Free cholesterol accumulation in the liver is relevant to the pathogenesis of nonalcoholic steatohepatitis. Acute free cholesterol accumulation induced the fusion of LDs, followed by degradation of the coat protein of LDs, perilipin 2, and association of apolipoprotein 100 to LDs in Rab18-dependent manner.
Abstract
Dysregulated hepatic cholesterol homeostasis with free cholesterol accumulation in the liver is relevant to the pathogenesis of nonalcoholic steatohepatitis, contributing to the chronicity of liver toxicity. Here we examined the effect of free cholesterol accumulation on the morphology and biochemical properties of lipid droplets (LDs) in cultured hepatocytes. Acute free cholesterol accumulation induced the fusion of LDs, followed by degradation of the coat protein of LDs, perilipin 2 (PLIN2; also called adipophilin or adipose differentiation–related protein), and association of apolipoprotein B 100 (ApoB 100) to LDs. The degradation of PLIN2 was inhibited by inhibitors of ubiquitination, autophagy, and protein synthesis. The results indicate that association of ApoB 100 with LDs is dependent on the activity of low–molecular weight GTP-binding protein Rab18 and highlight the role of LDs as targets of free cholesterol toxicity in hepatocytes.
INTRODUCTION
Emerging experimental and clinical data suggest that disturbed hepatic cholesterol homeostasis and free cholesterol accumulation in the liver are relevant to the pathogenesis of nonalcoholic steatohepatitis (NASH; Bashiri et al., 2013; Musso et al., 2013; Arguello et al., 2015). Critical roles for endoplasmic reticulum (ER) stress (Ashraf and Sheikh, 2015) and mitochondrial free cholesterol accumulation (Mari et al., 2006) have been suggested. However, the precise mechanisms of free cholesterol lipotoxicity in NASH are not well understood. Increasing evidence suggests that contact zones between cytoplasmic lipid droplets (LDs) and the ER and mitochondria play important roles in trafficking of lipids and regulation of lipid metabolism (Barbosa et al., 2015).
LDs are intracellular organelles that store neutral lipids within cells. Recent studies show that LDs are authentic organelles that are actively engaged in multiple functions through interaction with other organelles (Fujimoto and Parton, 2011; Krahmer et al., 2013; Pol et al., 2014; Sahini and Borlak, 2014). In hepatocytes, LDs act as a reservoir of neutral lipids for the formation of very low-density lipoprotein (VLDL), which is assembled by the lipidation of apolipoprotein B (ApoB) in the lumen of the ER (Lehner et al., 2012; Yao et al., 2013). The LD-coat protein perilipin 2 (PLIN2; formerly known as adipophilin or adipose differentiation–related protein) is reported to regulate VLDL secretion. Knockdown of PLIN2 induces close apposition between LDs and the ER, suggesting that the PLIN2 content of LDs regulates the interaction of LDs and the ER. However, little is known about the mechanisms that govern the transfer of lipids from LDs to the lumen of the ER (Lehner et al., 2012).
Rab low–molecular weight GTP-binding proteins are involved in various steps of intracellular traffic (Stenmark, 2009; Segev, 2011). Similar to PLIN2, Rab18 is specifically associated with LDs (Martin et al., 2005; Ozeki et al., 2005). However, Rab18 and PLIN2 are localized to different population of LDs (Ozeki et al., 2005). Overexpression of Rab18 induces a decrease in amount of PLIN2 and the close apposition of LDs to the ER-derived membrane in HepG2 and BALB/c 3T3 cells (Ozeki et al., 2005). In adipocytes, the stimulation of lipolysis increases the localization of Rab18 to LDs (Martin et al., 2005). These results suggest that Rab18 is involved in lipid movement from LDs to the ER. Rab18 is involved in insulin-mediated lipogenesis and β-adrenergic–induced lipolysis in adipocytes (Pulido et al., 2011). It is postulated that Rab18 facilitates the interaction of LDs with ER membranes and the exchange of lipids between these compartments. The role of Rab18 in hepatocytes remains to be elucidated.
In this study, we examined the effect of the acute accumulation of free cholesterol on LDs in cultured human hepatocytes. Accumulation of cholesterol induced LD fusion and degradation of PLIN2, followed by the association of ApoB with LDs. Our results also indicate the role of Rab18 in LD fusion to ER.
RESULTS
An acyl-CoA:cholesterol acyltransferase inhibitor induces fusion of LDs
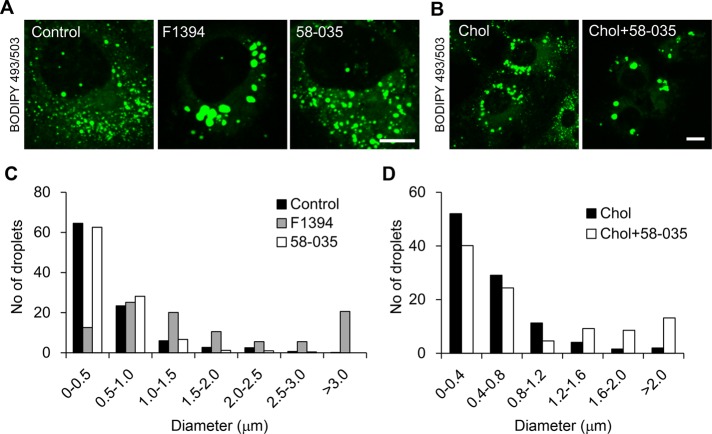
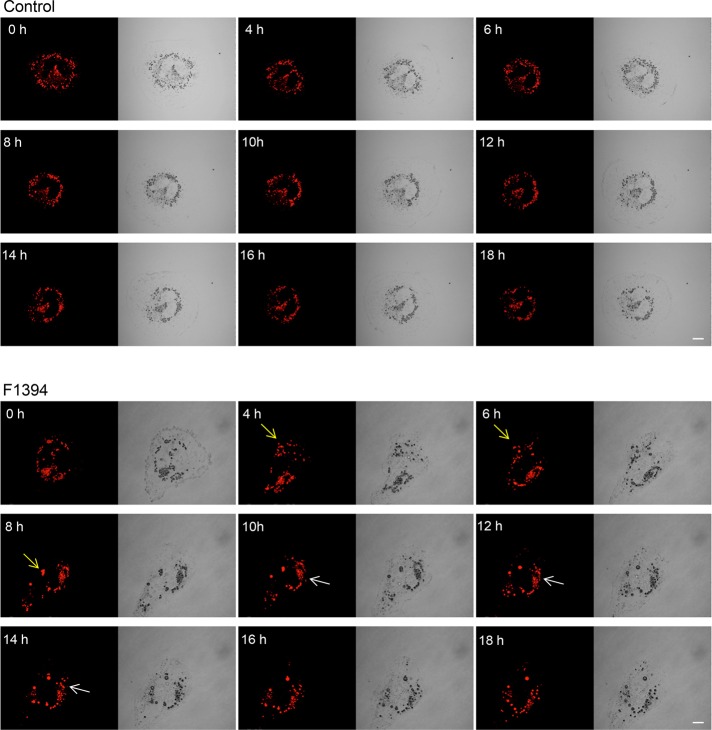
To study the effect of acyl-CoA:cholesterol acyltransferase (ACAT) inhibition on lipoprotein/cholesterol metabolism in human liver cells, we treated cultured human hepatocyte Huh7 cells with two ACAT inhibitors, (1s,2s)-2-[3-(2,2-dimethylpropyl)-3-nonylureido]aminocyclohexane-1-yl-3-[N-(2,2,5,5-tetramethyl-1,3-dioxane-4-carbonyl)amino]propionate (F1394; Kusunoki et al., 1995) and 3-[decyldimethylsilyl]-N-[2-(4-methylphenyl)-1-phenylethyl]pro panamide (58-035; Ross et al., 1984), and examined the effect on the morphology of lipid droplets (LDs). After incubation with the inhibitors, cells were labeled with BODIPY 493/503, which stains neutral lipids of LDs (Gocze and Freeman, 1994). Cell treatment with F1394 for 24 h resulted in LD enlargement with a concomitant decrease in the number of smaller (<0.5 μm) LDs (Figure 1, A and C). However, unlike F1394, 58-035 did not significantly affect the size of LDs, with the smaller LDs representing the majority of LDs (Figure 1, A and C). Of note, the combination of methyl-β-cyclodextrin (MβCD)/cholesterol in order to increase the cholesterol content and 58-035 increased the size of LDs (Figure 1, B and D). Figure 2 and Supplemental Movies S1 and S2 show the dynamics of Nile red–labeled LDs in the absence (Figure 2, control; Supplemental Movie S2) and presence (Figure 2, F1394; Supplemental Movie S1) of F1394. Figure 2 shows that F1394 induces fusion of LDs (white and yellow arrows).
FIGURE 1:
Treatment of Huh7 cells with ACAT inhibitors enlarges LDs. (A) Huh7 cells were cultured for 24 h in the absence (Control) or presence of 10 μg/ml F1394 or 10 μg/ml 58-035. Cells were then labeled with 2 μg/ml BODIPY 493/503 for 30 min. Bar, 10 μm. (B) Cells were treated with MβCD/cholesterol in LPDS (Chol) or MβCD/cholesterol plus 58-035 in LPDS (Chol + 58-035) as described in Materials and Methods. Cells were labeled with BODIPY 493/503. Bar, 10 μm. (C) The size distributions of LDs were quantified as described in Materials and Methods after 24-h treatment of Huh7 cells with 10 μg/ml F1394 or 10 μg/ml 58-035. (D) The size distributions of LDs were quantified after 24-h treatment of Huh7 cells with MβCD/cholesterol in LPDS (Chol) or MβCD/cholesterol plus 58-035 in LPDS (Chol + 58-035).
