Abstract
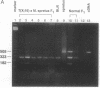
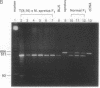
ZFX, an X chromosome-linked gene encoding a zinc-finger protein, has previously been shown to escape X inactivation in humans. Here we report studies of the inactivation status of the homolog, Zfx, on the mouse X chromosome. We took advantage of both the preferential inactivation of the normal X chromosome in females carrying the T(X;16)16H translocation and the high degree of nucleotide sequence variation between the laboratory strain of mouse [corrected] and Mus spretus genomes. An EcoRV restriction fragment difference between laboratory strain of mouse [corrected] and M. spretus was detected after amplification of Zfx transcripts using the reverse transcription-polymerase chain reaction. Using this allelic variation, we assessed expression of the two Zfx genes in females carrying the T(X;16)16H translocation (from laboratory strain of mouse [corrected]) and an intact X chromosome (from M. spretus). Such females exhibit Zfx transcription from the active laboratory strain of mouse [corrected] chromosome but not from the inactive M. spretus chromosome. These results indicate that the mouse Zfx gene is subject to X inactivation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Affara N. A., Chambers D., O'Brien J., Habeebu S. S., Kalaitsidaki M., Bishop C. E., Ferguson-Smith M. A. Evidence for distinguishable transcripts of the putative testis determining gene (ZFY) and mapping of homologous cDNA sequences to chromosomes X,Y and 9. Nucleic Acids Res. 1989 Apr 25;17(8):2987–2999. doi: 10.1093/nar/17.8.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth A., Skene B., Swift S., Lovell-Badge R. Zfa is an expressed retroposon derived from an alternative transcript of the Zfx gene. EMBO J. 1990 May;9(5):1529–1534. doi: 10.1002/j.1460-2075.1990.tb08271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M. A., Heidler S. A., Bell G. I., Seino S., Le Beau M. M., Westbrook C. A., Neuman W., Shapiro L. J., Mohandas T. K., Roessler B. J. Cloning of cDNAs for human phosphoribosylpyrophosphate synthetases 1 and 2 and X chromosome localization of PRPS1 and PRPS2 genes. Genomics. 1990 Nov;8(3):555–561. doi: 10.1016/0888-7543(90)90043-t. [DOI] [PubMed] [Google Scholar]
- Brown C. J., Willard H. F. Localization of a gene that escapes inactivation to the X chromosome proximal short arm: implications for X inactivation. Am J Hum Genet. 1990 Feb;46(2):273–279. [PMC free article] [PubMed] [Google Scholar]
- Brown C. J., Willard H. F. Noninactivation of a selectable human X-linked gene that complements a murine temperature-sensitive cell cycle defect. Am J Hum Genet. 1989 Oct;45(4):592–598. [PMC free article] [PubMed] [Google Scholar]
- Cattanach B. M., Williams C. E. Evidence of non-random X chromosome activity in the mouse. Genet Res. 1972 Jun;19(3):229–240. doi: 10.1017/s001667230001449x. [DOI] [PubMed] [Google Scholar]
- Chapman V. M., Kratzer P. G., Quarantillo B. A. Electrophoretic variation for X chromosome-linked hypoxanthine phosphoribosyl transferase (HPRT) in wild-derived mice. Genetics. 1983 Apr;103(4):785–795. doi: 10.1093/genetics/103.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman V. M., Kratzer P. G., Siracusa L. D., Quarantillo B. A., Evans R., Liskay R. M. Evidence for DNA modification in the maintenance of X-chromosome inactivation of adult mouse tissues. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5357–5361. doi: 10.1073/pnas.79.17.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Disteche C. M., Eicher E. M., Latt S. A. Late replication patterns in adult and embryonic mice carrying Searle's X-autosome translocation. Exp Cell Res. 1981 Jun;133(2):357–362. doi: 10.1016/0014-4827(81)90328-1. [DOI] [PubMed] [Google Scholar]
- Epstein C. J., Smith S., Travis B., Tucker G. Both X chromosomes function before visible X-chromosome inactivation in female mouse embryos. Nature. 1978 Aug 3;274(5670):500–503. doi: 10.1038/274500a0. [DOI] [PubMed] [Google Scholar]
- Fialkow P. J. X-chromosome inactivation and the Xg locus. Am J Hum Genet. 1970 Jul;22(4):460–463. [PMC free article] [PubMed] [Google Scholar]
