Abstract
The anaplastic lymphoma kinase (ALK) gene plays an important physiologic role in the development of the brain and can be oncogenically altered in several malignancies, including non-small-cell lung cancer (NSCLC) and anaplastic large cell lymphomas (ALCL). Most prevalent ALK alterations are chromosomal rearrangements resulting in fusion genes, as seen in ALCL and NSCLC. In other tumors, ALK copy-number gains and activating ALK mutations have been described. Dramatic and often prolonged responses are seen in patients with ALK alterations when treated with ALK inhibitors. Three of these—crizotinib, ceritinib, and alectinib—are now FDA approved for the treatment of metastatic NSCLC positive for ALK fusions. However, the emergence of resistance is universal. Newer ALK inhibitors and other targeting strategies are being developed to counteract the newly emergent mechanism(s) of ALK inhibitor resistance. This review outlines the recent developments in our understanding and treatment of tumors with ALK alterations.
INTRODUCTION
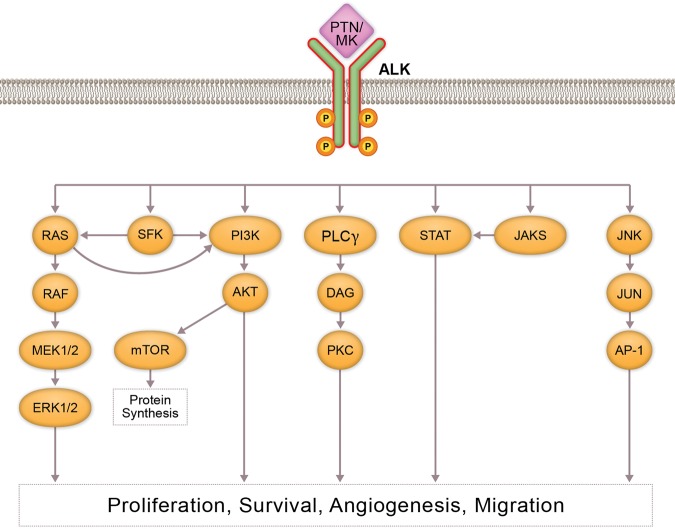
In 1994, a positional cloning strategy revealed a unique rearrangement resulting from the fusion of the nucleophosmin (NPM1) gene, located on 5q35, to a previously unidentified protein tyrosine kinase gene located on 2p23 in an anaplastic large-cell lymphoma (ALCL) cell line (Morris et al. 1994). This new protein, called anaplastic lymphoma kinase (ALK), is expressed normally in the brain, small intestine, and testis, but not in the normal lymphoid cells (Morris et al. 1994). ALK shows the greatest sequence similarity to the insulin receptor subfamily of transmembrane tyrosine kinases. ALK contains an extracellular domain that has a low-density lipoprotein receptor domain class A (LDLa) region sandwiched between two meprin, A-5 protein, multiple receptor protein-tyrosine phosphatase mu (MAM) regions, followed by a glycine-rich region, a transmembrane region, and an intracellular domain containing a tyrosine kinase region. ALK is considered an orphan receptor, even though pleiotrophin (PTN) and midkine (MDK), both secreted growth factors, are known to bind and activate ALK downstream signaling (Stoica et al. 2001, 2002; Lu et al. 2005). PTN binding to ALK activates the mitogen-activated protein kinase (MAPK) pathway, whereas MDK binding to ALK induces insulin receptor substrate 1 (IRS1) phosphorylation, resulting in MAPK and phosphoinositide 3-kinase (PI3K) activation (Fig. 1; Bowden et al. 2002; Powers et al. 2002; Kuo et al. 2007; Palmer et al. 2009). However, studies that followed showed either no ligand binding activity of PTN and MDK to ALK (Moog-Lutz et al. 2005; Mathivet et al. 2007) or that PTN's effect on ALK is not due to direct binding (Perez-Pinera et al. 2007). Furthermore, a recent study shows longer heparin chains induce ALK dimerization, activation, and downstream signaling, indicating heparin serves as ALK's ligand or coligand (Murray et al. 2015).
Figure 1.
Anaplastic lymphoma kinase (ALK) signaling: Pathway figure that depicts normal activation of ALK through ligand binding. Receptor phosphorylation stimulates downstream signaling through the PI3K/AKT, MAPK/ERK, and STAT3 pathways. These signaling cascades can contribute to increased cancer cell growth, survival, and metastasis.
ALK is thought to play a significant role in the development and function of the nervous system, where it controls the basic mechanisms of cell proliferation, survival, and differentiation in response to extracellular stimuli (Iwahara et al. 1997; Yao et al. 2013). The role of ALK in the development of model organisms, such as Drosophila, Caenorhabditis elegans, and zebrafish, has been well documented (Palmer et al. 2009). In Drosophila, binding of a secreted protein ligand jelly belly (Jeb) activates dALK during synaptogenesis and organization of the visceral musculature of the gut (Loren et al. 2003; Rohrbough and Broadie 2010). In C. elegans, ALK homolog, suppressor of constitutive dauer formation 2 (SCD-2) signals through the ligand hesitation behavior 1 (HEN-1) during neuromuscular junction development and regulates the dauer response to environmental stress (Liao et al. 2004; Reiner et al. 2008). In zebrafish, leukocyte tyrosine kinase (Ltk), closely related to ALK, is required for the establishment of iridophores and mutations in ltk show defects in pigmentation patterns (Lopes et al. 2008).