FIGURE 2:
Treatment of Huh7 cells with F1394 induces fusion of LDs. Huh7 cells were incubated in DMEM with 10% FCS plus 20 ng/ml Nile red in the absence (Control) or presence (F1394) of 10 μg/ml F1394. Time-lapse images were acquired using an Olympus FV1000 confocal microscope. Arrows indicate the fusion of LDs. See Supplemental Movies S1 and S2. Bars, 10 μm.
ACAT inhibitors increase cellular free cholesterol
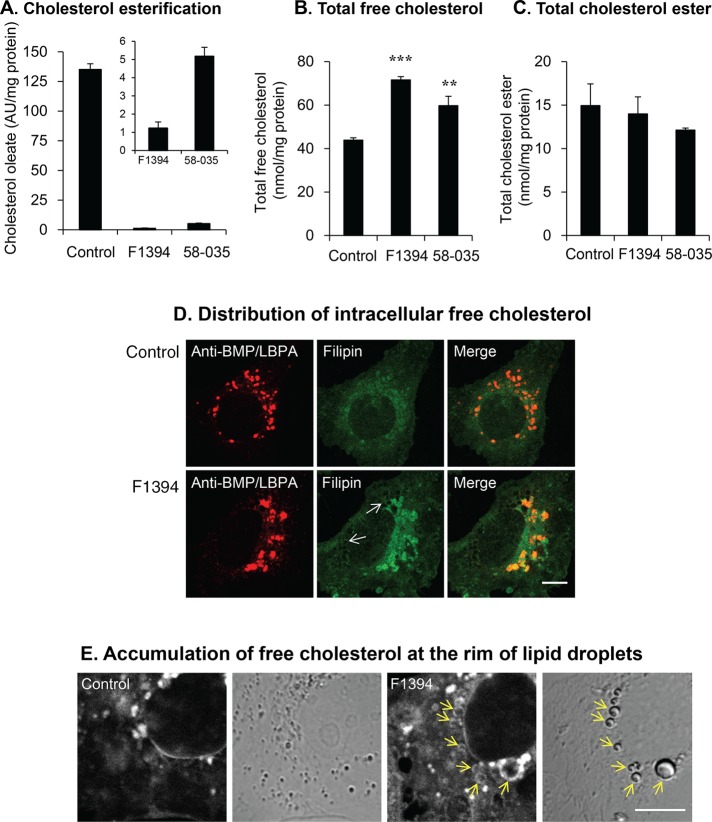
Figure 3A indicates that both F1394 and 58-035 significantly inhibited the esterification of cholesterol as monitored by the incorporation of [14C]oleate into cholesterol. The inset of Figure 3A shows that F1394 (99% inhibition) is a more potent inhibitor than 58-035 (95% inhibition). The total content of cellular cholesterol was quantified by gas chromatography (GC) analysis after 24 h of treatment with F1394 and 58-035 (Figure 3, B and C). Both inhibitors significantly increased cellular free cholesterol, with F1394 displaying a higher increase of 63% (p < 0.001) than 58-035, 36% (p < 0.02; Figure 3B). In contrast, total cholesterol ester was not significantly affected by these inhibitors (p > 0.1, Figure 3C). The cholesterol-binding dye filipin strongly labeled intracellular compartments when cells were incubated with 10 μg/ml F1394 for 24 h. Filipin labeling partially overlapped with anti-bis(monoacylglycero)phosphate (BMP)/lysobisphosphatidic acid (LBPA) antibody staining (Figure 3D), which is enriched in late endosomes (Kobayashi et al., 1998). In addition to late endosomes, filipin stained the rim of LDs (arrows) after F1394 treatment (Figure 3, D and E).
FIGURE 3:
ACAT inhibitors increase cellular free cholesterol. (A) Huh7 cells were treated as described in Figure 1A. Incorporation of [14C]oleate into cholesterol was measured as described in Materials and Methods. Results indicate mean ± SD (n = 3) of [14C]cholesteryl ester formed. (B) Cells were treated as in A. Total cellular free cholesterol was quantified using GC as described in Materials and Methods. ***p < 0.001, **p < 0.02. (C) Cells were treated as in A. Total cellular cholesterol ester was quantified using GC as described in Materials and Methods. (D) Cells were grown for 24 h in the absence (top) or presence (bottom) of 10 μg/ml F1394. Cells were then fixed and permeabilized by digitonin as described in Materials and Methods and doubly labeled with filipin and anti-BMP/LBPA antibody. Bar, 10 μm. (E) Cells were grown for 24 h in the absence (Control) or presence (F1394) of 10 μg/ml F1394. Cells were then fixed and permeabilized by freezing and thawing as described in Materials and Methods and stained with filipin. Bar, 10 μm.
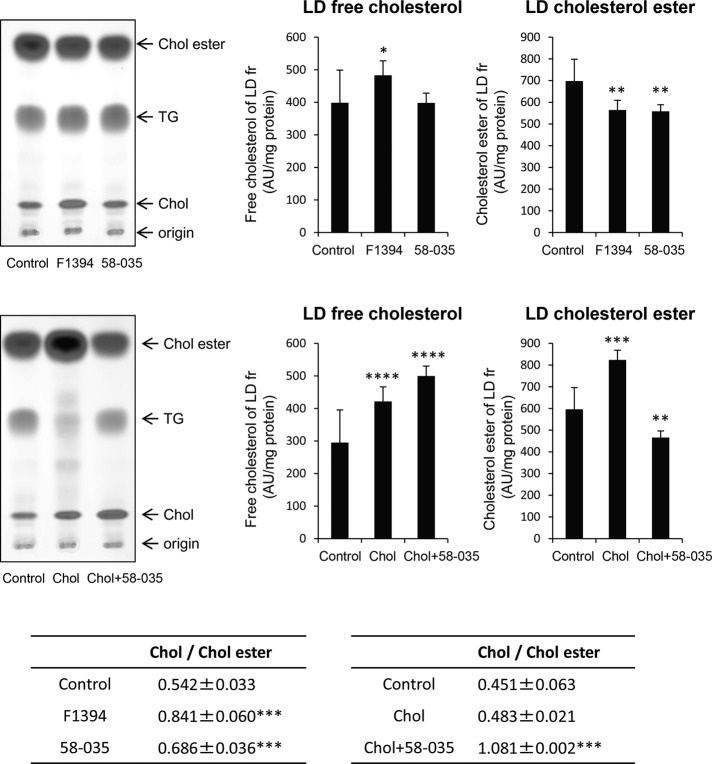
To make precise the link between LD enlargement and cholesterol content, we then examined by high-performance TLC (HPTLC) the neutral lipid content of the LD fractions obtained from Huh7 cells treated with ACAT inhibitors (Figure 4). We measured the intensity of cholesterol and cholesterol ester bands stained with ferric chloride/sulfuric acid/acetic acid and normalized the intensity with the amount of proteins. F1394 treatment slightly (p = 0.05) increased the amount of free cholesterol in LD fraction, whereas 58-035 did not. Both F1394 and 58-035 significantly (p < 0.05) decreased the content of cholesterol ester in the LD fraction. As a result, both F1394 and 58-035 increased the free cholesterol/cholesterol ester ratio (p < 0.01) in the LD fraction, with F1394 giving a higher ratio. Free cholesterol in the LD fraction was highly increased after cell treatment with cholesterol/MβCD complex (p < 0.001). However, cholesterol/MβCD complex also increased cholesterol ester (p < 0.01), and thus the cholesterol/cholesterol ester ratio in the LD fraction was not significantly affected by cholesterol/MβCD treatment. In contrast, cholesterol/cholesterol ester was highly increased (p < 0.01) when cells were treated with both cholesterol/MβCD and 58-035.
FIGURE 4:
F1394 increases free cholesterol/cholesterol ester ratio in LDs. Huh7 cells were cultured for 24 h in the absence (Control) or presence of 10 μg/ml F1394 or 10 μg/ml 58-035 or treated with MβCD/cholesterol in LPDS (Chol) or MβCD/cholesterol plus 58-035 in LPDS (Chol + 58-035) as described in Materials and Methods. LD fractions were prepared and lipids analyzed by HPTLC as described in Materials and Methods. Results indicate the mean ± SD (n = 3). *p = 0.05, **p < 0.05, ***p < 0.01, ****p < 0.001.
There are two genes encoding the two ACAT enzymes, ACAT1 and ACAT2 (Buhman et al., 2000). ACAT2 is the major ACAT isozyme in adult human liver (Parini et al., 2004). To study the specific role of ACAT inhibition in the free cholesterol increase, we prepared Huh7 knockout cells of ACAT1, ACAT2, and both ACAT1 and ACAT2. We confirmed that each single knockout inhibited cholesterol esterification but not as efficiently as the F1394 inhibitor (Supplemental Figure S1A). Unfortunately, double-knockout cells did not grow well and could not be surveyed for cholesterol content. Knockout of ACAT2 gave a cholesterol/cholesterol ester ratio for LDs of 0.70 (Supplemental Figure S1B). In addition, LD staining with BODIPY 493/503 of ACAT1- or ACAT2-knockout Huh7 cells showed a weaker and heterogeneous enlargement of LDs compared with F1394-treated cells (Supplemental Figure S1C).
