptides. To assay inner membrane translocation and processing (Chase), Δψmt was restored by addition of BSA (2 mg/ml; lanes 10–12 and 16–18) in the presence (lanes 17 and 18) and absence of Aβ peptides. For comparison, a complete one-step import reaction was performed (lanes 1–9). (C, D) Effect of Aβ peptides on cotranslational and posttranslational import. (C) As shown in the scheme of the experimental setup, cotranslational import was performed by incubating rabbit reticulocyte lysate, Su9(70)DHFR mRNA, [35S]methionine, and isolated energized mitochondria in the presence or absence of Aβ peptides (3.5 μM) as indicated at 30°C for 60 min. (D) In posttranslational import, first the translation of [35S]Su9(70)DHFR was performed using rabbit reticulocyte lysate, Su9(70)DHFR mRNA, and [35S]methionine for 30 min, and then isolated mitochondria were added in the presence of Aβ peptides (3.5 μM) for an additional 30 min to perform the mitochondrial import reaction. The translation was stopped by adding 8 mM cold methionine. All samples were analyzed by tricine-SDS–PAGE, followed by Western blot, digital autoradiography, and immunodecoration against Aβ peptides and Tim23. L, total amount of Aβ peptides added; m, mitochondrial mature form; Mock, control experiment in the absence of mitochondria; p, mitochondrial precursor protein; WB, Western blot.
Effects on the transport and processing reactions were tested by a two-step protocol that separated the binding of the precursor from the actual translocation process. The precursor protein [35S]Su9(70)‑DHFR was first incubated with mitochondria, where the Δψmt was dissipated by the addition of carbonyl cyanide m-chlorophenyl hydrazone (CCCP; 1 μM). In this way, the precursor protein was able to bind to the TOM machinery without being imported. After removing excess unbound precursor proteins, Δψmt was restored by taking away the CCCP by binding it to excess amounts of bovine serum albumin (BSA) and reenergizing the mitochondria, allowing the translocation and processing reaction to proceed. Of interest, an inhibition of protein import was observed only when Aβ peptides were present already in the first step of the experiment (Figure 3B, lanes 11 and 12), while adding the peptides directly in the second step, after the binding step was completed, did not show any effect on the import reaction (Figure 3B, lanes 17 and 18). This directly demonstrated that Aβ peptides did not negatively affect the later phases of the import reaction but instead interfered with the first steps of the import reaction that happen at the outer face of the OMM.
In the in vitro system used, the translation reaction to produce radiolabeled preproteins is separated from the actual import reaction, essentially resulting in a posttranslational translocation process. However, in cells, the mitochondrial import most likely represents a mixture of posttranslational and cotranslational reactions, depending on the individual properties of preproteins or even their mRNAs (Fox, 2012). To clarify whether Aβ peptides were able to inhibit mitochondrial import also during a cotranslational mechanism, we performed an import reaction in the reticulocyte lysate system used for producing the 35S-labeled preprotein Su9(70)‑DHFR. We incubated the reticulocyte labeling mix containing ribosomes, energized mitochondria, preprotein mRNA, and [35S]methionine for 60 min in the presence or absence of Aβ40 peptides (Figure 3C). After analyzing the samples by tricine-SDS–PAGE, we did not observe an import inhibition in the presence of Aβ40 but still a significant reduction of the formation of the mature form in the presence of Aβ42, although not as pronounced as in the posttranslational situation. As posttranslational control, we used the same experimental setup but first performed the translation reaction without mitochondria for 30 min, stopped the labeling by addition of nonlabeled (“cold”) methionine, and only then added the isolated energized mitochondria in the presence or absence of Aβ peptides (Figure 3D). In this case, we observed partial inhibition of mitochondrial import in the presence of Aβ40, whereas Aβ42 gave a strong inhibitory effect. The less severe inhibitory effect in the case of forced cotranslational import supports our conclusion that Aβ peptides do not directly affect mitochondria function but instead act at an earlier step of the import process. Of note, in a control experiment without mitochondria, we observed that Aβ peptides did not affect ribosomal translation rates (Supplemental Figure S1D).
Interaction of Aβ peptides with human mitochondria
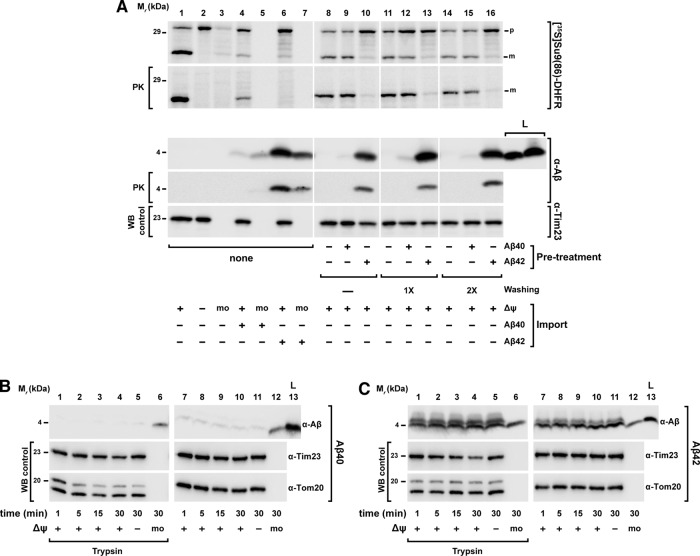
Because our experiments indicated the possibility of a direct association of Aβ peptides, in particular Aβ42, with mitochondria, despite any obvious deleterious effects on mitochondrial functions, we set out to analyze the inhibition properties on preprotein import in more detail. Because the degree of import inhibition seemed to correlate with the amount of Aβ peptides copurified with mitochondria, we checked whether Aβ peptides could also act in the absence of precursor proteins. We pretreated isolated mitochondria with Aβ peptides for 30 min, followed by several washing steps to remove excess unbound material. Then we performed a normal import reaction using the precursor protein [35S]Su9(86)‑DHFR without peptide addition (Figure 4A). Of interest, pretreatment of mitochondria with Aβ40 did not show any significant copurification of Aβ40 with the mitochondria and also did not affect a later import reaction (Figure 4A, lanes 9, 12 and 15). On the contrary, pretreatment of mitochondria with Aβ42 showed a strong, although not complete, inhibitory effect on the subsequent import reaction. Further, we were able to detect Aβ42 copurifying with the mitochondria even after extensive washing, confirming an association with mitochondria (Figure 4A, lanes 10, 13, and 16).
FIGURE 4:
Analysis of Aβ peptides interaction with human mitochondria. (A) Pretreatment of mitochondria with Aβ peptides. Isolated mitochondria were incubated with Aβ peptides (3.5 μM) at 30°C for 30 min. After several washing steps, mitochondria were reisolated and incubated in an energizing buffer with precursor protein [35S]Su9(86)DHFR for an import reaction in the absence of Aβ peptides (lanes 8–16). For comparison, the precursor protein [35S]Su9(86)DHFR was directly incubated with isolated and energized mitochondria and in mock samples (mo) in the presence or absence of Aβ peptides (lanes 1–7). Half of the samples were treated with PK (50 μg/ml) to digest nonimported precursor protein. (B, C) Mitochondrial association of Aβ peptides. Isolated and energized mitochondria and mock (mo) samples (lanes 6 and 12) were incubated with the same amount of Aβ40 (B) and Aβ42 (C) peptides (3.5 μM) for different times. The Δψmt was dissipated where indicated (lanes 5 and 11). Half of the samples were then treated with trypsin (100 μg/ml; lanes 1–6). All samples were processed by tricine-SDS–PAGE, followed by Western blot. As control, immunodecorations against mitochondrial Tom20, Tim23, and Tom40 proteins were performed. m, mature form; p, precursor protein; WB, Western blot.