The most prevalent genomic ALK aberrations in human cancer are chromosomal rearrangements, resulting in fusion genes. ALK fusions arise from fusion of the 3′ half of ALK, derived from Chromosome 2 that retains its kinase catalytic domain, and the 5′ portion of a different gene that provides its promoter. Multiple different 5′ partners have been identified. Wild-type ALK is normally activated through binding of ligands to its extracellular domain, resulting in dimerization and autophosphorylation of the kinase domain. Structural studies show that fusion with multiple 5′ partners helps bypass this requirement and increase oncogenic potential of ALK, as evidenced by NPM1-ALK (Fujimoto et al. 1996) and EML4-ALK (Wang et al. 2015) in non-small-cell lung cancer (NSCLC). Increased copy number and the presence of activating point mutations that result in kinase activation are also linked to oncogenic activity of ALK. These genetic alterations are found in multiple malignancies, including, but not limited to, lung cancer, neuroblastoma, rhabdomyosarcoma, renal cell carcinoma, inflammatory myofibroblastic tumor (IMT), and inflammatory breast cancer (Webb et al. 2009; Kelleher and McDermott 2010). Additionally, a recent report describes an alternative transcription initiation site leads to the detection of an oncogenic ALK isoform (ALKATI) in 11% of melanomas and other tumor types (Wiesner et al. 2015). ALKATI arises independently of other ALK genomic alterations, and in vivo and in vitro studies show that ALKATI-driven tumors are sensitive to crizotinib. Although activating mutations and copy-number changes of the ALK gene are currently being investigated for their role in tumor development and treatment response, the main clinical therapeutic implications of ALK lie in targeting of ALK fusions with tyrosine kinase inhibitors. In general, ALK fusions are mutually exclusive with mutations in EGFR, KRAS, and ERBB2 genes, indicative of these genes’ signaling through similar downstream pathways (Takahashi et al. 2010).
ALK MUTATIONS
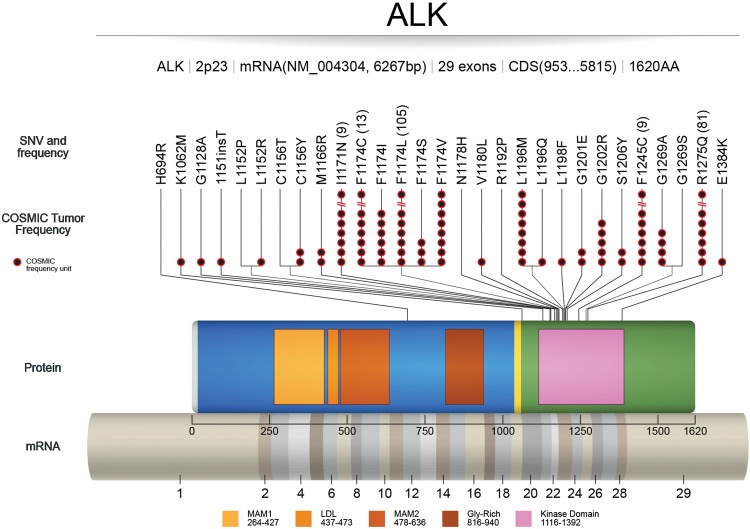
Gain-of-function mutations of ALK are described primarily in neuroblastoma. In addition, thyroid (Murugan and Xing 2011) and lung cancers (Wang et al. 2011) have been shown to carry activating ALK point mutations. Most of the mutations are located in the kinase domain, including two hotspot mutations: F1174 (mutated to C, I, L, S, or V) and R1275 (mutated to Q and L) (Franco et al. 2013). These two hotspot mutations represent 85% of all ALK mutations. All reported ALK mutations could be classified into three groups: ligand-independent mutations (F1174I, F1174S, F1174L, and R1275Q), ligand-dependent mutations (D1091N, T1151M, and A1234T), and a kinase-dead mutation (I1250T) (Hallberg and Palmer 2013). ALK mutations are frequently acquired within an ALK fusion gene as a result of crizotinib resistance (Choi et al. 2010), alectinib resistance (Kodama et al. 2014) in NSCLC, and lorlatinib resistance in ALCL (Mologni et al. 2015). Figure 2 and Table 1 describe functionally characterized ALK mutations in the literature.
Figure 2.
Activating mutations in ALK: COSMIC (tumor only) frequencies of ALK mutations with published literature on functional and/or therapeutic significance. mRNA sequence depicts full reference sequence of ALK (NM_004304) with exon numbers marked. ALK protein sequence (0–1620 amino acids) shows different functional domains (MAM1, LDL, MAM2, Gly-rich, and kinase domain) with starting and ending amino acid numbers (UniProt). COSMIC frequency units (black circles and numbers in parentheses) refer to the number of tumor samples with a particular single-nucleotide variant (SNV) found in COSMIC. The SNVs without the black circles are referenced in literature that is not recorded in COSMIC.
Table 1.
List of ALK mutations known to be oncogenic
| Alteration | Location | Functional significance | Tumor type | Reference(s) |
|---|---|---|---|---|
| H694R | Extracellular | Increased phosphorylation, promotes tumors in mice | Wang et al. 2011 | |
| K1062M | Juxtamembrane | Transforms cells, promotes tumors in mice | Neuroblastoma | Chen et al. 2008; Murugan and Xing 2011 |
| G1128A | Kinase domain, glycine-rich region | Increased phosphorylation | Neuroblastoma | Mosse et al. 2008; Bossi et al. 2010; Schonherr et al. 2011 |
| M1166R | Kinase domain | Increased phosphorylation | Neuroblastoma | Mosse et al. 2008; Chand et al. 2013 |
| F1174I | Kinase domain | Increased phosphorylation | Neuroblastoma | Chand et al. 2013; Mologni et al. 2015 |
| F1174L | Kinase domain | Disrupts auto-inhibitory function | Neuroblastoma | Chen et al. 2008; George et al. 2008; De Brouwer et al. 2010; Sasaki et al. 2010; Bresler et al. 2011; Heuckmann et al. 2011; Kodama et al. 2014; Infarinato et al. 2015; Zou et al. 2015 |
| F1174S | Kinase domain | Ligand-independent activity | Neuroblastoma | Martinsson et al. 2011 |
| L1198F | Kinase domain | Increased phosphorylation | Thyroid | Murugan and Xing 2011 |
| G1201E | Kinase domain | Increased phosphorylation | Thyroid, Skin | Murugan and Xing 2011 |
| F1245C | Kinase domain | Increased phosphorylation | Neuroblastoma | Schonherr et al. 2011; Infarinato et al. 2015 |
| R1275Q | Kinase domain | Disrupts auto-inhibitory function | Neuroblastoma | Chen et al. 2008; George et al. 2008; De Brouwer et al. 2010; Bagci et al. 2012; Carpenter and Mosse 2012 |
| E1384K | Kinase domain | Increased phosphorylation | Cervix | Wang et al. 2011 |
ALK Mutations in Neuroblastoma
Gain-of-function mutations are reported in both familial and sporadic neuroblastoma patients (George et al. 2008; Janoueix-Lerosey et al. 2008). In familial neuroblastoma, ALK is a predisposition gene, and germline mutations have been found in 50% of familial neuroblastoma cases (Mosse et al. 2008). Most frequent gain-of-function germline mutations of ALK are G1128A, R1192P, and R1275Q (Janoueix-Lerosey et al. 2008; Mosse et al. 2008). However, in sporadic neuroblastoma, only 7% of cases show activating ALK mutations (Mosse et al. 2008). Two hotspot mutations, F1174L and R1275Q, lead to ALK autophosphorylation and cytokine-independent growth (Chen et al. 2008; Janoueix-Lerosey et al. 2008). Furthermore, F1174L, the most recurrent mutation, predominantly occurs in MYCN-amplified tumors and potentiates MYCN oncogenic activity in neuroblastoma (De Brouwer et al. 2010; Berry et al. 2012). In vitro and in vivo studies with cell lines expressing ALK mutations show different sensitivity to ALK inhibitors. Tumors expressing the R1275Q mutation are sensitive to crizotinib and TAE684, whereas F1174L mutant cells exhibit sensitivity to these agents only at higher doses (George et al. 2008; Bresler et al. 2011).