Free cholesterol accumulation induces the degradation of PLIN2
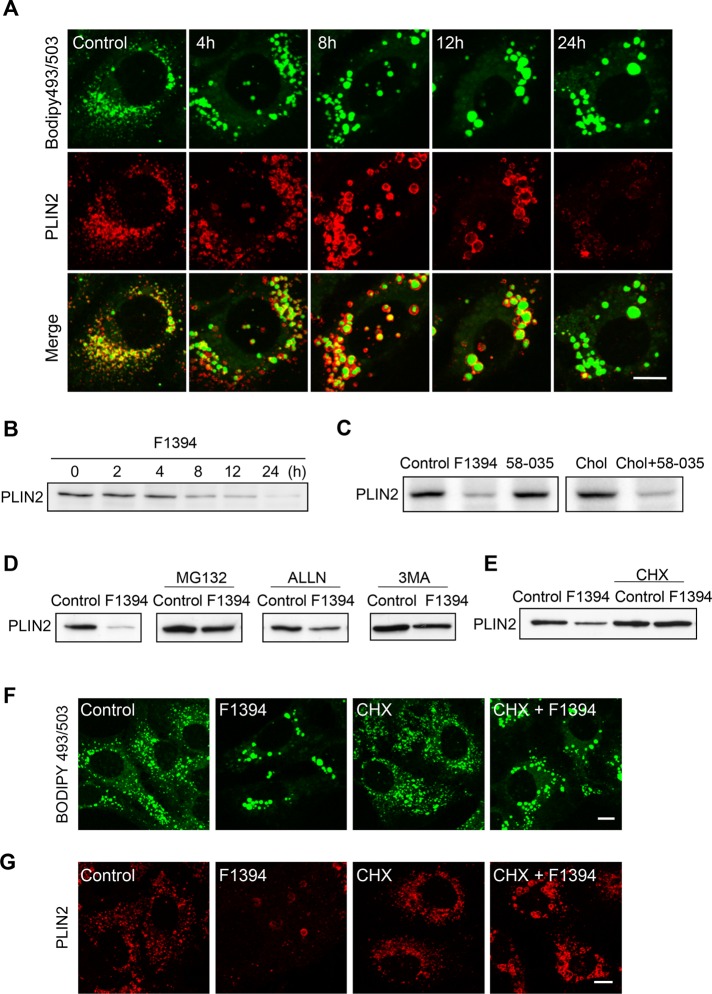
Because LD surface structure and lipolysis activity are regulated by PLIN proteins, a family of five genetically related coat proteins (Sztalryd and Kimmel, 2014), we decided to examine the distribution of adipocyte differentiation–related protein (PLIN2, also known as adipophilin), a major peripheral protein of LDs in Huh7 cells. PLIN2 was located at the periphery of enlarged LDs until 12 h after the addition of F1394 (Figure 5A). However, PLIN2 staining was drastically reduced after 24 h of cell treatment. Western blotting demonstrated that PLIN2 content decreased gradually during incubation with F1394 (Figure 5B). In contrast, 58-035 did not significantly decrease PLIN2 after 24 h of treatment compared with the control, whereas the combination of 58-035 and MβCD/cholesterol did so significantly (Figure 5C).
FIGURE 5:
Acute free cholesterol accumulation induces ubiquitin- and autophagy-mediated degradation of PLIN2. (A) F1394, 10 μg/ml, was added to Huh7 cells. At appropriate intervals, cells were fixed, permeabilized with digitonin, and doubly labeled with BODIPY 493/503 and anti-PLIN2 antibody as described in Materials and Methods. Bar, 10 μm. (B) Cells were treated as in A. Cellular PLIN2 was measured by Western blotting as described in Materials and Methods. (C) Cells were treated for 24 h with 10 μg/ml F1394, 10 μg/ml 58-035, MβCD/cholesterol in LPDS (Chol), or MβCD/cholesterol plus 58-035 in LPDS (Chol + 58-035) as described in Figure 1. Cellular PLIN2 was measured as in B. (D) Huh7 cells were treated with 10 μg/ml F1394 in the presence or absence of 10 μM MG132, 10 μM ALLN, or 10 mM 3MA. After 24-h incubation, PLIN2 in cells was revealed by Western blotting. (E) Huh7 cells were grown in the absence and presence of 10 μg/ml F1394, 10 μg/ml cycloheximide (CHX), or CHX plus F1394 for 18 h. PLIN2 was measured as in B. (F, G) Cells were treated with 10 μg/ml F1394, 10 μg/ml CHX, or CHX + F1394 for 24 h. Cells were then fixed and permeabilized with digitonin and labeled with BODIPY 493/503 (F) or anti-PLIN2 antibody (G) as described in Materials and Methods. Bars, 10 μm.
PLIN2 is degraded by the ubiquitin pathway during the regression of lipid-storing cells (Xu et al., 2005; Masuda et al., 2006). LDs can also be degraded through autophagosomes during nutrient starvation (Singh et al., 2009). We thus examined the effects of the ubiquitin inhibitors MG132 and N-acetyl-l-leucinyl-l-leucinyl-l-norleucinal (ALLN) and the autophagy inhibitor 3-methyladenine (3-MA; Ohsaki et al., 2006) on F1394-induced PLIN2 degradation. All three inhibitors partially inhibited degradation of PLIN2 (Figure 5D), suggesting that both the ubiquitin pathway and autophagy are involved in degradation of PLIN2 by F1394. Furthermore, degradation of PLIN2 was inhibited when protein synthesis was inhibited (Figure 5E). However, F1394 still induced enlargement of LDs as visualized by BODIPY 493/503 (Figure 5, F and G). These results indicate that F1394-induced enlargement of LDs does not require newly synthesized PLIN2 and that the enlargement is not the result of PLIN2 degradation. Supplemental Figure S2 examines whether PLIN1 substitutes for PLIN2 during F1394 treatment. Western blotting shows that F1394 did not alter PLIN1 content in Huh7 cells (Supplemental Figure S2A). Furthermore, staining with anti-PLIN1 antibody displays a similar ER staining pattern in both control and F1394-treated cells, suggesting that PLIN1 did not substitute for PLIN2 (Supplemental Figure S2B).
Acute accumulation of free cholesterol induces the fusion of LDs and ER
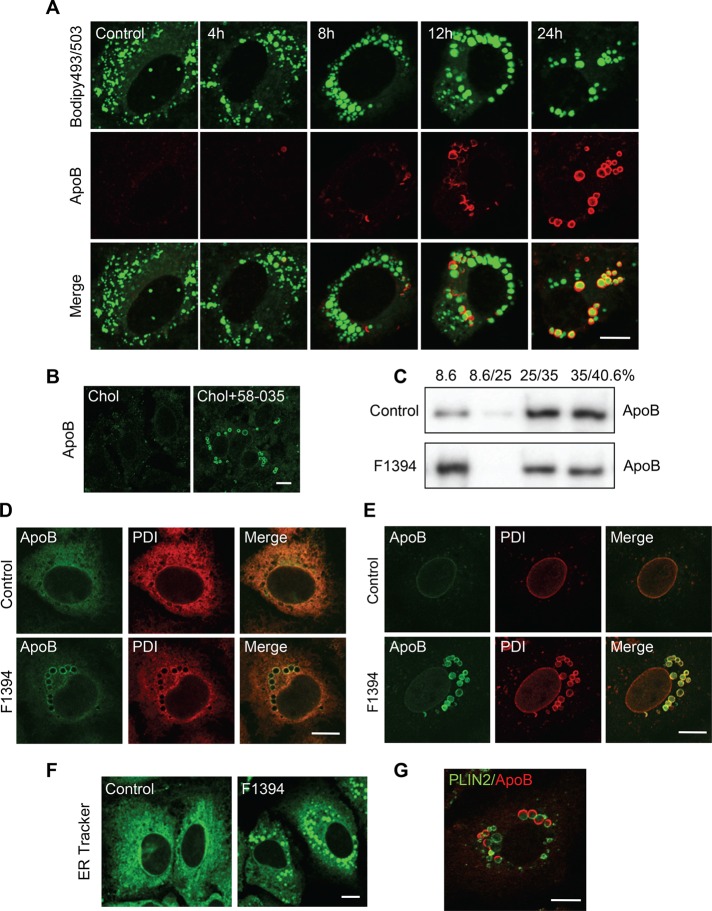
Of interest, the PLIN2 decrease in free cholesterol–accumulated cells was not accompanied by a decrease of BODIPY 493/503 staining (Figure 5A). Ohsaki et al. (2008) showed that knockdown of PLIN2 increases the association of ApoB 100 to LDs. Figure 6A examines the intracellular distribution of ApoB in F1394-treated cells. ApoB labeling increased (see later discussion for the permeabilization procedure) in a time-dependent manner, and ApoB was localized at the periphery of BODIPY 493/503 staining. Similar results were obtained when Huh7 cells were treated with the combination of MβCD/cholesterol and 58-035 (Figure 6B). Figure 6C shows the effect of F1394 treatment on the distribution of ApoB after sucrose density gradient fractionation. ApoB was increased in the very light 8.6% fraction (i.e., lipid enriched), supporting the idea that ApoB was associated with LDs by F1394 treatment. Furthermore, Western blotting of the subcellular fractions indicates that a considerable amount of cellular ApoB was not associated with LDs in both control and the F1394-treated cells (Figure 6C).
FIGURE 6:
Acute free cholesterol accumulation induces association of ApoB with LDs. (A) F1394,10 μg/ml, was added to Huh7 cells. At appropriate intervals, cells were fixed, permeabilized with digitonin, and doubly labeled with BODIPY 493/503 and anti-ApoB antibody as described in Materials and Methods. Bar, 10 μm. (B) Cells were treated with MβCD/cholesterol in LPDS (Chol) or MβCD/cholesterol plus 58-035 in LPDS (Chol + 58-035) as described in Figure 1. Cells were then fixed, and ApoB was labeled as described. (C) Huh7 cells were cultured for 24 h in the absence (Control) or presence of 10 μg/ml F1394. Cells were then homogenized, and PNS was separated by stepwise sucrose density gradient as described in Materials and Methods. The distribution of ApoB was measured by Western blotting. (D) Huh7 cells were grown for 24 h in the absence and presence of 10 μg/ml F1394. Cells were then fixed and permeabilized with 0.1% Triton X-100. Intracellular distribution of ApoB and PDI was examined as described in Materials and Methods. Bar, 10 μm. (E) Cells were treated as described and then fixed and permeabilized with 50 μg/ml digitonin, followed by double labeling with anti-ApoB and anti-PDI. Bar, 10 μm. (F) Cells were incubated as in A, followed by labeling with ER tracker. Bar, 10 μm. (G) Cells were treated as in D and fixed and permeabilized with 50 μg/ml digitonin, followed by double labeling with anti-PLIN2 and anti-ApoB. Bar, 10 μm.