- Fisher E. M., Beer-Romero P., Brown L. G., Ridley A., McNeil J. A., Lawrence J. B., Willard H. F., Bieber F. R., Page D. C. Homologous ribosomal protein genes on the human X and Y chromosomes: escape from X inactivation and possible implications for Turner syndrome. Cell. 1990 Dec 21;63(6):1205–1218. doi: 10.1016/0092-8674(90)90416-c. [DOI] [PubMed] [Google Scholar]
- GORMAN J. G., DI RE J., TREACY A. M., CAHAN A. The application of -Xga antiserum to the question of red cell mosaicism in female heterozygotes. J Lab Clin Med. 1963 Apr;61:642–649. [PubMed] [Google Scholar]
- Goodfellow P. J., Pritchard C., Tippett P., Goodfellow P. N. Recombination between the X and Y chromosomes: implications for the relationship between MIC2, XG and YG. Ann Hum Genet. 1987 May;51(Pt 2):161–167. doi: 10.1111/j.1469-1809.1987.tb01058.x. [DOI] [PubMed] [Google Scholar]
- Goodfellow P., Pym B., Mohandas T., Shapiro L. J. The cell surface antigen locus, MIC2X, escapes X-inactivation. Am J Hum Genet. 1984 Jul;36(4):777–782. [PMC free article] [PubMed] [Google Scholar]
- Johnson G. G., Kronert W. A., Bernstein S. I., Chapman V. M., Smith K. D. Altered turnover of allelic variants of hypoxanthine phosphoribosyltransferase is associated with N-terminal amino acid sequence variation. J Biol Chem. 1988 Jul 5;263(19):9079–9082. [PubMed] [Google Scholar]
- Kawasaki E. S., Clark S. S., Coyne M. Y., Smith S. D., Champlin R., Witte O. N., McCormick F. P. Diagnosis of chronic myeloid and acute lymphocytic leukemias by detection of leukemia-specific mRNA sequences amplified in vitro. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5698–5702. doi: 10.1073/pnas.85.15.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keitges E., Gartler S. M. Dosage of the Sts gene in the mouse. Am J Hum Genet. 1986 Oct;39(4):470–476. [PMC free article] [PubMed] [Google Scholar]
- Keitges E., Rivest M., Siniscalco M., Gartler S. M. X-linkage of steroid sulphatase in the mouse is evidence for a functional Y-linked allele. Nature. 1985 May 16;315(6016):226–227. doi: 10.1038/315226a0. [DOI] [PubMed] [Google Scholar]
- Konecki D. S., Brennand J., Fuscoe J. C., Caskey C. T., Chinault A. C. Hypoxanthine-guanine phosphoribosyltransferase genes of mouse and Chinese hamster: construction and sequence analysis of cDNA recombinants. Nucleic Acids Res. 1982 Nov 11;10(21):6763–6775. doi: 10.1093/nar/10.21.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman P., Gubbay J., Collignon J., Lovell-Badge R. Zfy gene expression patterns are not compatible with a primary role in mouse sex determination. Nature. 1989 Dec 21;342(6252):940–942. doi: 10.1038/342940a0. [DOI] [PubMed] [Google Scholar]
- Kratzer P. G., Gartler S. M. HGPRT activity changes in preimplantation mouse embryos. Nature. 1978 Aug 3;274(5670):503–504. doi: 10.1038/274503a0. [DOI] [PubMed] [Google Scholar]
- Krumlauf R., Chapman V. M., Hammer R. E., Brinster R., Tilghman S. M. Differential expression of alpha-fetoprotein genes on the inactive X chromosome in extraembryonic and somatic tissues of a transgenic mouse line. Nature. 1986 Jan 16;319(6050):224–226. doi: 10.1038/319224a0. [DOI] [PubMed] [Google Scholar]
- LYON M. F. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature. 1961 Apr 22;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- LYON M. F., SEARLE A. G., FORD C. E., OHNO S. A MOUSE TRANSLOCATION SUPPRESSING SEX-LINKED VARIEGATION. Cytogenetics. 1964;3:306–323. doi: 10.1159/000129820. [DOI] [PubMed] [Google Scholar]
- Labarca C., Paigen K. A simple, rapid, and sensitive DNA assay procedure. Anal Biochem. 1980 Mar 1;102(2):344–352. doi: 10.1016/0003-2697(80)90165-7. [DOI] [PubMed] [Google Scholar]
- Mardon G., Luoh S. W., Simpson E. M., Gill G., Brown L. G., Page D. C. Mouse Zfx protein is similar to Zfy-2: each contains an acidic activating domain and 13 zinc fingers. Mol Cell Biol. 1990 Feb;10(2):681–688. doi: 10.1128/mcb.10.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon G., Page D. C. The sex-determining region of the mouse Y chromosome encodes a protein with a highly acidic domain and 13 zinc fingers. Cell. 1989 Mar 10;56(5):765–770. doi: 10.1016/0092-8674(89)90680-6. [DOI] [PubMed] [Google Scholar]