ALK GENE REARRANGEMENTS
NPM1 was the first described fusion partner of ALK in 1994 in an ALCL cell line with a t(2;5) chromosomal rearrangement (Morris et al. 1994). Since then, several other ALK fusion partners have been described in multiple malignancies (Table 2). The role of ALK gene rearrangements as oncogenic drivers has been well established in preclinical models including transgenic mouse models (Solomon et al. 2009; Chen et al. 2010). In these models, ALK promotes the activation of downstream signaling pathways and other crucial aspects of malignant phenotypes like uncontrolled cellular proliferation and survival. The precise mechanisms that underlie the development of ALK gene rearrangements have yet to be elucidated. However, several steps are in play: the generation of double-strand DNA breaks, aberrant joining of the DNA ends, and selection of gene rearrangements that confer a survival advantage (Bunting and Nussenzweig 2013; Shaw and Engelman 2013). In general, different ALK fusion partners affect ALK homodimerization, as well as ALK signaling potential. In fact, the comparison of a number of ALK fusion proteins has suggested differences in transforming and tumorigenic potential (Bridge et al. 2001; Cools et al. 2002).
Table 2.
Known ALK chromosomal translocations that activate the tyrosine kinase domain
| Fusion | Chromosomal aberration | Tumor types | Reference(s) |
|---|---|---|---|
| ATIC-ALK | Inv(2)(p23;q35) | ALCL | Colleoni et al. 2000; Armstrong et al. 2004 |
| CAD-ALK | Inv(2)(p23;p22) | CRC | Amatu et al. 2015 |
| CLTC-ALK | t(2;17)(p23;q23) | IMT, ALCL, BCL | Bridge et al. 2001; Cools et al. 2002; De Paepe et al. 2003; McManus et al. 2004; Cerchietti et al. 2011 |
| DCTN1-ALK | t(2;12)(p23;q11) | IMT | Wang et al. 2012; Wiesner et al. 2014 |
| EML4-ALK | inv(2)(p21;p23) | NSCLC | Soda et al. 2007; Choi et al. 2008; Lin et al. 2009; Li et al. 2011 |
| FN1-ALK | t(2)(p23;q34) | Ovarian, IMT | Ren et al. 2012; Ouchi et al. 2015 |
| HIP1-ALK | t(2;7)(p23;q11.23) | NSCLC | Fang et al. 2014; Hong et al. 2014; Ou et al. 2014 |
| KIF5B-ALK | t(2;10)(p23;p11) | Lung | Takeuchi et al. 2009 |
| KLC1-ALK | t(2;14)(p23;q32.3) | Lung | Togashi et al. 2012 |
| NPM1-ALK | t(2;5)(p23;q35) | NHL, ALCL | Morris et al. 1994; Pulford et al. 2004; Palmer et al. 2009 |
| RANBP2-ALK | Inv(2)(p23;q11–13) | IMT | Ma et al. 2003 |
| SQSTM1-ALK | t(2;5)(p23;q35) | BCL | Takeuchi et al. 2011; d'Amore et al. 2013 |
| STRN-ALK | t(2)(p23;p22.2) | Thyroid | Kelly et al. 2014 |
| TFG-ALK | t(2;3)(p23;q21) | ALCL | Hernandez et al. 1999; Hernandez et al. 2002 |
| TPM3-ALK | t(2;1)(p23;q25) | ALCL, IMT | Lamant et al. 1999; Lawrence et al. 2000; Armstrong et al. 2004 |
| TPM4-ALK | t(2;19)(p23;q13.1) | ALCL, IMT | Lawrence et al. 2000; Meech et al. 2001 |
ALCL, anaplastic large-cell lymphoma; CRC, colorectal carcinoma; IMT, inflammatory myofibroblastic tumor; BCL, B-cell lymphoma; NSCLC, non-small-cell lung cancer; NHL, non-Hodgkin's lymphoma.