In this study, most of the immunofluorescence experiments were performed in cells fixed and permeabilized with 0.05% digitonin, which preferentially permeabilizes cholesterol-rich membrane (Elias et al., 1978; Ohsaki et al., 2008). When cells were fixed and permeabilized with 0.1% Triton X-100, ApoB was found to be distributed in the ER network and colocalized with the ER lumen marker protein disulfide isomerase (PDI) in control cells (Figure 6D). After F1394 treatment, ApoB was localized to LD-like structures in addition to the ER network. PDI was also observed in LD-like structures after F1394 treatment (Figure 6D). Of note, ApoB labeling was not observed in digitonin-treated control cells. In contrast, both ApoB and PDI were localized in LD-like structures after digitonin treatment of cells incubated with F1394 (Figure 6E). These results indicate that the ApoB-positive structure visualized in digitonin-permeabilized, F1394-treated cells is located in the ER lumen. The LD-like structures were also labeled with an ER tracker dye (glibenclamide BODIPY FL) that binds to the sulfonylurea receptors of ATP-sensitive K+ channels (Figure 6F). Sometimes, the PLIN2 labeling and ApoB labeling were complementary in a single LD and showed an “ApoB crescents” pattern as observed when proteasomes were inhibited in Huh7 cells (Ohsaki et al., 2006, 2008; Figure 6G). In contrast to the observation of Ohsaki et al. (2008), however, the majority of ApoB-positive structures appeared as circles in our experiments (Figure 6, A, B, D, and E). It was demonstrated that ApoB crescents were significantly reduced when ApoB lipidation is suppressed (Ohsaki et al., 2008). Supplemental Figure S3 examines the effect of the inhibitor of the microsomal triglyceride-transfer protein (MTP) BAY 13-9952 on ApoB labeling of digitonin-treated cells and the recovery of ApoB in LD fraction after sucrose density gradient centrifugation. In contrast to the result of the previous report (Ohsaki et al., 2008), ApoB staining was only partially suppressed, indicating that ApoB was not significantly lipidated. The recovery of ApoB to the LD fraction was 80% of control, supporting the immunofluorescence results.
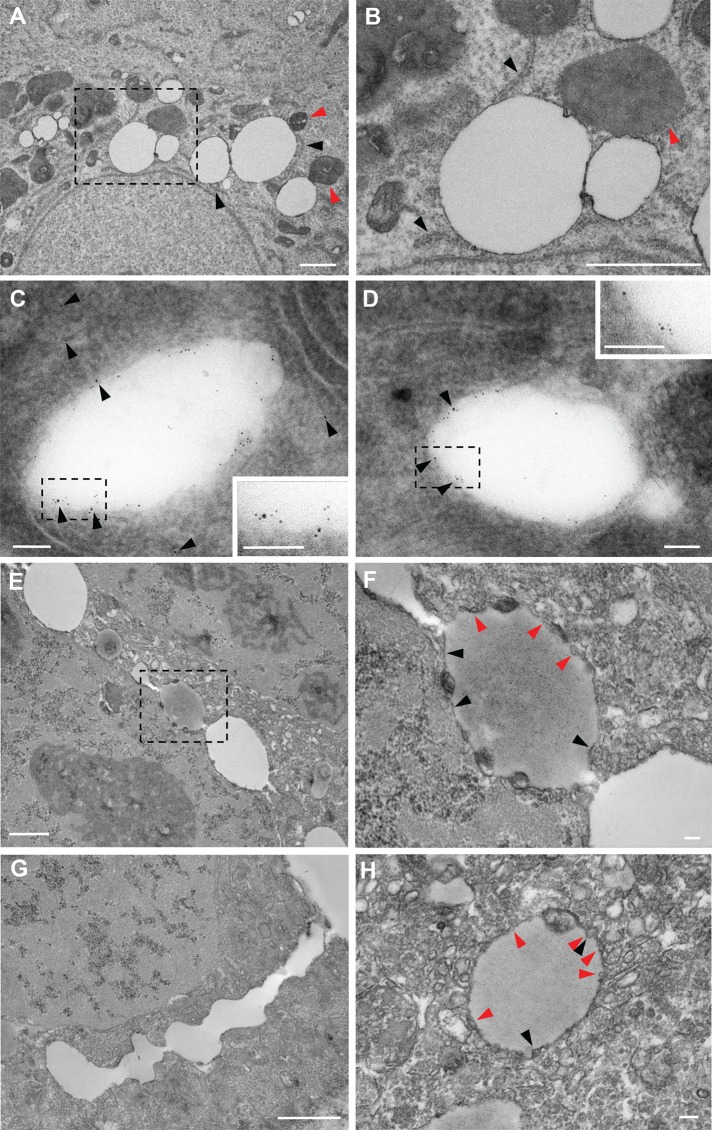
Electron microscopy showed LDs adjacent to the ER (black arrowheads in Figure 7, A and B) after 6 h of incubation with F1394. Mitochondria were often observed next to LDs (red arrowheads). Immunoelectron microscopy revealed ApoB (10 nm gold) both outside and at the rims of LDs (black arrowheads in Figure 7, C and D). Both ApoB (10 nm gold) and PLIN2 (5 nm gold) were observed in the same LDs after 6 h of F1394 treatment. Cells exhibited electron-lucent and -dense LDs connected to each other after 24 h of treatment with F1394 (Figure 7, E–G). Ohsaki et al. (2008) showed that enrichment of polyunsaturated fatty acid causes electron-dense LDs. Bilayer membranes were connected to LDs (black arrowheads in Figure 7, F and H). LDs were attached to and surrounded by a number of small vesicles (red arrowheads in Figure 7, F and H), suggesting the occurrence of active vesicle budding or fusion.
FIGURE 7:
Acute free cholesterol accumulation induces the association of LDs with the ER. (A, B) Huh7 cells were treated with 10 μg/ml F1394 for 6 h. Cells were then fixed and examined under an electron microscope as described in Materials and Methods. (B) Higher magnification image of a dashed square area in A. Black arrowheads indicate the cisterns of ER connected with LDs. Red arrowheads indicate the mitochondria connected with LDs. Bar, 1 μm. (C, D) Cells were treated as described and then fixed and doubly labeled with anti-PLIN2 (5 nm) and anti-ApoB antibodies (10 nm, black arrowheads) as described in Materials and Methods. Insets, enlarged image of dashed squares. Bar, 0.1 μm. (E–H) Huh7 cells were treated with 10 μg/ml F1394 for 24 h. Cells were then fixed and examined under electron microscope as described in Materials and Methods. Cells contain LDs with different electron densities (E). LDs were often connected (G). (F) Higher-magnification image from dashed square area in E. (H) Enlarged image of LD. Each LD was connected to bilayer membranes (black arrowheads in F and H) and surrounded by tiny vesicles (red arrows in F and H). Bars, 1 μm (E, G), 0.1 μm (F, H).
Our results are different from those of Ohsaki et al. (2008) on the association of ApoB to LDs during normal culture conditions: 1) Most of the ApoB staining labeled the entire LDs rather than appearing as “ApoB crescents.” 2) MTP inhibitor did not significantly inhibit the association of ApoB to LDs. 3) Both PLIN2 and ApoB are located to the rim of LDs. Because PDI staining and ER tracker dye were also associated with LDs and electron micrographs suggest budding/fusion of LDs, we speculate that F1394 favors the fusion of LDs to the ER.