- Mitchell M., Simon D., Affara N., Ferguson-Smith M., Avner P., Bishop C. Localization of murine X and autosomal sequences homologous to the human Y located testis-determining region. Genetics. 1989 Apr;121(4):803–809. doi: 10.1093/genetics/121.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins L. J., Grant S. G., Stephenson D. A., Chapman V. M. Multilocus molecular mapping of the mouse X chromosome. Genomics. 1988 Oct;3(3):187–194. doi: 10.1016/0888-7543(88)90078-x. [DOI] [PubMed] [Google Scholar]
- Müller G., Schempp W. Mapping the human ZFX locus to Xp21.3 by in situ hybridization. Hum Genet. 1989 Apr;82(1):82–84. doi: 10.1007/BF00288279. [DOI] [PubMed] [Google Scholar]
- Nagamine C. M., Chan K. M., Kozak C. A., Lau Y. F. Chromosome mapping and expression of a putative testis-determining gene in mouse. Science. 1989 Jan 6;243(4887):80–83. doi: 10.1126/science.2563174. [DOI] [PubMed] [Google Scholar]
- Nagamine C. M., Chan K., Hake L. E., Lau Y. F. The two candidate testis-determining Y genes (Zfy-1 and Zfy-2) are differentially expressed in fetal and adult mouse tissues. Genes Dev. 1990 Jan;4(1):63–74. doi: 10.1101/gad.4.1.63. [DOI] [PubMed] [Google Scholar]
- OHNO S., LYON M. F. CYTOLOGICAL STUDY OF SEARLE'S X-AUTOSOME TRANSLOCATION IN MUS MUSCULUS. Chromosoma. 1965 Jan 30;16:90–100. doi: 10.1007/BF00320564. [DOI] [PubMed] [Google Scholar]
- Page D. C., Disteche C. M., Simpson E. M., de la Chapelle A., Andersson M., Alitalo T., Brown L. G., Green P., Akots G. Chromosomal localization of ZFX--a human gene that escapes X inactivation--and its murine homologs. Genomics. 1990 May;7(1):37–46. doi: 10.1016/0888-7543(90)90516-w. [DOI] [PubMed] [Google Scholar]
- Page D. C., Mosher R., Simpson E. M., Fisher E. M., Mardon G., Pollack J., McGillivray B., de la Chapelle A., Brown L. G. The sex-determining region of the human Y chromosome encodes a finger protein. Cell. 1987 Dec 24;51(6):1091–1104. doi: 10.1016/0092-8674(87)90595-2. [DOI] [PubMed] [Google Scholar]
- Palmer M. S., Berta P., Sinclair A. H., Pym B., Goodfellow P. N. Comparison of human ZFY and ZFX transcripts. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1681–1685. doi: 10.1073/pnas.87.5.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUSSELL L. B. Genetics of mammalian sex chromosomes. Science. 1961 Jun 9;133(3467):1795–1803. doi: 10.1126/science.133.3467.1795. [DOI] [PubMed] [Google Scholar]
- Russell L. B., Cacheiro N. L. The use of mouse X-autosome translocations in the study of X-inactivation pathways and nonrandomness. Basic Life Sci. 1978;12:393–416. doi: 10.1007/978-1-4684-3390-6_27. [DOI] [PubMed] [Google Scholar]
- Schneider-Gädicke A., Beer-Romero P., Brown L. G., Nussbaum R., Page D. C. ZFX has a gene structure similar to ZFY, the putative human sex determinant, and escapes X inactivation. Cell. 1989 Jun 30;57(7):1247–1258. doi: 10.1016/0092-8674(89)90061-5. [DOI] [PubMed] [Google Scholar]
- Shapiro L. J., Mohandas T., Weiss R., Romeo G. Non-inactivation of an x-chromosome locus in man. Science. 1979 Jun 15;204(4398):1224–1226. doi: 10.1126/science.156396. [DOI] [PubMed] [Google Scholar]
- Sinclair A. H., Foster J. W., Spencer J. A., Page D. C., Palmer M., Goodfellow P. N., Graves J. A. Sequences homologous to ZFY, a candidate human sex-determining gene, are autosomal in marsupials. Nature. 1988 Dec 22;336(6201):780–783. doi: 10.1038/336780a0. [DOI] [PubMed] [Google Scholar]
- Takagi N., Endo S., Sugawara O. X chromosome inactivation in bone marrow cells of adult mice carrying Searle's X-autosome translocation: occurrence of the early-replicating inactive X chromosome. Cytogenet Cell Genet. 1984;38(1):62–69. doi: 10.1159/000132031. [DOI] [PubMed] [Google Scholar]
- Takagi N. Primary and secondary nonrandom X chromosome inactivation in early female mouse embryos carrying Searle's translocation T(X; 16)16H. Chromosoma. 1980;81(3):439–459. doi: 10.1007/BF00368155. [DOI] [PubMed] [Google Scholar]
- Wareham K. A., Lyon M. F., Glenister P. H., Williams E. D. Age related reactivation of an X-linked gene. 1987 Jun 25-Jul 1Nature. 327(6124):725–727. doi: 10.1038/327725a0. [DOI] [PubMed] [Google Scholar]
- Yen P. H., Allen E., Marsh B., Mohandas T., Wang N., Taggart R. T., Shapiro L. J. Cloning and expression of steroid sulfatase cDNA and the frequent occurrence of deletions in STS deficiency: implications for X-Y interchange. Cell. 1987 May 22;49(4):443–454. doi: 10.1016/0092-8674(87)90447-8. [DOI] [PubMed] [Google Scholar]