We therefore investigated more thoroughly the biochemical properties of the association of Aβ peptides with isolated mitochondria. First, we performed a standard mitochondrial import experiment using Aβ peptides to clarify whether they were taken up via the canonical import pathway. The import reaction was analyzed by tricine-SDS–PAGE, followed by Western blot using antiserum against Aβ peptides. As shown in Figure 4B, the smaller peptide, Aβ40, again did not show significant copurification with mitochondria even at longer incubation times. In contrast, with Aβ42, as seen before, a band of 4 kDa was visible in the samples containing mitochondria already at very short times (Figure 4C). The band intensity only slightly increased with longer incubation times. Owing to the small size and specific properties of the Aβ peptides, any processing event during a potential import reaction was not expected. However, for Aβ42, an additional band with a slightly higher molecular weight appeared in the presence of mitochondria, which is likely due to a different running behavior of the small peptide in the presence of large amounts of mitochondrial proteins or lipids. However, three observations argue strongly against a specific uptake of Aβ peptides via the mitochondrial import machinery: 1) there was no time dependence of the mitochondria-associated signals (e.g., Figure 4C, lanes 7–10), 2) the intensity of the copurifying Aβ signal was not influenced by Δψmt (Figure 4C, lane 11), and 3) both Aβ peptides showed a comparable signal also in the mock sample containing no mitochondria at all (Figure 4, B and C, lanes 6 and 12). Of interest, both the copurifying materials, as well as the peptides in the mock samples, were largely resistant to protease digestion (Figure 4, B and C, lane 6).
Because protection against proteases is a major hallmark of a successful mitochondrial import reaction (Ryan et al., 2001), we characterized the protease digestion behavior of Aβ peptides in more detail (Figure 5A). We incubated the Aβ peptides with isolated and energized mitochondria, followed by solubilization with 0.5% Triton X-100 (Figure 5A, lanes 5–8) or ultrasonication (Figure 5A, lanes 9–12). Under these conditions, the mitochondrial membranes are disrupted and would not be able to offer protection against external proteases. A titration with increasing amounts of trypsin was performed, and then all the samples underwent trichloroacetic acid (TCA) precipitation, tricine-SDS–PAGE, and detection of present Aβ peptides by Western blot. As shown in the control panels, lysis of mitochondria by both detergent and sonication was successful, as endogenous control proteins were efficiently degraded even at the lowest concentration of trypsin (5 μg/ml). In the mock samples, without mitochondria and used as control, we again found a significant protease resistance of both Aβ peptides (Figure 5A, lanes 1–4). The protease resistance of both Aβ peptides decreased in the presence of detergent or after sonication (Figure 5A, lanes 6–8 and 10–12). Aβ42 was found to be slightly more resistant than Aβ40 after detergent lysis but remained resistant to trypsin after ultrasound treatment. In the presence of mitochondria, the behavior of the two peptides was different. Because Aβ40 did not copurify or pellet with mitochondria, the analysis of Aβ40 susceptibility to protease digestion was not possible. In contrast, Aβ42 showed some copurification with the mitochondria and also a complete protease resistance that was neither affected by the presence of detergent nor by sonication. This intrinsic protease resistance and the detection of Aβ peptides in samples without mitochondria (mock) or after destruction of mitochondrial membranes suggest that in our experimental setup, Aβ peptides are more prone to form sedimentable aggregated material than to associate with the OMM. We also investigated whether the intrinsic resistance of Aβ peptides to protease digestion was specific to trypsin or could be extended to other proteases. We incubated equal amounts of Aβ peptides alone or with isolated mitochondria for 30 min at 30°C and subsequently treated all samples with the protease trypsin or PK for different times, followed by tricine-SDS–PAGE and Western blot (Figure 5B). As observed earlier, Aβ40 did not copurify with the mitochondria. However, in mock samples, Aβ40 was partially resistant to both proteases even after longer treatments. Aβ42 partially associated with mitochondria but was not completely digested by either protease (Figure 5B, lanes 5–12) in mitochondria as well as in mock samples, indicating that both Aβ peptides retain an intrinsic protease resistance that is independent of a mitochondrial association.
FIGURE 5:
Membrane interaction behavior of Aβ peptides. (A) Absent protease protection by mitochondria. Aβ peptides (3.5 μM) were incubated with or without (Mock) intact and energized mitochondria, followed by digestion with increasing amounts of trypsin (lanes 1–4). As controls, mitochondria were lysed by solubilization with 0.5% Triton X-100 (lanes 5–8) or by sonication (lanes 9–12) before addition of the protease. All samples underwent TCA precipitation. (B) Intrinsic protease resistance. Aβ peptides (3.5 μM) were incubated with or without (Mock) intact and energized mitochondria, followed by digestion with 100 μg/ml trypsin (lanes 5–8) and 100 μg/ml PK for different times as indicated. (C) Dependence of the interaction between Aβ peptides and isolated mitochondria on peripheral OMM receptors. Isolated mitochondria were pretreated with the indicated trypsin concentrations to digest exposed OMM proteins. After trypsin inactivation, isolated mitochondria were reisolated and incubated in an energized buffer with Aβ peptides (3.5 μM). (D) Alkaline extraction of Aβ peptides from mitochondria and mock samples. Aβ peptides (3.5 μM) were incubated in the presence or absence (Mock) of isolated and energized mitochondria. After reisolation, mitochondria and mock samples were subjected to alkaline extraction as described in Material and Methods. (E, F) Titration of Aβ peptide binding to mitochondria. Increasing concentrations of Aβ40 (E) and Aβ42 (F) peptides were incubated for 30 min in the presence or absence (Mock) of energized mitochondria and separated in insoluble (Pellet) and soluble (Supernatant) fractions. All samples were analyzed by tricine-SDS–PAGE and Western blot. As control, immunodecoration against the endogenous mitochondrial proteins SMAC (intermembrane space), MPP (matrix) and Tom40 (OMM) was carried out. L, loading control; P, pellet; S, supernatant; T, total; WB, Western blot.
The import of nuclear-encoded precursor proteins initially requires a specific interaction with receptor proteins at the surface of the OMM (Endo and Kohda, 2002). To analyze whether the interaction of Aβ peptides with mitochondria depends on the involvement of the OMM receptors, we pretreated isolated intact mitochondria with trypsin to digest any protein domains exposed on the cytosolic face of the outer membrane. Then we incubated the mitochondria with Aβ peptides (Figure 5C). We analyzed samples by tricine-SDS–PAGE, followed by Western blot. As control, Tom20 was degraded at the lowest trypsin concentration (5 μg/ml), whereas the inner membrane protein Tim23 was stable during both protease treatments, indicating the intactness of mitochondria. The amount of Aβ42 copurified with mitochondria did not show any difference between trypsin-pretreated mitochondria and untreated control samples, indicating that any potential interaction of Aβ42 with mitochondria is not based on a specific binding to the import-related receptor proteins of the TOM complex.
The previous experiments suggest that the association of Aβ peptides with mitochondria instead represents a nonspecific interaction with the OMM. We performed an alkaline extraction to assess the membrane interaction properties after incubating Aβ peptides with mitochondria (Figure 5D). During alkaline extraction, polypeptides that stably associate with membranes remain in the pellet fraction (P), whereas peripheral membrane proteins are found in the supernatant (S). As shown earlier, Aβ40 did not show a significant signal in the presence of mitochondria. However, the mock samples showed that minor amounts of Aβ40 accumulated in the pellet fraction, consistent with the generation of small amounts of protein aggregates. The Aβ42 peptides showed similar behavior in the mock samples. However, in the presence of mitochondria, a significant amount of copurified material was found in the supernatant fraction, excluding integration into the OMM and suggesting at most a peripheral association. The mitochondrial control proteins MPP (soluble) and Tom40 (membrane integrated) behaved as expected. A nonspecific interaction with the OMM, in particular for Aβ42, was also supported by a saturation titration experiment (Figure 5, E and F). Here we incubated increasing amounts of Aβ peptides with a constant amount of mitochondria and separated soluble and insoluble material by intermediate-speed centrifugation. With increased peptide concentration, most of the Aβ40 peptide remained in the supernatant, and only a minor amount appeared in the pellet fraction (Figure 5E) without being influenced by the presence of mitochondria. On the other hand, significant amounts of Aβ42 peptides accumulated in the pellet fraction both in the presence and absence of mitochondria (Figure 5F). In both cases, the amount of Aβ42 peptides recovered in the pellet fractions did not seem to be saturable, indicating again nonspecific mitochondrial association and a pronounced tendency to form sedimentable aggregate material.