Rab18 mediates the association of ApoB to LDs in acute cholesterol-loaded cells
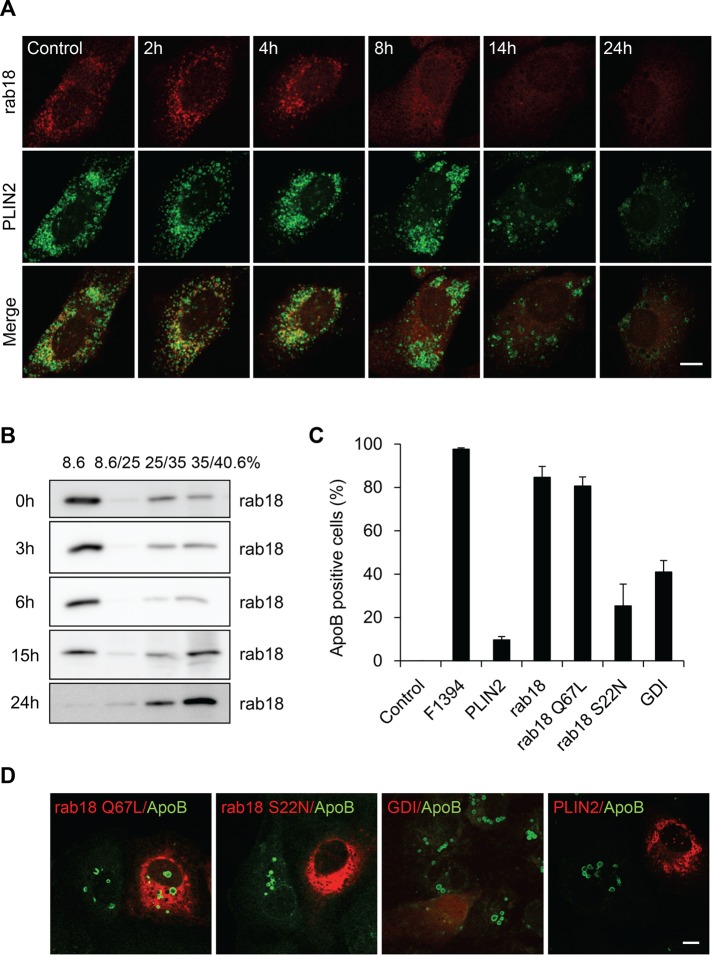
Rab low–molecular weight GTP-binding proteins are involved in the various steps of intracellular vesicular traffic (Stenmark, 2009; Segev, 2011). Rab18 is specifically associated with LDs (Martin et al., 2005; Ozeki et al., 2005) and induces close apposition of LDs to the ER-derived membrane (Ozeki et al., 2005). In adipocytes, the stimulation of lipolysis increased the localization of Rab18 to LDs (Martin et al., 2005). Ozeki et al. (2005) showed that endogenous Rab18 coexisted with PLIN2 in LDs, but the labeling intensity of the two proteins showed clear reciprocity in HepG2 cells. Figure 8A and Supplemental Figure S4 indicate that Rab18 was partially colocalized with PLIN2 in Huh7 cells as reported in HepG2 cells (Ozeki et al., 2005). F1394 treatment decreased the Rab18 labeling of LDs, and after 8 h, Rab18 was mainly localized in ER. Rab18 labeling was abolished after 24 h of treatment, as observed for PLIN2. The redistribution of Rab18 was confirmed by sucrose density gradient fractionation (Figure 8B). Rab18 was recovered mainly in the very light (8.6%) fraction in the control cells. The addition of F1394 decreased the intensity of Rab18 in the very light fraction. After 15 h, a significant amount of Rab18 was recovered in the heavy membrane fraction, where high-density membrane and soluble proteins are recovered. Rab18 was not recovered from the 8.6% fraction after 24 h of treatment with F1394. Unlike PLIN2, Rab18 was not degraded by F1394 treatment.
FIGURE 8:
Association of ApoB with LDs is dependent on the activity of Rab18. (A) F1394, 10 μg/ml, was added to Huh7 cells. At appropriate intervals, cells were fixed, permeabilized with digitonin, and doubly labeled with anti-PLIN2 and anti-Rab18 antibodies as described in Materials and Methods. Bar, 10 μm. (B) Cells were treated with F1394 as described and then homogenized, and PNS was separated by stepwise sucrose density gradient as described in Materials and Methods. The distribution of Rab18 was measured by Western blotting. (C, D) Huh7 cells overexpressing mCherry-PLIN2, mCherry-Rab18, mCherry-Rab18 S22N, mCherry-Rab18 Q67L, and mCherry-GDI were treated with F1394 as described in Materials and Methods, and the number of ApoB-positive cells was counted after immunofluorescence. Bar, 10 μm.
The precise role of Rab18 is not well understood. Rab proteins cycle between an active, GTP-binding form and an inactive, GDP-binding form. The GTP form binds the target membrane, whereas the GDP form is released from the target membrane. GDP-dissociation inhibitor (GDI) slows the rate of dissociation of GDP from Rab and thus inhibits the binding of Rab to the membrane. In this study, we examined the effect of overexpressing wild-type Rab18, constitutively active GTP-binding mutant (Q67L) Rab18, and dominant-negative GDP-binding mutant (S22N) Rab18, as well as of GDI, on the association of ApoB to LDs (Figure 8, C and D). The overexpression of Rab18 S22N and GDI inhibited the association of ApoB to LDs, whereas wild-type Rab18 and the Q67L mutant did not significantly affect the association. These results suggest that the active GTP form of Rab18 is involved in the association of ApoB to LDs. As reported (Ohsaki et al., 2008), the overexpression of PLIN2 also inhibited the association of ApoB to LDs.
Supplemental Figure S5, A and B, indicates that the overexpression of Rab18 Q67L and Rab18 S22N did not inhibit the F1394-induced enlargement of LDs. Supplemental Figure S5C shows that the overexpression of wild-type Rab18 slightly decreased endogenous PLIN2 as reported (Ozeki et al., 2005). Overexpression of green fluorescent protein (GFP)–PLIN2 also decreased endogenous PLIN2. However, overexpression of Rab18 Q67L and Rab18 S22N did not affect PLIN2 (Supplemental Figure S5, C–E). These results suggest that the inhibition of the association of ApoB to LDs by overexpression of Rab18 S22N is not because of the increase of PLIN2.
DISCUSSION
Acute accumulation of free cholesterol enlarges LDs and induces the degradation of PLIN2 in cultured human hepatocytes
Although both ACAT inhibitors F1394 and 58-035 significantly inhibited the esterification of cellular cholesterol in Huh7 cells after 24 h of treatment, LD enlargement was observed only with F1394. F1394 increased total cellular free cholesterol by 63%, whereas 58-035 increased it by 36%. F1394 also increased the free cholesterol/cholesterol ester ratio of the LD fraction to 0.841, whereas 58-035 increased this ratio to a lower level, 0.686. The LD size also increased when cells were treated with both MβCD/cholesterol and 58-035. In contrast, MβCD/cholesterol alone did not significantly affect the LD size. Both MβCD/cholesterol and MβCD/cholesterol plus 58-035 treatments increased the free cholesterol content of LD fraction. However, MβCD/cholesterol also increased the content of cholesterol esters in LDs. Our results suggest that the increase of the free cholesterol/cholesterol ester ratio of LDs plays an important role in controlling LD size.
Our results indicate that 95% inhibition of ACAT activity by 58-035 did not significantly affect LD structure in cultured hepatocytes. This finding is in marked contrast to that observed in macrophages, in which ACAT inhibitor does inhibit the accumulation of LDs (Namatame et al., 2004). Time-lapse measurement indicated that the fusion of LDs was accelerated by F1394-induced acute cholesterol loading in Huh7 cells.
The LD surface is covered with a phospholipid monolayer, of which phosphatidylcholine and phosphatidylethanolamine are major components (Tauchi-Sato et al., 2002; Bartz et al., 2007). The cylindrical shape of phosphatidylcholine stabilizes LDs and prevents LD coalescence, thus regulating the size of the LDs (Saito et al., 1999; Krahmer et al., 2011). The role of cholesterol in the size of LDs is not well understood (Yang et al., 2012). Free cholesterol is likely to reside on the LD surface layer (Prattes et al., 2000). Studies using emulsions containing triolein and egg phosphatidylcholine found that the presence of cholesterol induced the coalescence of the emulsions (Saito et al., 1999). It is speculated that cholesterol has a small polar group and thus decreases the mean spontaneous curvature of surface lipids. Our results suggest that the physical properties of cholesterol induce LD fusion in hepatocytes.
The enlargement of LDs was paralleled by the degradation of PLIN2. Similar to the enlargement of LDs, F1394 but not 58-035 induced the decrease of PLIN2. In addition, the combination of MβCD/cholesterol and 58-035 decreased PLIN2. These results suggest that the acute increase of free cholesterol triggers the degradation of PLIN2. The degradation of PLIN2 was inhibited by the inhibitors of ubiquitination, autophagy, and protein synthesis. Of interest, inhibition of the degradation of PLIN2 by protein synthesis inhibitor did not inhibit the enlargement of LDs (Figure 5, F and G), indicating that the degradation of PLIN2 did not induce the enlargement of LDs in our experimental conditions.
Cholesterol-dependent, ubiquitin-proteasome–mediated degradation of 3-hydroxy-3-methylglutaryl-CoA (HMG CoA) reductase, the key regulatory enzyme in the mevalonate pathway, is well known (Ravid et al., 2000; Jo and Debose-Boyd, 2010). Another biosynthetic enzyme of cholesterol, squalene monooxygenase, was also shown to undergo cholesterol-accelerated proteasomal degradation (Gill et al., 2011). Unlike HMG CoA reductase, the degradation of squalene monooxygenase is not mediated by Insig, 24,25-dihydrolanosterol, or side-chain oxysterols but instead is mediated by cholesterol itself. It is of interest that, similar to the degradation of PLIN2, the degradation of HMG-CoA reductase requires ongoing protein synthesis (Chun et al., 1990).
Proteasome-dependent degradation of PLIN2 has been reported to occur during regression of lipid-storing cells (Xu et al., 2005; Masuda et al., 2006). The N-terminal region of PLIN2 is suggested to mediate proteasomal degradation (Orlicky et al., 2008). However, the detailed mechanism underlying proteasome-dependent degradation of PLIN2 is not yet clarified. Further studies are required to understand the mechanism of cholesterol-mediated, proteasome-dependent degradation of PLIN2.
Accumulation of free cholesterol induces association of ApoB 100 with LDs
Ohsaki et al. (2006) showed that association of ApoB with LDs is induced when proteasome or autophagy is inhibited. They also showed that the association of ApoB with LDs is inhibited by overexpression of PLIN2 and accelerated by knockdown of PLIN2 (Ohsaki et al., 2008). The present study indicates that the association of ApoB to LDs is accompanied by the degradation of PLIN2 induced by acute free cholesterol loading and that the association is inhibited by PLIN2 overexpression. Similar to Ohsaki et al. (2006, 2008), we sometimes observed complementary labeling of PLIN2 and ApoB even in a single LD, showing an “ApoB crescents” pattern. However, in contrast to their results, the majority of ApoB-positive LDs exhibited circular labeling. They reported that abnormal lipidation of ApoB occurred in the cholesterol-rich lumen of the ER (Ohsaki et al., 2008). Under acute free cholesterol accumulation, the association of ApoB with LDs was not significantly inhibited by the inhibitors of MTP, indicating that ApoB was not lipidated. The observations that the association of other ER proteins with LDs (like PDI) and the colocalization of ApoB and PLIN2 at the rim of LDs suggest that LDs fuse with the ER under our experimental conditions.