ALK Gene Rearrangements in Different Tumor Types
In the majority of ALCL cases, ALK is activated through chromosomal rearrangement (Medeiros and Elenitoba-Johnson 2007). ALCL is a type of T-cell non-Hodgkin's lymphoma, and abnormalities of the ALK gene are common in this disease (Medeiros and Elenitoba-Johnson 2007). As many as 50% of all adult cases of ALCL are ALK-positive, and up to 90% of all pediatric ALCL patients are ALK-positive (Gustafson et al. 2009; Damm-Welk et al. 2015). The most frequent translocation in ALCL is NPM1-ALK, accounting for ∼75%–80% of all ALK-positive ALCL (Pulford et al. 2004). Additionally, TPM3-ALK has been found in 12%–18% of ALCL. Other fusion proteins are found at much lower frequency (<2%), and include TFG-ALK, CLTC1-ALK, and ATIC-ALK. Importantly, 5-yr survival for ALK-positive ALCL patients is 70%–80%, as compared with 15%–45% for ALK-negative ALCL patients (Roskoski 2013). In lung cancer, the EML4-ALK fusion was identified in 2007 NSCLCs in patients within NSCLC (Soda et al. 2007) and occurs at a frequency of ∼6.7% (Perner et al. 2008). This fusion is important diagnostically, as EML4-ALK is mutually exclusive with EGFR and KRAS mutations (Horn and Pao 2009). Moreover, ALK rearrangements with constitutive kinase activity occur in 2%–7% of all NSCLCs, and are associated with young age, male gender, and no or light smoking history (Shaw et al. 2009; Kwak et al. 2010). More than a dozen different variants of EML4-ALK have been identified in NSCLC. Other less frequent ALK fusions identified in lung cancer are SEC31A-ALK, HIP1-ALK, KIF5B-ALK, and KLC1-ALK.
Inflammatory myofibroblastic tumors (IMTs) are soft-tissue mesenchymal neoplasms. Fifty percent of IMTs show chromosomal translocations involving the 2p23 region, resulting in TPM3/4-ALK fusions (Griffin et al. 1999). Several other ALK fusion partners have been identified in IMT with <5% frequency, including TPM4 (Griffin et al. 1999; Lawrence et al. 2000), CLTC (Bridge et al. 2001), CARS (Cools et al. 2002), and RANBP2 (Ma et al. 2003). ALK fusions are associated with better prognosis in IMT (Chun et al. 2005). In thyroid cancer, translocations involving ALK are detected in 2.2% of papillary thyroid cancer (PTC) patients (Chou et al. 2015). Various ALK fusions (EML4-ALK, GFPT1-ALK, TFG-ALK, and STRN-ALK) are reported in thyroid cancer patient tumors (Kelly et al. 2014; Ji et al. 2015). Translocations of ALK identified by fluorescence in situ hybridization (FISH) show strong immunohistochemistry (IHC) positivity in these tumors.
In addition, ALK rearrangements occur in 10% of spitzoid tumors with DCTN1-ALK and TPM3-ALK being the most common (Busam et al. 2014; Wiesner et al. 2014). IHC confirms the expression of chimeric ALK protein, followed by downstream activation of AKT, ERK, and S6 proteins. Crizotinib is able to block these ALK fusion-induced downstream activities.
ALK COPY-NUMBER VARIANTS
ALK gene amplification has been detected in variety of tumors (http://cancergenome.nih.gov/). In some reports, ALK gene amplification or increase in copy number due to polysomy of Chromosome 2 does not always correspond to the overexpression of ALK protein or increased downstream signaling. However, copy-number gain in NSCLC cell lines is associated with increased sensitivity to ALK inhibitors, such as crizotinib (Miyake et al. 2002).
ALK Copy-Number Variants in Different Tumor Types
Neuroblastoma cell lines and primary neuroblastoma tissues show ALK gene amplification that correlates with ALK protein overexpression and promotion of tumorigenesis in neuroblastoma (Miyake et al. 2002; Osajima-Hakomori et al. 2005). Constitutively active ALK, due to gene amplification, results in hyper-phosphorylation of downstream SHCC protein in neuroblastoma cell lines (Miyake et al. 2002). A follow-up in vitro study revealed phosphorylation of SHCC and other downstream activities (MAPK) were suppressed, which was associated with induction of apoptosis in neuroblastoma cell lines upon siRNA-mediated down-regulation of amplified ALK, suggesting targeting of ALK may be an appropriate therapeutic approach for tumors with ALK amplification (Osajima-Hakomori et al. 2005). Likewise, ALK gene amplification was detected in 9.4% (8/85) of primary neuroblastoma tissues (Osajima-Hakomori et al. 2005). FISH analysis showed ALK copy-number increase is a recurrent genetic event in neuroblastic tumors (39.1%, 96/245), however ALK gene amplification was seen at a lower frequency (1.2%, 3/246) in neuroblastomas (Wang et al. 2013). ALK protein expression by IHC (50.5%, 51/101) in these tumors is associated with a worsened patient prognosis.
A 2011 report revealed that a little more than 10% (11/107) of NSCLC patients exhibit ALK amplification and 63% (68/110) had copy-number gains, although it was observed in a small percentage of cells and was not associated with increased tissue expression of ALK, nor was it thought to be a significant tumor driving event for tumors (Salido et al. 2011). Another report characterized 191 patient samples and found 11% to have at least six copies of ALK (Khadija et al. 2012, ASCO abstract #10556). In the same study, >70% of NSCLC cell lines that gained three or four ALK copies also showed crizotinib sensitivity.
Amplification of ALK has been detected in ∼11% of esophageal cancer (Schoppmann et al. 2013). However, there was no association of ALK amplification to either ALK protein expression or downstream phospho-STAT3 expression in these tumors. Copy-number gain of ALK was detected in 3.4% (26/756) (Bavi et al. 2013) and 37% (25/68) (Pietrantonio et al. 2014) of colorectal cancer patients. Copy-number increase does not correlate with protein expression in the above studies, but was associated with poor prognosis and may predict lack of benefit from anti-EGFR treatment in colorectal cancer patients. ALK copy number increases due to polysomy of Chromosome 2 or its gene amplification has been reported in multiple breast cancer studies and correlates with poor prognosis. However, in most of the cases this does not correlate with increased protein expression as measured by IHC. Studies include amplification of ALK in 13.3% (130/980) of breast cancer patients (Siraj et al. 2015), copy-number gain of ALK in 62% (82/133) of breast cancer cases (Hanna et al. 2015), and copy-number gain of ALK in 47.2% (17/36) of inflammatory breast cancer (Kim et al. 2015). Mild increase in ALK copy number due to Chromosome 2 aneuploidy is reported in 64% (16/25) of inflammatory breast cancer cases (Krishnamurthy et al. 2013). However, there was no increased ALK mRNA or protein expression in these tumors. Increased copy number, correlating with high ALK protein expression, has been reported in 25% of rhabdomyosarcoma cases and is associated with poor prognosis in rhabdomyosarcoma patients (van Gaal et al. 2012; Yoshida et al. 2013; Lee et al. 2014). In vitro studies show antitumor activity with ALK inhibitors in ALK-positive rhabdomyosarcoma cell lines (van Gaal et al. 2013; Megiorni et al. 2015).