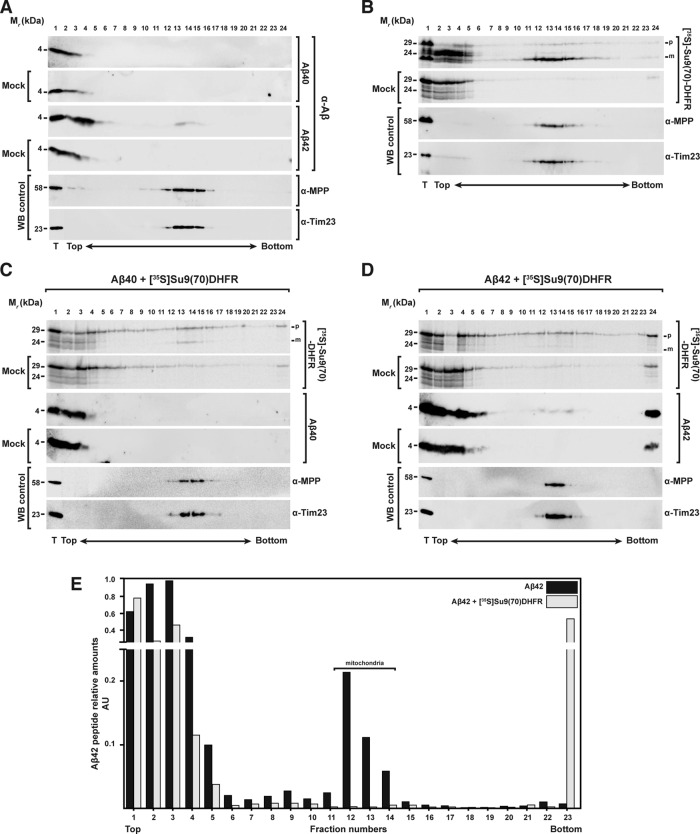
From the foregoing results, it was not possible to clearly distinguish between Aβ peptides associated with the OMM and Aβ peptides prone to aggregation that are able to sediment with mitochondria by conventional differential centrifugation methods used in a standard import assay. Thus we decided to analyze the behavior of Aβ peptides during the mitochondrial import using rate-zonal density gradient centrifugation. After performing an import reaction with the precursor protein [35S]Su9(70)‑DHFR in the presence or absence of Aβ peptides, we separated samples by centrifugation through a sucrose gradient (20–50%). Fractions from top to bottom were collected and analyzed by Western blot or autoradiography for the presence of the imported precursor protein or Aβ peptides (Figure 6). As controls, we carried out the same experiment in the absence of mitochondria (mock) or in the absence of Aβ peptides (Figure 6B). From the sedimentation behavior of mitochondrial marker MPP and Tim23, isolated mitochondria were concentrated mostly around the middle of the gradient (Figure 6E, fractions 12–14). Most of Aβ40 accumulated as monomer or as small, low-density, and SDS-soluble aggregates at the top of the gradient, and no cosedimentation with the mitochondria was observed (Figure 6A, top). This observation is consistent with the behavior in our differential centrifugation experiments (see earlier discussion). However, Aβ42 behaved significantly differently (Figure 6A, middle). In the presence of isolated mitochondria, a small percentage of Aβ42 (20% of the total Aβ42 added to the experiment; Figure 6E) was found in the gradient fractions together with the mitochondrial markers, suggesting direct interaction with mitochondria. In the mock samples, most of the Aβ42 accumulated on the top of the gradient as for Aβ40. In the control samples containing only the precursor protein, [35S]Su9(70)‑DHFR showed localization of the mature form (m) in the same fractions as the bulk mitochondria (Figure 6B). As expected, in the presence of Aβ40, the amount of mature form was partially reduced (Figure 6C), whereas Aβ42 treatment resulted in complete disappearance of the mature form, demonstrating again complete inhibition of mitochondrial import (Figure 6D). Of interest, the presence of precursor proteins changed the behavior of Aβ42, as the amount of mitochondria-associated material decreased while the amount in the bottom fractions, representing aggregates, increased (Figure 6, D and E). In addition, in the presence of Aβ42, a considerable amount of the precursor protein was found in the aggregate fraction at the bottom of the gradient, indicating the formation of coaggregates between Aβ peptides and mitochondrial precursor proteins (Figure 6, D, lane 23, and E).
FIGURE 6:
Analysis of the interaction between Aβ peptides and mitochondrial precursor proteins with mitochondria through density gradient centrifugation. (A) Sucrose gradient centrifugation of 3.5 μM Aβ40 (top) and Aβ42 (bottom) incubated with and without (Mock) isolated and energized mitochondria. (B) As control, a sucrose gradient of precursor protein [35S]Su9(70)DHFR incubated with or without (Mock) isolated and energized mitochondria in the absence of Aβ peptides was performed. (C, D) Sucrose gradients with or without (Mock) mitochondria incubated with precursor protein [35S]Su9(70)DHFR in the presence of Aβ40 (C) or Aβ42 (D). Density gradient fractionations were performed as reported in Materials and Methods. Samples were analyzed by tricine-SDS–PAGE and Western blot. As control, immunodecorations against MPP and Tim23 were used. (E) Quantification of the Aβ42 band intensities incubated with mitochondria in the absence (A) or presence (D) of precursor protein [35S]Su9(70)DHFR. Each value is the ratio between the intensity of the Aβ42 band in each fraction and the total sample (T). m, mature form of the preprotein; p, precursor form; WB, Western blot.
Taken together, these data do not support previous conclusions about uptake of Aβ peptides into the organelle, as the typical criteria for mitochondrial import were not fulfilled: specificity for mitochondria, dependence on surface import receptors and Δψmt, and acquisition of protease resistance. However, we observed some degree of nonspecific association with the mitochondrial surface in the case of Aβ42. This peptide also exhibited a strong tendency to form aggregates, independently of the presence of mitochondria. Of interest, in the presence of mitochondrial precursor proteins, the association of Aβ42 with the mitochondria was reduced, whereas at the same time, increased formation of sedimentable preprotein–Aβ42 conglomerates was observed.