Rab18 activity is required for the association of ApoB with LDs
Rab18 is a low–molecular weight GTPase that specifically associates with LDs (Martin et al., 2005; Ozeki et al., 2005). Rab GTPases regulate many steps of membrane traffic by switching between an inactive, GDP form and an active, GTP form (Stenmark, 2009; Segev, 2011). However, the precise role of Rab18 in hepatocytes is not well understood.
Acute accumulation of free cholesterol relocated Rab18 from LDs to ER in Huh7 cells. Cholesterol loading–mediated association of ApoB with LDs was inhibited by overexpression of the GDP form (Rab18 Q67L) of Rab18 but not by wild-type or the GTP form (Rab18 S22N) of the protein, indicating that Rab18 activity is required for the association of ApoB with LDs. Consistently, overexpression of GDI, which stabilizes the GDP form of Rab proteins, also inhibited the association of ApoB with LDs.
Overexpression of Rab18 has been reported to induce close apposition of LDs to the ER-derived membrane in HepG2 and BALB/c 3T3 cells (Ozeki et al., 2005). Our results suggest that Rab18 plays a role in lipid traffic from LDs to the ER in cholesterol-loaded hepatocytes.
Conclusion
Our results show that the following sequential events occur when free cholesterol is loaded into cultured hepatocytes (Supplemental Figure S6):
Fusion of LDs, leading to LD enlargement, as well as to fusion of LDs to the ER.
Degradation of PLIN2, which is sensitive to inhibitors of ubiquitination, autophagy, and protein synthesis.
Rab18-dependent association of ApoB 100 with LDs.
Free cholesterol accumulation in the liver is relevant to the pathogenesis of NASH (Bashiri et al., 2013; Musso et al., 2013; Arguello et al., 2015). Emerging evidence indicates a physical and metabolic connection linking LDs with the ER (Barbosa et al., 2015). The present results highlight the role of LDs as targets of free cholesterol toxicity in hepatocytes.
MATERIALS AND METHODS
Cell line and reagents
The human hepatocellular carcinoma cell line Huh7 was obtained from the Japanese Collection of Research Biosources Cell Bank (Tokyo, Japan). Cells were cultured in DMEM supplemented with 10% fetal calf serum (FCS), 100 U/ml penicillin, and 100 μg/ml streptomycin. F1394 was provided by Fujirebio (Tokyo, Japan; Kusunoki et al., 1995). MβCD/cholesterol complex (cholesterol water soluble), 58-035, MG132, ALLN, and 3-MA were purchased from Sigma-Aldrich (St. Louis, MO). Mouse monoclonal anti-human ApoB 100 (clone 5F8) was from MONOSAN (Uden, Netherlands). Goat polyclonal anti-human ApoB 100 was from Rockland (Gilbertsville, PA). Mouse anti-PLIN1 and mouse anti-PLIN2 were purchased from Progen (Brisbane, Australia). BODIPY 493/503, ER-Tracker Green, and mouse anti-PDI BODIPY FL were from Invitrogen (Carlsbad, CA). [1-14C]Oleic acid was from PerkinElmer (Waltham, MA). Filipin was purchased from Polysciences (Warrington, PA). Anti-BMP (also called LBPA) monoclonal antibody was prepared as described (Kobayashi et al., 1998). Anti-Rab18 rabbit antibody was prepared as reported (Ozeki et al., 2005). The antibody does not react with Rab1, Rab2, Rab3, Rab5, Rab7, Rab9, and Rab10 by immunofluorescence microscopy and Western blotting using cells transfected with the respective Rab cDNAs.
Treatment of cells with ACAT inhibitors
Huh7 cells were grown in DMEM supplemented with 10% FCS, 100 U/ml penicillin, and 100 μg/ml streptomycin in the presence of F1394 (10 μg/ml) or 58-035 (10 μg/ml). Alternatively, cells were grown in DMEM supplemented with 5% lipoprotein-depleted serum (LPDS), 100 U/ml penicillin, and 100 μg/ml streptomycin in the presence of MβCD/cholesterol complex at a final cholesterol concentration of 0.1 mM or MβCD/cholesterol and 58-035.
Treatment of cells with MTP inhibitor
Huh7 cells were grown for 24 h in DMEM supplemented with 10% FCS, 100 U/ml penicillin, and 100 μg/ml streptomycin in the presence of 100 nM BAY13-9952 (Bayer Healthcare). Culture medium was then changed to DMEM supplemented with 10% FCS, 100 U/ml penicillin, and 100 μg/ml streptomycin in the presence of 100 nM BAY13-9952 and 10 μg/ml F1394, and cells were further grown for 24 h.
Fluorescence microscopy
Cells grown on coverslips were washed with phosphate-buffered saline (PBS) and then fixed for 30 min with 3% paraformaldehyde in PBS. The cells were washed with PBS and quenched with 50 mM NH4Cl for 5 min. After being washed with PBS, cells were permeabilized by treatment with 50 μg/ml digitonin or 0.1% Triton X-100 for 10 min. Alternatively, the coverslips were dipped in liquid nitrogen for 3 s (freeze-thaw). The specimens were blocked with 0.1% bovine serum albumin in PBS for 30 min. After 1 h of treatment with the first antibody, cells were washed and labeled with the fluorescent second antibody for 1 h. When required, 2 μg/ml BODIPY 493/503 was added to the second antibody. The stained cells were washed with PBS, mounted in Mowiol, and examined under a Zeiss LSM 510 or Zeiss LSM 700 confocal microscope equipped with C-Apochromat 63×W (1.2 numerical aperture [NA]) or Plan-Apochromat 63× (1.40 NA) oil objectives (Carl Zeiss, Jena, Germany). Size and number of lipid droplets were quantified using MetaMorph software (Molecular Devices, Sunnyvale, CA).
To follow the dynamics of lipid droplets, Huh7 cells were incubated in DMEM with 10% (vol/vol) fetal bovine serum plus 20 ng/ml Nile red at 37°C in a humidified chamber equilibrated at 5% CO2. Time-lapse sequences were recorded at 15-min intervals on an Olympus FV1000 confocal microscope (Olympus, Tokyo, Japan) using a Plan-Apo 60× (1.10 NA) oil immersion objective.
Thin-layer chromatography
Lipids were extracted (Bligh and Dyer, 1959) from Huh7 cells and applied to HPTLC plates (Merck, Darmstadt, Germany). The plates were developed in hexane/diethyl ether/acetic acid (80:20:2). Lipids were stained with a mixture of ferric chloride/sulfuric acid/acetic acid (Lowry, 1968) and then heated and quantified by scanning densitometry using LAS 2000 (Fujifilm, Tokyo, Japan).
Cholesterol esterification assay
Cells were treated overnight with ACAT inhibitors in complete medium. After a DMEM wash, cells were labeled for 4 h with [1-14C]oleic acid (0.5 μCi/ml) in DMEM plus 1% fetal bovine serum in the presence of the inhibitors. After a PBS wash, the cell layer was scraped. Aliquots of cell extract were taken for protein quantification, and lipids were extracted and then separated on HPTLC as described. Lipids were identified by comparison with radioactive lipid standards and visualized with a Fujifilm Imaging Plate (BAS-SR2025). The relative radioactivity was quantified using the BAS5000 Bioimaging Analyzer (Fujifilm).
Analysis of cholesterol content by gas chromatography
Lipids were extracted from cells, and stigmasterol (5 μg) was added as internal standard. Lipids were separated by HPTLC as described. Free cholesterol spots were revealed after spraying primuline solution (0.1% [wt/vol] primuline solution/water/acetone 1:1:8 [vol/vol/vol]), scraped, and then extracted with 2 ml of methanol, 1 ml of water, and 2 ml of hexane. After centrifugation, the upper hexane phase was collected, dried under nitrogen, and analyzed by GC.
In the case of esterified cholesterol (EC) spots, stigmasterol (2.5 μg) was added as internal standard during the methanol:water:hexane extraction. Saponification of EC samples was performed with 5 N methanolic KOH (2 ml) for 1 h at 80°C. The reaction was stopped on ice and neutralized with 1 N HCl (1.5 ml). Then extraction was performed with 2 ml of hexane, and 0.5 ml of water was added to favor phase separation. After centrifugation, the upper hexane phase was collected, dried under nitrogen, and used for GC analysis.
Cholesterol was analyzed with a Shimadzu GC-14AH gas chromatograph (Shimadzu, Kyoto, Japan). An Alltech Econo-Cap EC-5 (Analytical Columns, New Addington, UK) capillary column (30 m × 0.32 mm, 0.25 mm) was used with helium as carrier gas and an oven temperature program from 290 to 320°C at 2°C /min, held isothermal at 320°C for 10 min. Cholesterol was quantified using stigmasterol as the internal standard.
Western blot analysis
To detect PLIN2, Huh7 cells were lysed for 3 min in ice-cold RIPA buffer containing protease inhibitors. Cells were centrifuged at 15,000 rpm for 5 min, and the supernatant was electrophoresed on 10% SDS–polyacrylamide gel. A 6% gel was used to detect ApoB. The gels were electrotransferred to a polyvinylidene fluoride membrane using a Bio-Rad semidry transfer system (Bio-Rad, Hercules, CA). For detection of PLIN2 or ApoB, anti-PLIN2 or anti-ApoB was used. After incubation with horseradish peroxidase–conjugated second antibodies, the membranes were developed using ECL Plus (GE Healthcare, Pittsburgh, PA).