THERAPEUTIC IMPLICATIONS
Therapeutic implications for ALK gene alterations are predominantly associated with ALK gene fusions, which predict tumor response to ALK inhibitors. Crizotinib, ceritinib, and recently alectinib are FDA approved for the treatment of patients with metastatic NSCLC whose tumors are positive for ALK fusions. Early phase 1 studies showed crizotinib yielded sustained responses in ALK-fusion-positive metastatic NSCLC patients (Kwak et al. 2010; Shaw et al. 2011; Camidge et al. 2012). Two phase 3 studies, which led to FDA approval of crizotinib, further confirmed that crizotinib was superior to standard first-line pemetrexed + cisplatin chemotherapy in patients with previously untreated advanced ALK-rearranged NSCLC. In one study (PROFILE 1007), crizotinib showed overall response rate (ORR) of 65% as compared with 20% with either pemetrexed or docetaxel in patients who had failed one prior platinum-based regimen (Shaw et al. 2013). In another study (PROFILE 1014), progression-free survival (PFS) and ORR were significantly improved with crizotinib than with first-line therapy with platinum-pemetrexed (median, 10.9 mo vs. 7 mo) (Solomon et al. 2014). Furthermore, crizotinib was associated with disease control in ALK-fusion-positive NSCLC patients who had brain metastasis (Costa et al. 2015). In addition, crizotinib also showed therapeutic response in ALK-fusion-positive IMT patients (Butrynski et al. 2010) and pediatric patients with anaplastic large cell lymphoma and IMT (Mosse et al. 2013).
Although crizotinib has shown excellent activity in patients with NSCLC who are ALK-fusion-positive, durable responses remain uncommon, with a median PFS of 13 mo, because of the development of resistance that leads to disease progression. Although the mechanism of resistance is still being delineated, acquired secondary mutations in the ALK kinase domain (F1174L, F1174C, L1196M, I1171T, G1202R, S1206Y, G1269S, and G1269A) or ALK gene amplification (Choi et al. 2010; Sasaki et al. 2010; Heuckmann et al. 2011; Doebele et al. 2012b; Katayama et al. 2012) are known to be associated with resistance. Resistance can also be mediated by activation of alternative ALK-independent survival pathways that hamper the effectiveness of crizotinib, including the epidermal growth factor pathway, insulin-like growth factor pathway, RAS/SRC signaling, and AKT/mTOR signaling, among others (Doebele et al. 2012b; Katayama et al. 2012; Crystal et al. 2014; Ji et al. 2014; Mengoli et al. 2016). Tables 3 and 4 show ALK alterations that are either sensitive to ALK inhibitors (Table 3) or resistant to ALK inhibitors (Table 4).
Table 3.
Therapy-sensitive ALK alterations
| Drug (type) | Sensitive variant(s) | Tumor type | Level of evidencea | Reference(s) |
|---|---|---|---|---|
| Crizotinib (ALK TKI) | Fusion | NSCLC | 1A | Kwak et al. 2010; Camidge et al. 2012; Shaw et al. 2013; Solomon et al. 2014 |
| RANBP2-ALK | IMT | 3A | Butrynski et al. 2010 | |
| NPM1-ALK EMLA-ALK |
Neuroblastoma, lung | 3B | Christensen et al. 2007 | |
| F1174L R1275Q |
IMT | 3B | Lovly et al. 2011; Di Paolo et al. 2015 | |
| Ceritinib (ALK TKI) | Fusion | Lung | 1A | Shaw et al. 2014 |
| Fusion | Thyroid | 3A | Godbert et al. 2015 | |
| EML4-ALK/I1171T EML4-ALK/V1180L EML4-ALK/L1196M EML4-ALK/S1206Y EML4-ALK/G1269A |
3B | Friboulet et al. 2014; Katayama et al. 2014 | ||
| Alectinib (ALK TKI) | Fusion | Lung | 1A | Seto et al. 2013; Gadgeel et al. 2014 |
| Amplification | Neuroblastoma | 3B | Sakamoto et al. 2011 | |
| L1196M G1269A C1156Y F1174L 1151Tins L1152R |
NSCLC | 3B | Sakamoto et al. 2011; Kodama et al. 2014; Yoshimura et al. 2016 | |
| EML4-ALK/1151Tins EML4-ALK/L1152R EML4-ALK/C1156Y EML4-ALK/F1174L EML4-ALK/L1196M EML4-ALK/S1206Y EML4-ALK/G1269A |
NSCLC | 3B | Sakamoto et al. 2011; Kodama et al. 2014 | |
| Lorlatinib (ALK TKI) | NPM1-ALK/C1156F NPM1-ALK/I1171T NPM1-ALK/I1171N NPM1-ALK/F1174I NPM1-ALK/N1178H NPM1-ALK/E1201K NPM1-ALK/D1203N |
ALCL | 3B | Mologni et al. 2015 |
| F1174L F1245C R1275Q |
Neuroblastoma | 3B | Infarinato et al. 2015 | |
| 1151Tins C1156T L1196M G1202R G1269A |
3B | Johnson et al. 2014; Zou et al. 2015 | ||
| EML4-ALK/L1196M EML4-ALK/G1269A |
Lung | 3B | Zou et al. 2015 | |
| Brigatinib (ALK TKI) | EML4-ALK/L1196M NPM1-ALK/L1196Q |
NSCLC | 3B | Katayama et al. 2011; Ceccon et al. 2013 |
| ASP3026 (ALK TKI) | NPM1-ALK | ALCL | 3B | George et al. 2014 |
| EML4-ALK/L1196M | NSCLC | 3B | Mori et al. 2014 | |
| Entrectinib (ALK TKI) | EML4-ALK CAD-ALK |
Colorectal | 3B | Amatu et al. 2015, Lee et al. 2015 |
| X-396 (ALK TKI) | EML4-ALK/C1156Y EML4-ALK/L1196M |
Lung | 3B | Lovly et al. 2011 |
| F1174L R1275Q |
Neuroblastoma | 3B | Di Paolo et al. 2015 | |
| Retaspimycin (HSP90 inhibitor) | EML4-ALK | NSCLC | 3A | Sequist et al. 2010; Normant et al. 2011 |
| Tanespimycin (HSP90 inhibitor) | EML4-ALK/V1180L | NSCLC | 3B | Katayama et al. 2014 |
| NPM1-ALK TPR-ALK |
ALCL | 3B | Bonvini et al. 2002 | |
| RANBP2-ALK/F1174L | IMT | 3B | Sasaki et al. 2010 |
Only clinically available drugs are listed in the table.