Preprotein import competence is reduced by the formation of Aβ–preprotein coaggregates
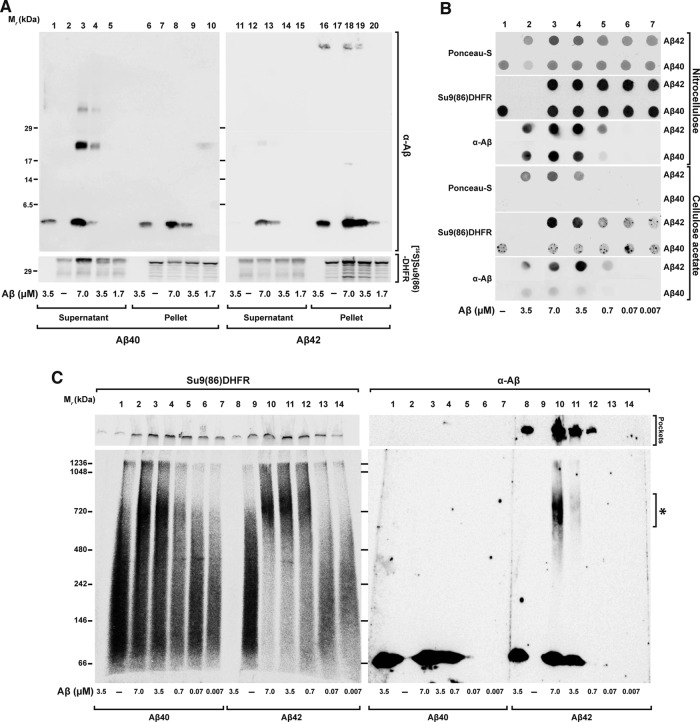
Because aggregate formation is an intrinsic pathological property of Aβ peptides (Thal et al., 2015), we reasoned that a reduction of preprotein solubility by aggregation in the presence of Aβ peptides might contribute to the inhibitory effect on the import reaction. We therefore analyzed a coaggregation by three types of assays: 1) high-speed centrifugation followed by tricine-SDS–PAGE, 2) a filter retardation assay, and 3) BN–PAGE. These techniques provide direct information about the aggregation behavior of precursor polypeptides in the presence of the Aβ peptides and partially characterize the nature of the aggregates. After incubation of radiolabeled precursor proteins with Aβ peptides, samples were centrifuged at high speed (45,000 rpm; 124,500 × g) to separate the insoluble high–molecular weight aggregates from the soluble proteins. The resulting pellets and supernatants were analyzed by Western blot and immunodecoration against Aβ peptides, as well as by autoradiography to detect the precursor polypeptides (Figure 7A). The precursor protein alone partially fractionated to the pellet, suggesting an intrinsic aggregation propensity (Figure 7A, lanes 7 and 17). However, in the presence of increasing concentrations of Aβ42, the amount of [35S]Su9(86)‑DHFR found in the pellet was significantly increased (Figure 7A, lanes 18–20). In contrast, Aβ40 had less severe effects on the distribution of precursor polypeptides in the centrifugation assay (Figure 7A, lanes 8–10), where most precursor protein remained soluble in the supernatant (Figure 7A, lanes 3–5). Aβ42 itself was mostly found in the pellet fraction, suggesting a strong propensity to form insoluble aggregates (Figure 7A, lanes 16 and 18–20). In the pellet fraction, but not in the supernatant, an additional band was detected for Aβ42 at the top part of the tricine gel, corresponding to the loading pockets. This suggested that Aβ42 formed high–molecular weight aggregates that were insensitive to SDS solubilization. For Aβ40, part of the peptides sedimented as insoluble aggregates (Figure 7A, lanes 6 and 8–10), and part remained soluble in the supernatant (Figure 7A, lanes 1 and 3–5). In the supernatant fraction, Aβ40 showed two bands around 20 and 35 kDa in addition to the predominant band at 4 kDa (Figure 7A, lanes 3 and 4). These bands were present only when Aβ40 was incubated with the precursor proteins but not with the peptides alone. Similar bands were also detected with Aβ42 but in much lower amounts (Figure 7A, lanes 12 and 13).
FIGURE 7:
Coaggregation between Aβ peptides and mitochondrial precursor protein. Precursor protein [35S]Su9(86)-DHFR was incubated for 30 min at 30°C in import buffer in the presence or absence of the indicated amounts of Aβ peptides. After incubation, samples were analyzed by the following techniques, (A) Tricine-SDS–PAGE. Soluble fractions (Supernatant) were separated from the insoluble (Pellet) by centrifugation for 40 min at 123,000 × g at 4°C. Samples were analyzed by tricine-SDS–PAGE. (B) Filter retardation assay. Samples were filtered directly through cellulose acetate and nitrocellulose membranes using a dot blot filtration unit as described in Material and Methods. Proteins bound to both membranes were stained with Ponceau S. Bound Aβ peptides were detected by immunodecoration and the precursor protein by digital autoradiography. (C) BN–PAGE. Samples were loaded on native PAGE as described in Materials and Methods and analyzed by Western blot. The precursor protein signal was detected by digital autoradiography and the Aβ peptides by immunodecoration.
In the filter retardation assay, different amounts of Aβ peptides were incubated with the [35S]Su9(86)‑DHFR (Figure 7B) or [35S]OTC (Supplemental Figure S3) and subsequently filtered through nitrocellulose or cellulose acetate membranes. With the cellulose acetate membrane, which does not have an intrinsic protein-binding affinity, inclusions or aggregates >0.2 μm are trapped under these conditions, whereas smaller complexes pass through and are washed away (Heiser et al., 2000). Because most of the added protein should be retained on a nitrocellulose membrane, this type of membrane was used as loading control. Precursor proteins were detected by autoradiography and the presence of Aβ peptides by immunodecoration. The total amount of retained polypeptides was also evaluated by Ponceau red staining of the membranes. As expected from their intrinsic aggregation propensities, Aβ42, but not Aβ40, showed a signal on cellulose acetate membranes when similar concentrations were loaded (Figure 7B). Whereas the precursor protein [35S]Su9(86)‑DHFR alone showed a weak signal on cellulose acetate membrane, a strong signal was detected when it was incubated together with Aβ42 (Figure 7B). The formation of the precursor protein aggregates increased with the amount of Aβ42 peptides added. [35S]OTC showed similar behavior (Supplemental Figure S3).
We also applied the samples on BN–PAGE to characterize the complex formation between Aβ peptides and precursor proteins under native condition. After incubation of the [35S]Su9(86)‑DHFR with different concentrations of Aβ peptides, the complete samples were separated by BN–PAGE gradient gel (5–16.5%) and then analyzed by Western blot and autoradiography. The precursor protein [35S]Su9(86)‑DHFR alone distributed over a large size range without forming a defined band, typical behavior for a soluble protein in native PAGE (Figure 7C, lanes 2 and 9). In the presence of Aβ40, some of the precursor proteins shifted to a higher–molecular weight zone of the gel in a concentration-dependent manner (Figure 7C, lanes 3–7). In the presence of Aβ42, the signals of the precursor protein almost exclusively shifted to an area around 720 kDa (Figure 7C, lanes 10–13). Of interest, immunodecoration with anti-Aβ serum showed that some Aβ42 material accumulated at the same molecular weight range (Figure 7C, lanes 10 and 11). In addition, Aβ42 also exhibited a signal at the highest part of the membrane, related to the loading pockets in the gel, representing large, insoluble aggregate material (Figure 7C lanes 8 and 10–12). The fact that in native conditions, the precursor protein band together with Aβ42 band shifted to the same area strongly suggests a direct interaction between the precursor protein and Aβ42. The large size of the complex, comprising multiple copies of both molecules, was consistent with the formation of Aβ42–preprotein coaggregates.
Taken together, the data obtained from three different technical approaches clearly confirmed a coaggregation phenomenon between the precursor proteins and Aβ peptides that reduced precursor protein solubility. Because solubility of the precursor proteins is required for efficient mitochondrial import, formation of coaggregates between precursor proteins and Aβ peptides interferes with insertion of precursor protein inside the TOM channel. This represents the initial step of an import reaction that was found to be defective in our experiments in the presence of Aβ peptides. Of note, the two Aβ peptides analyzed showed different effects on coaggregate formation, correlating well with the observed preprotein inhibition efficiency, their aggregation propensity, and also the pathological effect in AD patients.
DISCUSSION
Pathological properties of intracellular Aβ peptides, in particular in correlation with mitochondrial dysfunction, have been observed on many occasions in the context of AD. Aβ peptides 1) localize to mitochondria from postmortem AD brains and several experimental models of the disease (Pagani and Eckert, 2011), 2) physically interact with some mitochondrial components (Lustbader et al., 2004), and 3) exert harmful effects on mitochondrial function (Kaminsky et al., 2015). Because in situ production of Aβ peptides in mitochondria seems unlikely (Sannerud and Annaert, 2009), our study addressed the possible mechanisms of Aβ peptide interaction with mitochondria, as well as the correlation between a mitochondrial localization of Aβ peptides and the mitochondrial dysfunctions observed in AD.
Under in vitro conditions, we observed a clear-cut and strong inhibitory effect of Aβ peptides on mitochondrial import. The inhibitory effect of Aβ42 was significantly stronger than that of the related Aβ40, correlating well with the stronger pathogenic effect of Aβ42 in human AD patients (Eckman and Eckman, 2007). Of note, the Aβ42 concentration that resulted in a significant inhibition of mitochondrial import was comparable to the concentration of the peptide that was previously found in AD brains, 2 μM for Aβ42 (Roher et al., 2009). Our experiments also shed a light on the biochemical details of the inhibitory mechanism, in particular on the stage of the import process that was affected. The inhibitory effect occurred immediately and did not require a prolonged preincubation period. Although previous work reported that treatment of mitochondria with Aβ peptides resulted in a reduction of Δψmt (Kaminsky et al., 2015), in our model system, we did not observe any changes in Δψmt in the time frame of the import experiments, excluding an Aβ‑related reduction of the membrane potential as a cause for the import inhibition. Neither did we observe changes in the size and composition of the precursor protein translocase complexes (TOM and TIM) nor of the metabolic complexes of the respiratory chain. The possibility of direct physical damage on mitochondrial membranes, the oxidative phosphorylation system, or the preprotein import machinery by Aβ peptides is therefore very unlikely.