Isolation of lipid droplet fraction from Huh7 cells
Huh7 cells were cultured in presence of F1394 for 24 h and washed with PBS. Cells were scraped and homogenized in ice-cold homogenization buffer (HB; 250 mM sucrose [8.5%], 3 mM imidazole, pH 7.4) containing protease inhibitors, and then a postnuclear supernatant (PNS) was prepared. The PNS was adjusted to 40.6% sucrose solution in 3 mM imidazole, pH 7.4, loaded at the bottom of tube, and overlaid sequentially with 35 and 25% sucrose solutions in 3 mM imidazole, pH 7.4, and then in HB. The gradient was centrifuged for 60 min at 35,000 rpm using an MLS 55 rotor on a Beckman Coulter (Brea) Optima TL100 ultracentrifuge. The LD fraction was collected from the top of tube. Enrichment of LDs in the fraction was assessed by the enrichment of PLIN2 in control cells.
Transmission electron microscopy
Transmission electron microscopy was performed as reported previously (Makino et al., 2006). For ultrastructural examination of F1394-treated cells, the cells were grown on Aclar plastic sheets (Nissin EM, Tokyo, Japan) with or without 10 μg/ml F1394 for 1 d, fixed in 4% paraformaldehyde and 2.5% glutaraldehyde in 60 mM PIPES, 25 mM HEPES, 10 mM EGTA, and 2 mM MgCl2, pH 6.9 (PHEM buffer), postfixed with 1% osmium tetraoxide, and then stained with 1% tannic acid. The samples were then dehydrated in the graded series of ethanol, en bloc stained with uranyl acetate in 70% ethanol, embedded in Araldite resin, and sectioned with an ultramicrotome (EM UC6; Leica, Vienna, Austria). Ultrathin sections were stained with uranyl acetate and lead stain solution (Sigma-Aldrich Japan, Tokyo, Japan) and observed under a transmission electron microscope (1200EX II; JEOL, Tokyo, Japan). Electron micrographs recorded on imaging plates were scanned and digitized by an FDL 5000 imaging system (Fujifilm). For immunoelectron microscopy, cells were grown on culture dishes and fixed with 4% paraformaldehyde and 0.05% glutaraldehyde in PHEM buffer. The cells were scraped off the dishes, pelleted, and embedded in gelatin. Cell pellets were infused with 20% polyvinylpyrrolidone and 1.84 M sucrose and frozen in liquid nitrogen. Ultrathin sections were cut and picked up in 1.15 M sucrose and 1% methylcellulose. Labeling of PLIN2 was performed with mouse anti-PLIN2 antibody and 5 nm colloidal gold conjugated–anti-mouse immunoglobulin G antibody (Amersham Biosciences, Amersham, United Kingdom), and ApoB was labeled with goat anti-ApoB antibody and 10 nm gold-conjugated secondary antibody (British BioCell International, Cardiff, United Kingdom). After labeling, the sections were stained and embedded in 1.6% methylcellulose and TI-blue staining solution (Nissin EM). Both sections for conventional and immunoelectron microscopy were observed under a transmission electron microscope (JEOL 1200EX II) with the help of the Support Unit for Neuromorphological Analysis in RIKEN. Electron micrographs recorded on imaging plates were scanned and digitized by an FDL 5000 imaging system (Fujifilm).
Construct
Full-length PLIN2 and Rab18 were amplified by PCR out of a human cDNA library and cloned into pENTR/SD/d-TOPO (Invitrogen). Using a linker, KpnI and HindIII sites were inserted just before the attR1 recombination site of pcDNA-DEST40. AcGFP was amplified by PCR using pAcGFP (Clontech) as a template, introduced between the KpnI and HindIII sites, and generated pDEST-AcGFP. pDEST-mCherry was also prepared. The PLIN2 and Rab18 genes were introduced into pDEST-AcGFP or pDEST-mCherry by LR recombination (Invitrogen). GFP- and mCherry-Rab18 S22N were constructed by introducing the EcoRI site into GFP-Rab18 or mCherry-Rab18 by PCR amplification with 5′-CCGGAATTCCCTGCTCTTGAGGTTCACAGA-3′ and 5′-CCGGAATTCTTGCCCAC-CCCACTCTCGCCG-3′. GFP- and mCherry-Rab18 Q67L were constructed by introducing the XhoI site into GFP- and mCherry-Rab18 by PCR amplification with 5′-CCGCTCGAGAGGTTTAGAACATTAACTCCC-3′ and 5′-CCGCTCGAGACCAGCAGTATCCCAT-ATTGC-3′. mCherry-GDIα was prepared by introducing full-length mouse GDIα (Takahashi et al., 2007) into pDEST-mCherry. The DNA sequence was verified.
Transfection
Huh7 cells were transfected with 1 μg of plasmid DNA and 6 μl of Polyfect transduction reagent (Qiagen, Venlo, Netherlands) using the manufacturer’s protocol. After 24 h of incubation, cells were treated with 10 μg/ml F1394 and incubated for an additional 24 h.
Creation of clustered regularly interspaced short palindromic repeats–mediated cell lines
Huh7 ACAT1 or ACAT2 stable knockouts were generated using a clustered regularly interspaced short palindromic repeats (CRISPR) technique (Ran et al., 2013). The sgRNAs were designed using the CRISPR.mit.edu design software. Oligonucleotides (5′-caccgGCCTCTCAAAGTCTTATGTG-3′ and 5′-aaacCACATAAGACTTTGA-GAGGCc-3′ for ACAT1; 5′-caccgGCAGTCCAGTCAATAGGGAT-3′ and 5′-aaacATCCCTATTGACTGGACTGCc-3′ for ACAT2) were annealed and cloned into the BbsI site of the pSpCas9(BB)-2A-puro vector (plasmid 48139; Addgene). The plasmids were introduced into Huh7 cells using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA). After 3 d, single cells were selected with puromycin, and individual clones were expanded to cell lines. Genomic DNA was extracted from the cell lines using GenElute Mammalian Genomic DNA Miniprep Kits (Sigma-Aldrich). PCR-amplified products were directly sequenced using the Big Dye Terminator Cycle Sequencing v3.1 Kit (PE Applied Biosystems, Foster City, CA). Sequence analysis revealed that Huh7 ACAT1 knockout had a 1–base pair insertion and Huh7 ACAT2 knockout had 2– and 13–base pair heterogeneous deletions next to PAM.
Statistical analysis
The p values were calculated using paired Student’s t test, with p < 0.05 being statistically significant. Values are mean ± SD.
Supplementary Material
Acknowledgments
This work was supported by the Integrated Lipidology Program of RIKEN and Grants in Aid for Scientific Research 23590251 and 15K08167 (to M.M.) and 22390018 and 25293015 (to T.K.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Abbreviations used:
- 3-MA
3-methyladenine
- ACAT
acyl-CoA:cholesterol acyltransferase
- ALLN
N-acetyl-l-leucinyl-l-leucinyl-l-norleucinal; ApoB, apolipoprotein B
- ER
endoplasmic reticulum
- F1394
(1s,2s)-2-[3-(2,2-dimethylpropyl)-3-nonylureido]aminocyclohexane-1-yl-3-[N-(2,2,5,5-tetramethyl-1,3-dioxane-4-carbonyl)amino]propionate
- GDI
GDP-dissociation inhibitor
- HMG CoA
3-hydoxy-3-methylglutaryl-CoA
- LD
lipid droplet
- MβCD
methyl-β-cyclodextrin
- MTP
microsomal triglyceride-transfer protein
- NASH
nonalcoholic steatohepatitis
- PBS
phosphate-buffered saline
- PDI
protein disulfide isomerase
- PLIN
perilipin
- PNS
postnuclear supernatant
- VLDL
very low-density lipoprotein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-10-0730) on August 31, 2016.