TKI, tyrosine kinase inhibitor; NSCLC, non-small-cell lung cancer; NB, neuroblastoma; ALCL, anaplastic large-cell lymphoma; IMT, inflammatory myofibroblastic tumor; CRC, colorectal cancer.
aDefinition of level of evidence based on Meric-Bernstam et al. 2015.
Table 4.
Therapy resistant ALK alterations
| Drug (type) | Resistant variant(s) | Tumor type | Level of evidencea | Reference(s) |
|---|---|---|---|---|
| Crizotinib (ALK TKI) | Amplification-EML4-ALK | NSCLC | 3A | Doebele et al. 2012b |
| 1151insT L1152R C1156Y F1174V G1202R S1206Y G1269A |
NSCLC | 3A | Choi et al. 2010; Doebele et al. 2012b; Katayama et al. 2012; Ou et al. 2015; Zou et al. 2015 | |
| EML4-ALK/1151insT EML4-ALK/L1152R EML4-ALK/C1156Y EML4-ALK/L1196M EML4-ALK/L1198P EML4-ALK/D1203N |
3B | Choi et al. 2010; Doebele et al. 2012b; Katayama et al. 2012; Ou et al. 2015; Zou et al. 2015 | ||
| NPM1-ALK | ALCL | 3B | Ceccon et al. 2013 | |
| RANBP2-ALK | IMT | 3A | Sasaki et al. 2010 | |
| Ceritinib (ALK TKI) | EML4-ALK/1151Tins EML4-ALK/L1152R EML4-ALK/C1156Y EML4-ALK/F1174C EML4-ALK/G1202R |
3B | Friboulet et al. 2014 | |
| Alectinib (ALK TKI) | EML4-ALK/I1171Tins EML4-ALK/V1180L EML4-ALK/G1202R |
NSCLC | 3B | Katayama et al. 2014; Kodama et al. 2014 |
Only clinically available drugs are listed in the table.
TKI, tyrosine kinase inhibitor; NSCLC, non-small-cell lung cancer; NB, neuroblastoma; ALCL, anaplastic large-cell lymphoma; IMT, inflammatory myofibroblastic tumor; CRC, colorectal cancer.
aDefinition of level of evidence based on Meric-Bernstam et al. 2015.
Even though different resistance mechanisms are being discovered, most crizotinib-resistant tumors continue to depend on ALK signaling and are sensitive to more potent, structurally distinct, second-generation ALK inhibitors, such as ceritinib, alectinib, brigatinib, and lorlatinib. A preclinical study showed alectinib was able to block the resistant gatekeeper mutation (L1196M) in ALK-fusion-positive NSCLC cells (Sakamoto et al. 2011). Two phase 1/2 studies showed alectinib was well tolerated. The first study conducted in ALK inhibitor-naïve patients with ALK-rearranged NSCLC showed objective response of 93.5% (43 of 46) that included two patients with complete response and 41 patients with partial response (Seto et al. 2013). Another study that tested efficacy of alectinib in patients with crizotinib-resistant ALK-rearranged NSCLC showed objective response of 55% (24 of 44), with a confirmed complete response (CR) in one patient and confirmed partial response (PR) in 14 patients (Gadgeel et al. 2014). In a multicenter phase 2 study in crizotinib-resistant ALK-fusion-positive NSCLC patients, alectinib showed a 48% response rate with 33 of 69 patients demonstrating confirmed PRs (Shaw et al. 2016b).
In preclinical studies, ceritinib efficiently inhibits several ALK secondary mutations developed in the setting of crizotinib therapy (Friboulet et al. 2014). In a phase 1 study, 114 ALK-rearranged, crizotinib-naïve (80) and -resistant (34) NSCLC patients received ceritinib (Shaw et al. 2014). The overall response rate was 58%, with one patient achieving a CR, 65 patients a PR, and 25 patients with stable disease. This study also had six of seven patients with a confirmed response who carried ALK gene amplification or mutations (L1196M, S1206Y) after crizotinib therapy. In another phase 1 study with 20 ALK-rearranged patients (19 NSCLC and 1 IMT) ceritinib treatment resulted in an ORR of 55% (Nishio et al. 2015). A recent multicenter phase 1 study (ASCEND-1) tested the activity of ceritinib in 246 ALK-rearranged NSCLC patients (Kim et al. 2016). The ORR was 72% (60 of 83 ALK inhibitor [ALKi] naïve patients) and 56% (92 of 163 ALK inhibitor pretreated patients). A still ongoing phase 2 (ASCEND-2) study that evaluated ceritinib activity in 140 ALK-rearranged NSCLC patients who failed crizotinib in addition to other regimens showed investigator assessed RR of 38.6% (Crino et al. 2016).