Most of the mitochondrial proteins are synthesized at the cytosolic ribosomes and then imported inside the mitochondria. Concerning the cellular environment, nascent mitochondrial precursor polypeptides may associate with the import machinery while still being synthesized on the ribosome (cotranslational import) or may be first released from the ribosome after translation is completed and only then interact with the mitochondria (posttranslational import). Most likely, also depending on the individual properties of the preproteins, the in vivo situation is represented by a mixture of the two processes (Verner, 1993; Mukhopadhyay et al., 2004). As discussed earlier, in particular Aβ42 showed a strong inhibitory effect in standard posttranslational import experiments. After performing import in a cotranslational manner, we found that the mitochondrial import was still inhibited by Aβ peptides, although with less efficiency than under posttranslational conditions. We conclude that the import-inhibitory effect of Aβ peptides mainly affects a very early step of the process when newly synthesized mitochondrial polypeptides are exposed to the cytosolic environment.
To date, scarce information has been available about direct effects of Aβ peptides on mitochondrial protein biogenesis. Using flow cytometry, it was demonstrated that after long-term exposure to Aβ peptides, differentiated PC12 cells exhibited a reduction of newly synthesized, mitochondrially targeted green fluorescent protein (Sirk et al., 2007). These results are generally in line with our observations. Owing to the long exposure to potentially toxic molecules, however, these experiments could not distinguish whether the import inhibition was a direct or an indirect consequence of the presence of Aβ peptides. The immediate inhibitory effect of Aβ peptides on the import reaction in healthy mitochondria, as observed in our experiments, essentially rules out that the inhibition was caused indirectly by long-term accumulation of functional defects in the affected mitochondria. One previous study also used isolated mitochondria pretreated for a short time with Aβ peptides but did not detect a deficiency of mitochondrial import (Hansson Petersen et al., 2008). However, the discrepancy can be explained by the use of an insufficient amount of Aβ peptides in that study to observe significant import inhibition. Of interest, inhibition of mitochondrial protein biogenesis was suggested as a potential cause for Huntington’s disease (Yano et al., 2014). It was observed that a mutant form of the protein huntingtin partially inhibited mitochondrial import via physical association with the TIM23 translocase complex. The concentration of huntingtin sufficient to obtain inhibition was comparable to the Aβ peptide concentration used in our model.
A recent study proposed that Aβ peptides indirectly interfered with the processing of imported precursor proteins to the mature and active forms (Mossmann et al., 2014), which is an important late step of the mitochondrial import reaction. The authors found that inhibition of PreP (or its yeast homologue, Cym1) by Aβ peptides (Alikhani et al., 2011) resulted in the accumulation of prepeptides in the mitochondrial matrix, which in turn interfered with the activity of the processing peptidase MPP, which is required for the maturation of mitochondrial precursor proteins. This is in strong contrast to our study, which showed that Aβ peptides acted at an early step of the import reaction. Two observations from our study directly argue against a mitochondrial processing defect caused by Aβ peptides. First, the precursor form visible in import experiments after Aβ peptide inhibition was always sensitive to digestion by external proteases, indicating that the preproteins never crossed the mitochondrial membrane. Second, using two-step import experiments, which separated the binding from the translocation and processing reaction, we observed an inhibitory effect of Aβ peptides only in the first step, which is independent of the membrane potential, but not in the second translocation step into the matrix, which would comprise the processing reaction. Although Mossmann et al. (2014) found an impaired precursor protein processing activity in the presence of Aβ peptides using soluble mitochondrial extracts from yeast, as well as in total brain extracts from a murine AD model, the relevance of the claimed processing inhibition for the in vivo situation is questionable. Of interest, they also observed minor accumulation of precursor polypeptides after cellular expression of Aβ in intact yeast cells and also in brain extracts from AD patients. Given that cytosolic accumulation of unprocessed precursor forms is the typical hallmark of a defective overall import process, this observation is consistent with our results of a direct inhibitory effect of Aβ peptides on preprotein import but not processing.
Although a previous experiment indicated specific and complete import of Aβ peptides into mitochondria (Hansson Petersen et al., 2008), we revisited this question by analyzing the biochemical properties of the interaction of Aβ peptides with isolated and energized mitochondria. Also in our experiments, Aβ42 exhibited some cosedimentation with mitochondria during the differential centrifugation procedure typically used to reisolate mitochondria after an import experiment. In addition, Aβ42, but also Aβ40, showed some degree of resistance against added proteases, and both observations superficially argue for a successful import reaction. However, our analysis clearly showed that both Aβ peptides were not taken up by mitochondria, because they did not satisfy the required criteria for a mitochondrial import reaction. Of greatest importance, the sedimentation behavior and partial protease resistance of Aβ42 were largely maintained in the absence of mitochondria (mock samples), correlating with its intrinsic tendency to form aggregates. In line with our results are data from the literature showing that both Aβ peptides extracted from AD brains and synthetic Aβ peptides spiked into brain homogenates acquired detergent insolubility and resistance to protease digestion (Soto and Castano, 1996; Xiao et al., 2014). Taken together, these results exclude a complete import of Aβ peptides into mitochondria but not a peripheral association between Aβ peptides, in particular Aβ42, with the OMM. Our experiments indicated that the presence of mitochondria promotes both aggregation propensity and protease resistance of Aβ42 (Murphy, 2007; Henry et al., 2015).
Generally, Aβ peptides have an intrinsic tendency to self-assemble into a range of different aggregates also under the conditions that we applied in our mitochondrial import assay (Snyder et al., 1994; Stine et al., 2003; Thal et al., 2015). Using density gradient centrifugation to separate protein aggregates from cell organelles such as mitochondria (Sehlin et al., 2012), we observed that a fraction of the Aβ42 peptide added to the experiment directly associated with mitochondria. Of interest, the presence of precursor proteins changed the behavior of Aβ42, as the amount of mitochondria-associated material decreased, whereas the aggregated forms increased. In addition, in the presence of Aβ42, a considerable amount of the precursor protein was found in the aggregate fraction, indicating the formation of coaggregates. We propose that this coaggregation of precursor proteins and Aβ peptides is the main reason for the strong inhibitory effect of mitochondrial protein import. Formation of high–molecular weight aggregates and concomitant reduction of the solubility would significantly reduce the import competence of precursor proteins. Several further observations support this coaggregation model. Correlated with the much stronger import-inhibitory effect of Aβ42 compared with Aβ40, the coaggregation phenomenon was particularly pronounced in the presence of Aβ42. The solubility of the precursor proteins was reduced in the presence of Aβ42, as assayed by a centrifugation assay. Together with Aβ42, precursor proteins formed large aggregates that were retarded in a filtration assay. In native PAGE experiments, precursor protein signals were shifted to a high–molecular weight complex in the range of 700 kDa that copurified with Aβ42. Of interest, recent results showing negative consequences of coaggregation between cytosolic enzymes and Aβ peptides support this AD‑specific pathological mechanism (Itakura et al., 2015). Our work therefore adds an important aspect concerning the deleterious consequences of coaggregation processes during the etiology of neurodegenerative diseases. Many amyloid diseases involve coaggregation of different protein species (Penke et al., 2012; Sarell et al., 2013), although the pathological mechanisms are not always entirely clear. It is conceivable that amyloidogenic β-sheet peptides interact with many different endogenous proteins, resulting in their sequestration and functional impairment (Olzscha et al., 2011).
Considering the intracellular space as a crowded environment, Aβ peptides likely undergo multiple, largely nonspecific interactions with any protein and lipid components of the cytosol. The import-competent state of mitochondrial preproteins is represented by an incompletely folded conformation that is prone to irregular interactions with Aβ peptides and subsequent aggregation. Already during the onset of the disease, at a point at which the concentration of Aβ peptides is increasing, the formation of coaggregates with newly synthesized mitochondrial precursor polypeptides might progressively interfere with the import process. This would eventually result in reduction or even loss of mitochondrial enzyme activities, in turn leading to the pleiotropic nature of mitochondrial dysfunction observed in AD patients and respective disease models (Wang et al., 2007; Kaminsky et al., 2015). Hence the observed strong inhibitory effect on mitochondrial protein import, in particular in the case of the pathogenic Aβ42, strongly supports the hypothesis of a direct mitochondrial toxicity of Aβ peptides on mitochondria in AD.