REFERENCES
- Arguello G, Balboa E, Arrese M, Zanlungo S. Recent insights on the role of cholesterol in non-alcoholic fatty liver disease. Biochim Biophys Acta. 2015;1852:1765–1778. doi: 10.1016/j.bbadis.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Ashraf NU, Sheikh TA. Endoplasmic reticulum stress and oxidative stress in the pathogenesis of non-alcoholic fatty liver disease. Free Radic Res. 2015;49:1405–1418. doi: 10.3109/10715762.2015.1078461. [DOI] [PubMed] [Google Scholar]
- Barbosa AD, Savage DB, Siniossoglou S. Lipid droplet-organelle interactions: emerging roles in lipid metabolism. Curr Opin Cell Biol. 2015;35:91–97. doi: 10.1016/j.ceb.2015.04.017. [DOI] [PubMed] [Google Scholar]
- Bartz R, Li WH, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RG, Liu P, Chapman KD. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res. 2007;48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- Bashiri A, Tavallaee G, Li L, Ng DS. Emerging role of cellular cholesterol in the pathogenesis of nonalcoholic fatty liver disease. Curr Opin Lipid. 2013;24:275–276. doi: 10.1097/MOL.0b013e328361633e. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Buhman KK, Accad M, Novak S, Choi RS, Wong JS, Hamilton RL, Turley S, Farese RV., Jr Resistance to diet-induced hypercholesterolemia and gallstone formation in ACAT2-deficient mice. Nat Med. 2000;6:1341–1347. doi: 10.1038/82153. [DOI] [PubMed] [Google Scholar]
- Chun KT, Bar-Nun S, Simoni RD. The regulated degradation of 3-hydroxy-3-methylglutaryl-CoA reductase requires a short-lived protein and occurs in the endoplasmic reticulum. J Biol Chem. 1990;265:22004–22010. [PubMed] [Google Scholar]
- Elias PM, Goerke J, Friend DS, Brown BE. Freeze-fracture identification of sterol-digitonin complexes in cell and liposome membranes. J Cell Biol. 1978;78:577–596. doi: 10.1083/jcb.78.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto T, Parton RG. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb Perspect Biol. 2011;3:a004838. doi: 10.1101/cshperspect.a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S, Stevenson J, Kristiana I, Brown AJ. Cholesterol-dependent degradation of squalene monooxygenase, a control point in cholesterol synthesis beyond HMG-CoA reductase. Cell Metab. 2011;13:260–273. doi: 10.1016/j.cmet.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Gocze PM, Freeman DA. Factors underlying the variability of lipid droplet fluorescence in MA-10 Leydig tumor cells. Cytometry. 1994;17:151–158. doi: 10.1002/cyto.990170207. [DOI] [PubMed] [Google Scholar]
- Jo Y, Debose-Boyd RA. Control of cholesterol synthesis through regulated ER-associated degradation of HMG CoA reductase. Crit Rev Biochem Mol Biol. 2010;45:185–198. doi: 10.3109/10409238.2010.485605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Stang E, Fang KS, de Moerloose P, Parton RG, Gruenberg J. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 1998;392:193–197. doi: 10.1038/32440. [DOI] [PubMed] [Google Scholar]
- Krahmer N, Farese RV, Jr, Walther TC. Balancing the fat: lipid droplets and human disease. EMBO Mol Med. 2013;5:905–915. doi: 10.1002/emmm.201100671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N, Guo Y, Wilfling F, Hilger M, Lingrell S, Heger K, Newman HW, Schmidt-Supprian M, Vance DE, Mann M, et al. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 2011;14:504–515. doi: 10.1016/j.cmet.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusunoki J, Aragane K, Yamaura T, Ohnishi H. Studies on acyl-CoA: cholesterol acyltransferase (ACAT) inhibitory effects and enzyme selectivity of F-1394, a pantotheic acid derivative. Jpn J Pharmacol. 1995;67:195–203. doi: 10.1254/jjp.67.195. [DOI] [PubMed] [Google Scholar]
- Lehner R, Lian J, Quiroga AD. Lumenal lipid metabolism: implications for lipoprotein assembly. Arterioscler Thromb Vasc Biol. 2012;32:1087–1093. doi: 10.1161/ATVBAHA.111.241497. [DOI] [PubMed] [Google Scholar]
- Lowry RR. Ferric chloride spray detector for cholesterol and cholesteryl esters on thin-layer chromatograms. J Lipid Res. 1968;9:397. [PubMed] [Google Scholar]
- Makino A, Ishii K, Murate M, Hayakawa T, Suzuki Y, Suzuki M, Ito K, Fujisawa T, Matsuo H, Ishitsuka R, Kobayashi T. d-threo-1-Phenyl-2-decanoylamino-3-morpholino-1-propanol alters cellular cholesterol homeostasis by modulating the endosome lipid domains. Biochemistry. 2006;45:4530–4541. doi: 10.1021/bi052104y. [DOI] [PubMed] [Google Scholar]
- Mari M, Caballero F, Colell A, Morales A, Caballeria J, Fernandez A, Enrich C, Fernandez-Checa JC, Garcia-Ruiz C. Mitochondrial free cholesterol loading sensitizes to TNF- and Fas-mediated steatohepatitis. Cell Metab. 2006;4:185–198. doi: 10.1016/j.cmet.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Martin S, Driessen K, Nixon SJ, Zerial M, Parton RG. Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J Biol Chem. 2005;280:42325–42335. doi: 10.1074/jbc.M506651200. [DOI] [PubMed] [Google Scholar]
- Masuda Y, Itabe H, Odaki M, Hama K, Fujimoto Y, Mori M, Sasabe N, Aoki J, Arai H, Takano T. ADRP/adipophilin is degraded through the proteasome-dependent pathway during regression of lipid-storing cells. J Lipid Res. 2006;47:87–98. doi: 10.1194/jlr.M500170-JLR200. [DOI] [PubMed] [Google Scholar]
- Musso G, Gambino R, Cassader M. Cholesterol metabolism and the pathogenesis of non-alcoholic steatohepatitis. Progress Lipid Res. 2013;52:175–191. doi: 10.1016/j.plipres.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Namatame I, Tomoda H, Ishibashi S, Omura S. Antiatherogenic activity of fungal beauveriolides, inhibitors of lipid droplet accumulation in macrophages. Proc Natl Acad Sci USA. 2004;101:737–742. doi: 10.1073/pnas.0307757100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y, Cheng J, Fujita A, Tokumoto T, Fujimoto T. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol Biol Cell. 2006;17:2674–2683. doi: 10.1091/mbc.E05-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y, Cheng J, Suzuki M, Fujita A, Fujimoto T. Lipid droplets are arrested in the ER membrane by tight binding of lipidated apolipoprotein B-100. J Cell Sci. 2008;121:2415–2422. doi: 10.1242/jcs.025452. [DOI] [PubMed] [Google Scholar]
- Orlicky DJ, Degala G, Greenwood C, Bales ES, Russell TD, McManaman JL. Multiple functions encoded by the N-terminal PAT domain of adipophilin. J Cell Sci. 2008;121:2921–2929. doi: 10.1242/jcs.026153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki S, Cheng J, Tauchi-Sato K, Hatano N, Taniguchi H, Fujimoto T. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J Cell Sci. 2005;118:2601–2611. doi: 10.1242/jcs.02401. [DOI] [PubMed] [Google Scholar]
- Parini P, Davis M, Lada AT, Erickson SK, Wright TL, Gustafsson U, Sahlin S, Einarsson C, Eriksson M, Angelin B, et al. ACAT2 is localized to hepatocytes and is the major cholesterol-esterifying enzyme in human liver. Circulation. 2004;110:2017–2023. doi: 10.1161/01.CIR.0000143163.76212.0B. [DOI] [PubMed] [Google Scholar]
- Pol A, Gross SP, Parton RG. Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J Cell Biol. 2014;204:635–646. doi: 10.1083/jcb.201311051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prattes S, Horl G, Hammer A, Blaschitz A, Graier WF, Sattler W, Zechner R, Steyrer E. Intracellular distribution and mobilization of unesterified cholesterol in adipocytes: triglyceride droplets are surrounded by cholesterol-rich ER-like surface layer structures. J Cell Sci. 2000;113:2977–2989. doi: 10.1242/jcs.113.17.2977. [DOI] [PubMed] [Google Scholar]
- Pulido MR, Diaz-Ruiz A, Jimenez-Gomez Y, Garcia-Navarro S, Gracia-Navarro F, Tinahones F, Lopez-Miranda J, Fruhbeck G, Vazquez-Martinez R, Malagon MM. Rab18 dynamics in adipocytes in relation to lipogenesis, lipolysis and obesity. PLoS One. 2011;6:e22931. doi: 10.1371/journal.pone.0022931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Doolman R, Avner R, Harats D, Roitelman J. The ubiquitin-proteasome pathway mediates the regulated degradation of mammalian 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem. 2000;275:35840–35847. doi: 10.1074/jbc.M004793200. [DOI] [PubMed] [Google Scholar]
- Ross AC, Go KJ, Heider JG, Rothblat GH. Selective inhibition of acyl coenzyme A:cholesterol acyltransferase by compound 58-035. J Biol Chem. 1984;259:815–819. [PubMed] [Google Scholar]
- Sahini N, Borlak J. Recent insights into the molecular pathophysiology of lipid droplet formation in hepatocytes. Progress Lipid Res. 2014;54:86–112. doi: 10.1016/j.plipres.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Saito H, Kawagishi A, Tanaka M, Tanimoto T, Okada S, Komatsu H, Handa T. Coalescence of lipid emulsions in floating and freeze-thawing processes: examination of the coalescence transition state theory. J Colloid Interface Sci. 1999;219:129–134. doi: 10.1006/jcis.1999.6452. [DOI] [PubMed] [Google Scholar]
- Segev N. GTPases in intracellular trafficking: an overview. Semin Cell Dev Biol. 2011;22:1–2. doi: 10.1016/j.semcdb.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Sztalryd C, Kimmel AR. Perilipins: lipid droplet coat proteins adapted for tissue-specific energy storage and utilization, and lipid cytoprotection. Biochimie. 2014;96:96–101. doi: 10.1016/j.biochi.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Murate M, Fukuda M, Sato SB, Ohta A, Kobayashi T. Cholesterol controls lipid endocytosis through Rab11. Mol Biol Cell. 2007;18:2667–2677. doi: 10.1091/mbc.E06-10-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J Biol Chem. 2002;277:44507–44512. doi: 10.1074/jbc.M207712200. [DOI] [PubMed] [Google Scholar]
- Xu G, Sztalryd C, Lu X, Tansey JT, Gan J, Dorward H, Kimmel AR, Londos C. Post-translational regulation of adipose differentiation-related protein by the ubiquitin/proteasome pathway. J Biol Chem. 2005;280:42841–42847. doi: 10.1074/jbc.M506569200. [DOI] [PubMed] [Google Scholar]
- Yang H, Galea A, Sytnyk V, Crossley M. Controlling the size of lipid droplets: lipid and protein factors. Curr Opin Cell Biol. 2012;24:509–516. doi: 10.1016/j.ceb.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Yao Z, Zhou H, Figeys D, Wang Y, Sundaram M. Microsome-associated lumenal lipid droplets in the regulation of lipoprotein secretion. Curr Opin Lipid. 2013;24:160–170. doi: 10.1097/MOL.0b013e32835aebe7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.