Brain metastasis is a common problem in advanced NSCLC patients and 10%–40% NSCLC patients develop brain metastasis during the course of their disease (Huang and Ouyang 2013). Brain metastasis occurs in 30%–50% of ALK-positive NSCLC patients who are naïve to ALK inhibitors (Doebele et al. 2012a; Simoff et al. 2013; Rangachari et al. 2015). Despite the inability of most of the chemotherapy regimens to cross the blood–brain barrier, pemetrexed treatment in NSCLC patients shows some effectiveness against brain metastasis (Bearz et al. 2010). Crizotinib has shown systemic and intracranial disease control in NSCLC patients that are ALK-positive (Costa et al. 2015). However, crizotinib-resistant patients develop new or show progression of preexisting intracranial lesions (Costa et al. 2015). Recent studies showed that second-generation agents ceritinib (Kim et al. 2014; Crino et al. 2016), alectinib (Gadgeel et al. 2014; Shaw et al. 2016b), and brigatinib (Kerstein et al. 2015) are active in intracranial diseases.
Clinical data with crizotinib, ceritinib, and alectinib showed that G1202R is a common ALK resistance mutation. However, there are other mutations that differ in sensitivity to the various anti-ALK kinase inhibitors (Table 4). Several next-generation ALK inhibitors are being developed to treat previously treated ALK inhibitor resistant mutations. The list includes brigatinib (Huang et al. 2016; Siaw et al. 2016), lorlatinib (Gainor et al. 2016; Shaw et al. 2016a), and X-396 (Di Paolo et al. 2015). Both brigatinib and lorlatinib inhibit several known resistant mutations, and lorlatinib showed effective inhibition against the G1202R mutation (Gainor et al. 2016). These results suggest that understanding the resistance mechanism and selecting the appropriate tyrosine kinase inhibitor (TKI) may be required to optimize response to therapy. Interestingly, a preclinical study showed that the inhibition of autophagy with chloroquine may restore crizotinib sensitivity (Ji et al. 2014).
In addition to targeting ALK directly, there are pharmacological strategies that allow for its indirect targeting. Specifically, there has been some success with inhibiting ALK indirectly by targeting heat-shock proteins, namely HSP90, in lung cancer. Inhibition of HSP90, a chaperone protein that stabilizes a wide variety of proteins, including ALK, has shown some preclinical efficacy in crizotinib-resistant ALK fusions (EML4-ALK and NPM1-ALK), including secondary resistant mutants in lung cancer models (Sang et al. 2013). In addition, several drug combinations, including ALK inhibitors and other receptor tyrosine kinase (RTK) inhibitors or HSP90 inhibitors, are being explored in preclinical/clinical studies: IGF1R (Lovly et al. 2014); MEK (Tanizaki et al. 2012; Crystal et al. 2014; Hrustanovic et al. 2015); and HSP90 (Sang et al. 2013). As recent preclinical data indicate, the immune checkpoint proteins are induced in ALK-positive NSCLC tumors (Ota et al. 2015; Hong et al. 2016); thus, combination therapies of checkpoint (PD-1/PD-L1, CTLA-4) and ALK inhibitors are being explored in the clinical setting for ALK-positive NSCLC patients (NCT02393625, NCT01998126).
Chemotherapy also remains a viable option in patients with ALK translocations. In terms of chemotherapy for NSCLC, pemetrexed-based chemotherapy may be more effective than other nonpemetrexed combinations (Camidge et al. 2011).
CONCLUSIONS
ALK rearranged tumors represent a specific subset of tumors that can be effectively targeted with currently available ALK inhibitors. Hence testing for ALK alterations in tumors known to have this molecular aberration is now an obligatory part of the diagnosis. FISH, next-generation sequencing (NGS) of tumor tissue, and sequencing of circulating tumor cells offer alternative and often complementary ways of detecting tumors with ALK alterations. The almost inevitable emergence of resistance during TKI therapy requires rebiopsy of the tumor at relapse to identify resistance mechanisms that could be targeted with newer ALK inhibitors and other novel therapeutic strategies, including HSP90 inhibitors and pemetrexed-based chemotherapy. Checkpoint inhibitors either as single agents or combination with ALK inhibitors are being evaluated in clinical trials.
ADDITIONAL INFORMATION
Funding
This work was supported in part by 1U01 CA180964 from the Sheikh Khalifa Bin Zayed Al Nahyan Institute for Personalized Cancer Therapy; Cancer Prevention and Research Institute of Texas (CPRIT) grant RP150535, Precision Oncology Decision Support Core; National Center for Advancing Translational Sciences (NCATS) grant UL1 TR000371 (Center for Clinical and Translational Sciences); the Bosarge Foundation; and the MD Anderson Cancer Center Support grant (P30 CA016672).
Competing Interest Statement
Funda Meric-Bernstam has had an honoraria role with Genentech and Roche Diagnostics; a consulting or advisory role for Genentech, Novartis, Roche, Inflection Biosciences, and Celgene; and research funding from Novartis, AstraZeneca, Taiho Pharmaceutical, Genentech, Calithera Biosciences, Debiopharm Group, Bayer, Aileron Therapeutics, PUMA Biotechnology, Verastem, and CytomX Therapeutics. G.B.M. has had a consulting role with Adventist Health, Allostery, AstraZeneca, Catena Pharmaceuticals, Critical Outcome Technologies, ImmunoMET, Isis Pharmaceuticals, Lilly, Medimmune, Novartis, Precision Medicine, Provista Diagnostics, Signalchem Lifesciences, Symphogen, Takeda/Millenium Pharmaceuticals, Tau Therapeutics, and Tarveda; stock options in Catena Pharmaceuticals, ImmunoMet, and Spindle Top Ventures; licensed technology in the HRD Assay, Myriad Genetics; and research funding from Adelson Medical Research Foundation, AstraZeneca, Breast Cancer Research Foundation, Critical Outcome Technologies, Illumina, Karus, Komen Research Foundation, Nanostring, and Takeda/Millenium Pharmaceuticals. J.M. has had a consulting role with MedImmune and Ziopharm; he serves on the board of directors and owns stock in Merrimack Pharmaceuticals and has patents/royalties in Imclone and Lilly.