MATERIALS AND METHODS
Preparation of Aβ peptides and mitochondrial treatment
The Escherichia coli–expressed human recombinant Aβ peptides 1-40 (Ultra Pure HFIP; A-1153-2) and 1-42 (Ultra Pure HFIP; A-1163-2) used in this study were purchased from AJ Roboscreen (Leipzig, Germany). Working solutions of both peptides were prepared as described (Stine et al., 2003). Briefly, the lyophilized peptides were dissolved in 100% 1,1,1,3,3,3-hexafluoro-2-propanol (Sigma-Aldrich, Munich, Germany) and distributed in low-binding microcentrifuge tubes (VWR, Darmstadt, Germany). The solvent was allowed to evaporate overnight at room temperature, and the Aβ peptide aliquots were stored at −80°C. Immediately before use, each aliquot was warmed to room temperature, followed by resuspension of the peptide film to a stock of 5 mM in dimethyl sulfoxide (DMSO; AppliChem, Darmstadt, Germany) to remove any preexisting aggregated structures and provide a homogeneous, nonaggregated peptide preparation. After being well mixed, the Aβ peptide DMSO stock was freshly diluted with ice-cold distilled water to a final concentration of 100 μM. This dilution was mixed and used immediately. All experiments with Aβ peptides were performed in super-clear tubes (VWR). In some of experiments, Aβ peptides were precipitated with 72% TCA, followed by tricine-SDS–PAGE, Western blot, and immunodecoration to improve the running behavior of small peptides.
Cell culture and isolation of mitochondria
HeLa cells were cultured in RPMI 1640 medium with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in a saturated humidity atmosphere containing 5% CO2. All chemicals were bought from Thermo Fisher Scientific (Waltham, MA). Mitochondria were isolated from HeLa cells as described (Becker et al., 2012). Briefly, after harvesting and washing in phosphate-buffered saline (PBS), cells were incubated for 40 min on ice with HMS-A buffer (0.22 M mannitol, 0.07 M sucrose, 0.02 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.4, 1 mM EDTA, 0.2% BSA, 1 mM phenylmethylsulfonyl fluoride [PMSF]). Then cells were homogenized with a glass/Teflon homogenizer (B. Braun, Melsungen, Germany), followed by differential centrifugation steps to isolated mitochondria. The mitochondria were washed and resuspended in HMS-B buffer (0.22 M mannitol, 0.07 M sucrose, 0.02 M HEPES, pH 7.4, 1 mM EDTA, 1 mM PMFS).
Import of radiolabeled preproteins into isolated mitochondria
The import of radiolabeled precursor proteins was performed essentially as described (Becker et al., 2012). Radiolabeled preproteins were synthesized by in vitro transcription/translation using the mMESSAGE mMACHINE transcription kit (Life Technologies) and rabbit reticulocyte lysate (Promega, Madison, WI) in the presence of [35S]methionine/cysteine (PerkinElmer, Waltham, MA). For the import reaction, mitochondria were diluted in import buffer (20 mM HEPES-KOH, pH 7.4, 250 mM sucrose, 5 mM magnesium acetate, 80 mM potassium acetate, 5 mM KPi, pH 7.4, 7.5 mM glutamate, 5 mM malate, 1 mM dithiothreitol, 2 mM ATP) to a final concentration of 50 μg/100 μl. In mock samples, Aβ peptides were incubated under the same buffer conditions but without added mitochondria. Where indicated, Δψmt was dissipated by adding a mixture of 8 μM antimycin A (Sigma-Aldrich), 0.5 μM valinomycin, and 2 μM oligomycin (Sigma-Aldrich). All of the import reactions were performed at 30°C and stopped by addition of 50 μM valinomycin and placement of the samples on ice. Nonimported, protease-accessible mitochondrial proteins were digested by incubation with 100 μg/ml trypsin (Biochrom, Berlin, Germany) for 30 min on ice and terminated by adding 800 μg/ml trypsin inhibitor (Sigma-Aldrich) and 1 mM PMFS (Carl Roth, Karlsruhe, Germany). Then mitochondria were washed in import buffer without substrates. Where indicated, samples were treated with 25 μg/ml PK; Carl Roth) on ice for 30 min before the addition of 1 mM PMSF. After centrifugation for 10 min at 12,000 × g and 4°C, mitochondrial pellets were analyzed by tricine-SDS–PAGE, Western blot, digital autoradiography, and immunodecoration.
For two-step import reactions, Δψmt was first depleted with 1 μM CCCP. Mitochondria were incubated with radiolabeled preprotein for 30 min at 30°C. After washing, the mitochondria were reincubated for 30 min at 30°C in energized import buffer supplemented with 2 mg/ml BSA to restore the membrane potential in the presence or absence of 3.5 μM Aβ peptides. After reisolation of mitochondria, imported proteins were separated by tricine-SDS–PAGE and detected by immunodecoration and digital autoradiography.
BN–PAGE
To analyze mitochondrial protein complexes and Aβ peptide aggregation states under native conditions, samples were analyzed by BN–PAGE (Wittig et al., 2006). Isolated mitochondria, as well as coaggregates containing Aβ peptides and radiolabeled preproteins, were solubilized in BN-lysis buffer (20 mM Tris-HCl, pH 7.4, 50 mM NaCl, 10% glycerol, 1 mM EDTA, 1% digitonin, 1 mM PMFS). BN gel loading buffer (100 mM Bis-Tris, pH 7.0, 500 mM ε-amino-n-caproic acid, 5% [wt/vol] Coomassie brilliant blue G250) was added, and the samples were loaded on 5–16.5% BN gels. Native unstained protein standard (Thermo Fisher Scientific) was used to estimate molecular weights of protein complexes. After running overnight, gels were equilibrated in SDS buffer (1% [wt/vol] SDS, 0.19 M glycine, 25 mM Tris) and blotted on polyvinylidene fluoride (PVDF) membrane (Carl Roth), followed by immunodecoration and digital autoradiography.
Sodium carbonate extraction
After incubation of isolated and intact mitochondria with 3.5 μM Aβ peptides, further incubation in 0.1 M Na2CO3 solution (pH 11) was performed on ice for 30 min. Then, after withdrawal of a total sample, an ultracentrifugation step was done in a Beckman TLA-55 at 45,000 rpm (123,000 × g) for 40 min at 4°C. The pellets were resuspended in tricine sample buffer and the supernatants were precipitated with 72% TCA, followed by tricine-SDS–PAGE, Western blot, and immunodecoration.
Sucrose density gradient centrifugation
After incubation with Aβ peptides (35 μM) and/or [35S]Su9(70)‑DHFR, isolated mitochondria and mock samples were loaded on a continuous sucrose gradient (25–50%) and centrifuged in a Beckman SW41 rotor at 33,000 rpm (135,000 × g) for 1 h at 4°C. Then 500-μl fractions were collected from the top of each gradient, followed by 72% TCA precipitation. Protein pellets were resuspended in tricine loading buffer, separated by tricine-SDS–PAGE, and analyzed by Western blot and immunodecoration.
Membrane potential measurement in isolated mitochondria
The Δψmt was analyzed by the potential-sensitive fluorescent dye TMRE (Thermo Fisher). After incubation with Aβ peptides, isolated mitochondria were resuspended in potential buffer (0.6 M sorbitol, 0.1% BSA, 10 mM MgCl2, 20 mM KPi, pH 7.2, 5 mM malate, 10 mM glutamate) and incubated with 1 μM TMRE for 30 min at 30°C on ice. After washing away of excess of TMRE, the TMRE fluorescence was measured in a microplate reader (excitation 540 nm, emission 585 nm; Infinite M200 PRO; TECAN, Männedorf, Switzerland).
Filter retardation assay
To visualize the formation of aggregates and coaggregates, a modified filter retardation assay (Scherzinger et al., 1997) was used. After incubation of radiolabeled precursor proteins with different amounts of Aβ peptides for 30 min at 30°C in energized import buffer, samples were filtered directly through cellulose acetate membrane (0.2-μm pore size; GE Healthcare, Freiburg, Germany) or nitrocellulose membrane (GE Healthcare) using a dot blot filtration unit (SCIE-PLAS, Cambridge, United Kingdom). Proteins retarded on the membranes were analyzed by immunodecoration and digital autoradiography.
Miscellaneous methods
All chemicals using in this study were from Carl Roth or Sigma-Aldrich. Standard techniques were used for tricine-SDS–PAGE, Western blot, and immunodecoration. After performing tricine-SDS–PAGE, samples were transferred on PVDF membrane (Carl Roth) followed by blocking in TBS/Tween (0.9% NaCl, 10 mM Tris/HCl, pH 7.4, 0.25% Tween 20) with 5% milk and immunodecoration with antibodies appropriately diluted in TBS/Tween. Signal detection was performed by enhanced chemiluminescence (Serva Light Eos Ultra; Serva, Heidelberg, Germany). The antibodies used were as follows: Aβ 6E10 (SIG-39320; Covance, Princeton, NJ); Tim23 (611222; BD Bioscience, Heidelberg, Germany), Tom 20 (SC-11415; Santa Cruz Biotechnology, Dallas TX), Tom 40 (SC-11414; Santa Cruz Biotechnology), SMAC (SC-22766; Santa Cruz Biotechnology), MPP (HPA021648; Sigma‑Aldrich), Complex-I (459100; Invitrogen), Complex-II (459200; Thermo Fisher Scientific), Complex III (SC-23986; Santa Cruz Biotechnology), Complex‑IV (3E11; Cell Signaling, Frankfurt, Germany), F1β (A21351; Thermo Fisher Scientific), rabbit immunoglobulin G (IgG)–peroxidase (A6154; Sigma-Aldrich) and mouse IgG–peroxidase (A4416; Sigma-Aldrich). Digital autoradiography was performed using a FLA5100 phosphorimaging system (Fujifilm, Düsseldorf, Germany). Quantitative analysis was done by ImageJ 64 (National Institutes of Health, Bethesda, MD) and GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA).
Supplementary Material
Acknowledgments
We are grateful to U. Gerken for expert technical support and J. Walter from the University of Bonn (Bonn, Germany) for critical comments. This work was supported by a fellowship from the European Commission Framework Programs (Marie Curie Action 7th Framework Program–International Incoming Fellowship to G.C.) and a research grant from the Deutsche Forschungsgemeinschaft (VO657/5-2 to W.V.).
Abbreviations used:
- BN–PAGE
blue-native gel electrophoresis
- DHFR
dihydrofolate reductase
- MPP
mitochondrial processing peptidase
- OMM
outer mitochondrial membrane
- TCA
trichloroacetic acid
- TIM
translocase of the inner membrane
- TMRE
tetramethylrhodamine ethyl ester
- TOM
translocase of the outer membrane.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-05-0313) on September 14, 2016.
REFERENCES
- Agostinho P, Pliassova A, Oliveira CR, Cunha RA. Localization and trafficking of amyloid-beta protein precursor and secretases: impact on Alzheimer’s disease. J Alzheimers Dis. 2015;45:329–347. doi: 10.3233/JAD-142730. [DOI] [PubMed] [Google Scholar]
- Alikhani N, Guo L, Yan S, Du H, Pinho CM, Chen JX, Glaser E, Yan SS. Decreased proteolytic activity of the mitochondrial amyloid-beta degrading enzyme, PreP peptidasome, in Alzheimer’s disease brain mitochondria. J Alzheimers Dis. 2011;27:75–87. doi: 10.3233/JAD-2011-101716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D, Richter J, Tocilescu MA, Przedborski S, Voos W. Pink1 kinase and its membrane potential (Deltapsi)-dependent cleavage product both localize to outer mitochondrial membrane by unique targeting mode. J Biol Chem. 2012;287:22969–22987. doi: 10.1074/jbc.M112.365700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB. Amyloid beta-protein (Abeta) assembly: Abeta 40 and Abeta 42 oligomerize through distinct pathways. Proc Natl Acad Sci USA. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckman CB, Eckman EA. An update on the amyloid hypothesis. Neurol Clin. 2007;25:669–682. doi: 10.1016/j.ncl.2007.03.007. vi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Kohda D. Functions of outer membrane receptors in mitochondrial protein import. Biochim Biophys Acta. 2002;1592:3–14. doi: 10.1016/s0167-4889(02)00259-8. [DOI] [PubMed] [Google Scholar]
- Fox TD. Mitochondrial protein synthesis, import, and assembly. Genetics. 2012;192:1203–1234. doi: 10.1534/genetics.112.141267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouras GK, Tampellini D, Takahashi RH, Capetillo-Zarate E. Intraneuronal beta-amyloid accumulation and synapse pathology in Alzheimer’s disease. Acta Neuropathol. 2010;119:523–541. doi: 10.1007/s00401-010-0679-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Hansson Petersen CA, Alikhani N, Behbahani H, Wiehager B, Pavlov PF, Alafuzoff I, Leinonen V, Ito A, Winblad B, Glaser E, Ankarcrona M. The amyloid beta-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proc Natl Acad Sci USA. 2008;105:13145–13150. doi: 10.1073/pnas.0806192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Head E, Lott IT. Down syndrome and beta-amyloid deposition. Curr Opin Neurol. 2004;17:95–100. doi: 10.1097/00019052-200404000-00003. [DOI] [PubMed] [Google Scholar]
- Heiser V, Scherzinger E, Boeddrich A, Nordhoff E, Lurz R, Schugardt N, Lehrach H, Wanker EE. Inhibition of huntingtin fibrillogenesis by specific antibodies and small molecules: implications for Huntington’s disease therapy. Proc Natl Acad Sci USA. 2000;97:6739–6744. doi: 10.1073/pnas.110138997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry S, Vignaud H, Bobo C, Decossas M, Lambert O, Harte E, Alves ID, Cullin C, Lecomte S. Interaction of Abeta(1-42) amyloids with lipids promotes “off-pathway” oligomerization and membrane damage. Biomacromolecules. 2015;16:944–950. doi: 10.1021/bm501837w. [DOI] [PubMed] [Google Scholar]
- Itakura M, Nakajima H, Kubo T, Semi Y, Kume S, Higashida S, Kaneshige A, Kuwamura M, Harada N, Kita A, et al. Glyceraldehyde-3-phosphate dehydrogenase aggregates accelerate amyloid-beta amyloidogenesis in Alzheimer disease. J Biol Chem. 2015;290:26072–26087. doi: 10.1074/jbc.M115.669291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett JT, Berger EP, Lansbury PT., Jr The C-terminus of the beta protein is critical in amyloidogenesis. Ann NY Acad Sci. 1993;695:144–148. doi: 10.1111/j.1749-6632.1993.tb23043.x. [DOI] [PubMed] [Google Scholar]
- Kaminsky YG, Tikhonova LA, Kosenko EA. Critical analysis of Alzheimer’s amyloid-beta toxicity to mitochondria. Front Biosci. 2015;20:173–197. doi: 10.2741/4304. [DOI] [PubMed] [Google Scholar]
- Kummer MP, Heneka MT. Truncated and modified amyloid-beta species. Alzheimers Res Ther. 2014;6:28. doi: 10.1186/alzrt258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFerla FM, Green KN, Oddo S. Intracellular amyloid-beta in Alzheimer’s disease. Nat Rev Neurosci. 2007;8:499–509. doi: 10.1038/nrn2168. [DOI] [PubMed] [Google Scholar]
- Lustbader JW, Cirilli M, Lin C, Xu HW, Takuma K, Wang N, Caspersen C, Chen X, Pollak S, Chaney M, et al. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer’s disease. Science. 2004;304:448–452. doi: 10.1126/science.1091230. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Gleichmann M, Cheng A. Mitochondria in neuroplasticity and neurological disorders. Neuron. 2008;60:748–766. doi: 10.1016/j.neuron.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawuenyega KG, Sigurdson W, Ovod V, Munsell L, Kasten T, Morris JC, Yarasheski KE, Bateman RJ. Decreased clearance of CNS beta-amyloid in Alzheimer’s disease. Science. 2010;330:1774. doi: 10.1126/science.1197623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossmann D, Vogtle FN, Taskin AA, Teixeira PF, Ring J, Burkhart JM, Burger N, Pinho CM, Tadic J, Loreth D, et al. Amyloid-beta peptide induces mitochondrial dysfunction by inhibition of preprotein maturation. Cell Metab. 2014;20:662–669. doi: 10.1016/j.cmet.2014.07.024. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, Ni L, Weiner H. A co-translational model to explain the in vivo import of proteins into HeLa cell mitochondria. Biochem J. 2004;382:385–392. doi: 10.1042/BJ20040065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RM. Kinetics of amyloid formation and membrane interaction with amyloidogenic proteins. Biochim Biophys Acta. 2007;1768:1923–1934. doi: 10.1016/j.bbamem.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Murphy MP, LeVine H., 3rd Alzheimer’s disease and the amyloid-beta peptide. J Alzheimers Dis. 2010;19:311–323. [Google Scholar]
- Musiek ES, Holtzman DM. Three dimensions of the amyloid hypothesis: time, space and “wingmen.”. Nat Neurosci. 2015;18:800–806. doi: 10.1038/nn.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- Pagani L, Eckert A. Amyloid-beta interaction with mitochondria. Int J Alzheimers Dis. 2011;2011:925050. doi: 10.4061/2011/925050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penke B, Toth AM, Foldi I, Szucs M, Janaky T. Intraneuronal beta-amyloid and its interactions with proteins and subcellular organelles. Electrophoresis. 2012;33:3608–3616. doi: 10.1002/elps.201200297. [DOI] [PubMed] [Google Scholar]
- Piaceri I, Rinnoci V, Bagnoli S, Failli Y, Sorbi S. Mitochondria and Alzheimer’s disease. J Neurol Sci. 2012;322:31–34. doi: 10.1016/j.jns.2012.05.033. [DOI] [PubMed] [Google Scholar]
- Roher AE, Esh CL, Kokjohn TA, Castano EM, Van Vickle GD, Kalback WM, Patton RL, Luehrs DC, Daugs ID, Kuo YM, et al. Amyloid beta peptides in human plasma and tissues and their significance for Alzheimer’s disease. Alzheimers Dement. 2009;5:18–29. doi: 10.1016/j.jalz.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MT, Voos W, Pfanner N. Assaying protein import into mitochondria. Methods Cell Biol. 2001;65:189–215. doi: 10.1016/s0091-679x(01)65012-x. [DOI] [PubMed] [Google Scholar]
- Sannerud R, Annaert W. Trafficking, a key player in regulated intramembrane proteolysis. Semin Cell Dev Biol. 2009;20:183–190. doi: 10.1016/j.semcdb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Sarell CJ, Stockley PG, Radford SE. Assessing the causes and consequences of co-polymerization in amyloid formation. Prion. 2013;7:359–368. doi: 10.4161/pri.26415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates GP, Davies SW, Lehrach H, Wanker EE. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- Sehlin D, Englund H, Simu B, Karlsson M, Ingelsson M, Nikolajeff F, Lannfelt L, Pettersson FE. Large aggregates are the major soluble Abeta species in AD brain fractionated with density gradient ultracentrifugation. PLoS One. 2012;7:e32014. doi: 10.1371/journal.pone.0032014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selfridge JE, Lezi E, Lu J, Swerdlow RH. Role of mitochondrial homeostasis and dynamics in Alzheimer’s disease. Neurobiol Dis. 2013;51:3–12. doi: 10.1016/j.nbd.2011.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirk D, Zhu Z, Wadia JS, Shulyakova N, Phan N, Fong J, Mills LR. Chronic exposure to sub-lethal beta-amyloid (Abeta) inhibits the import of nuclear-encoded proteins to mitochondria in differentiated PC12 cells. J Neurochem. 2007;103:1989–2003. doi: 10.1111/j.1471-4159.2007.04907.x. [DOI] [PubMed] [Google Scholar]
- Snyder SW, Ladror US, Wade WS, Wang GT, Barrett LW, Matayoshi ED, Huffaker HJ, Krafft GA, Holzman TF. Amyloid-beta aggregation: selective inhibition of aggregation in mixtures of amyloid with different chain lengths. Biophys J. 1994;67:1216–1228. doi: 10.1016/S0006-3495(94)80591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C, Castano EM. The conformation of Alzheimer’s beta peptide determines the rate of amyloid formation and its resistance to proteolysis. Biochem J. 1996;314:701–707. doi: 10.1042/bj3140701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine WB, Jr, Dahlgren KN, Krafft GA, LaDu MJ. In vitro characterization of conditions for amyloid-beta peptide oligomerization and fibrillogenesis. J Biol Chem. 2003;278:11612–11622. doi: 10.1074/jbc.M210207200. [DOI] [PubMed] [Google Scholar]
- Swerdlow RH, Burns JM, Khan SM. The Alzheimer’s disease mitochondrial cascade hypothesis: progress and perspectives. Biochim Biophys Acta. 2014;1842:1219–1231. doi: 10.1016/j.bbadis.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thal DR, Walter J, Saido TC, Fandrich M. Neuropathology and biochemistry of Abeta and its aggregates in Alzheimer’s disease. Acta Neuropathol. 2015;129:167–182. doi: 10.1007/s00401-014-1375-y. [DOI] [PubMed] [Google Scholar]
- Truscott KN, Wiedemann N, Rehling P, Muller H, Meisinger C, Pfanner N, Guiard B. Mitochondrial import of the ADP/ATP carrier: the essential TIM complex of the intermembrane space is required for precursor release from the TOM complex. Mol Cell Biol. 2002;22:7780–7789. doi: 10.1128/MCB.22.22.7780-7789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verner K. Co-translational protein import into mitochondria: an alternative view. Trends Biochem Sci. 1993;18:366–371. doi: 10.1016/0968-0004(93)90090-a. [DOI] [PubMed] [Google Scholar]
- Wang X, Su B, Perry G, Smith MA, Zhu X. Insights into amyloid-beta-induced mitochondrial dysfunction in Alzheimer disease. Free Radic Biol Med. 2007;43:1569–1573. doi: 10.1016/j.freeradbiomed.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Weller RO, Massey A, Kuo YM, Roher AE. Cerebral amyloid angiopathy: accumulation of A beta in interstitial fluid drainage pathways in Alzheimer’s disease. Ann NY Acad Sci. 2000;903:110–117. doi: 10.1111/j.1749-6632.2000.tb06356.x. [DOI] [PubMed] [Google Scholar]
- Wirths O, Bayer TA. Intraneuronal Abeta accumulation and neurodegeneration: lessons from transgenic models. Life Sci. 2012;91:1148–1152. doi: 10.1016/j.lfs.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Wirths O, Multhaup G, Bayer TA. A modified beta-amyloid hypothesis: intraneuronal accumulation of the beta-amyloid peptide–the first step of a fatal cascade. J Neurochem. 2004;91:513–520. doi: 10.1111/j.1471-4159.2004.02737.x. [DOI] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schägger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- Xiao X, Yuan J, Qing L, Cali I, Mikol J, Delisle MB, Uro-Coste E, Zeng L, Abouelsaad M, Gazgalis D, et al. Comparative study of prions in iatrogenic and sporadic Creutzfeldt-Jakob disease. J Clin Cell Immunol. 2014;5:240. doi: 10.4172/2155-9899.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano H, Baranov SV, Baranova OV, Kim J, Pan Y, Yablonska S, Carlisle DL, Ferrante RJ, Kim AH, Friedlander RM. Inhibition of mitochondrial protein import by mutant huntingtin. Nat Neurosci. 2014;17:822–831. doi: 10.1038/nn.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Thompson R, Zhang H, Xu H. APP processing in Alzheimer’s disease. Mol Brain. 2011;4:3. doi: 10.1186/1756-6606-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.