Referees
Jonathan H. Schatz
Anonymous
REFERENCES
- Amatu A, Somaschini A, Cerea G, Bosotti R, Valtorta E, Buonandi P, Marrapese G, Veronese S, Luo D, Hornby Z, et al. 2015. Novel CAD-ALK gene rearrangement is drugable by entrectinib in colorectal cancer. Br J Cancer 113: 1730–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong F, Duplantier MM, Trempat P, Hieblot C, Lamant L, Espinos E, Racaud-Sultan C, Allouche M, Campo E, Delsol G, et al. 2004. Differential effects of X-ALK fusion proteins on proliferation, transformation, and invasion properties of NIH3T3 cells. Oncogene 23: 6071–6082. [DOI] [PubMed] [Google Scholar]
- Bagci O, Tumer S, Olgun N, Altungoz O. 2012. Copy number status and mutation analyses of anaplastic lymphoma kinase (ALK) gene in 90 sporadic neuroblastoma tumors. Cancer Lett 317: 72–77. [DOI] [PubMed] [Google Scholar]
- Bavi P, Jehan Z, Bu R, Prabhakaran S, Al-Sanea N, Al-Dayel F, Al-Assiri M, Al-Halouly T, Sairafi R, Uddin S, et al. 2013. ALK gene amplification is associated with poor prognosis in colorectal carcinoma. Br J Cancer 109: 2735–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearz A, Garassino I, Tiseo M, Caffo O, Soto-Parra H, Boccalon M, Talamini R, Santoro A, Bartolotti M, Murgia V, et al. 2010. Activity of Pemetrexed on brain metastases from non-small cell lung cancer. Lung Cancer 68: 264–268. [DOI] [PubMed] [Google Scholar]
- Berry T, Luther W, Bhatnagar N, Jamin Y, Poon E, Sanda T, Pei D, Sharma B, Vetharoy WR, Hallsworth A, et al. 2012. The ALKF1174L mutation potentiates the oncogenic activity of MYCN in neuroblastoma. Cancer Cell 22: 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonvini P, Gastaldi T, Falini B, Rosolen A. 2002. Nucleophosmin-anaplastic lymphoma kinase (NPM-ALK), a novel Hsp90-client tyrosine kinase: down-regulation of NPM-ALK expression and tyrosine phosphorylation in ALK+ CD30+ lymphoma cells by the Hsp90 antagonist 17-allylamino,17-demethoxygeldanamycin. Cancer Res 62: 1559–1566. [PubMed] [Google Scholar]
- Bossi RT, Saccardo MB, Ardini E, Menichincheri M, Rusconi L, Magnaghi P, Orsini P, Avanzi N, Borgia AL, Nesi M, et al. 2010. Crystal structures of anaplastic lymphoma kinase in complex with ATP competitive inhibitors. Biochemistry 49: 6813–6825. [DOI] [PubMed] [Google Scholar]
- Bowden ET, Stoica GE, Wellstein A. 2002. Anti-apoptotic signaling of pleiotrophin through its receptor, anaplastic lymphoma kinase. J Biol Chem 277: 35862–35868. [DOI] [PubMed] [Google Scholar]
- Bresler SC, Wood AC, Haglund EA, Courtright J, Belcastro LT, Plegaria JS, Cole K, Toporovskaya Y, Zhao H, Carpenter EL, et al. 2011. Differential inhibitor sensitivity of anaplastic lymphoma kinase variants found in neuroblastoma. Sci Transl Med 3: 108ra114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge JA, Kanamori M, Ma Z, Pickering D, Hill DA, Lydiatt W, Lui MY, Colleoni GW, Antonescu CR, Ladanyi M, et al. 2001. Fusion of the ALK gene to the clathrin heavy chain gene, CLTC, in inflammatory myofibroblastic tumor. Am J Pathol 159: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Nussenzweig A. 2013. End-joining, translocations and cancer. Nat Rev Cancer 13: 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busam KJ, Kutzner H, Cerroni L, Wiesner T. 2014. Clinical and pathologic findings of Spitz nevi and atypical Spitz tumors with ALK fusions. Am J Surg Pathol 38: 925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butrynski JE, D'Adamo DR, Hornick JL, Dal Cin P, Antonescu CR, Jhanwar SC, Ladanyi M, Capelletti M, Rodig SJ, Ramaiya N, et al. 2010. Crizotinib in ALK-rearranged inflammatory myofibroblastic tumor. N Engl J Med 363: 1727–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camidge DR, Kono SA, Lu X, Okuyama S, Baron AE, Oton AB, Davies AM, Varella-Garcia M, Franklin W, Doebele RC. 2011. Anaplastic lymphoma kinase gene rearrangements in non–small cell lung cancer are associated with prolonged progression-free survival on pemetrexed. J Thorac Oncol 6: 774–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camidge DR, Bang YJ, Kwak EL, Iafrate AJ, Varella-Garcia M, Fox SB, Riely GJ, Solomon B, Ou SH, Kim DW, et al. 2012. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol 13: 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter EL, Mosse YP. 2012. Targeting ALK in neuroblastoma—preclinical and clinical advancements. Nat Rev Clin Oncol 9: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccon M, Mologni L, Bisson W, Scapozza L, Gambacorti-Passerini C. 2013. Crizotinib-resistant NPM-ALK mutants confer differential sensitivity to unrelated Alk inhibitors. Mol Cancer Res 11: 122–132. [DOI] [PubMed] [Google Scholar]
- Cerchietti L, Damm-Welk C, Vater I, Klapper W, Harder L, Pott C, Yang SN, Reiter A, Siebert R, Melnick A, et al. 2011. Inhibition of anaplastic lymphoma kinase (ALK) activity provides a therapeutic approach for CLTC-ALK-positive human diffuse large B cell lymphomas. PLoS One 6: e18436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chand D, Yamazaki Y, Ruuth K, Schonherr C, Martinsson T, Kogner P, Attiyeh EF, Maris J, Morozova O, Marra MA, et al. 2013. Cell culture and Drosophila model systems define three classes of anaplastic lymphoma kinase mutations in neuroblastoma. Dis Model Mech 6: 373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, Wang L, Soda M, Kikuchi A, Igarashi T, et al. 2008. Oncogenic mutations of ALK kinase in neuroblastoma. Nature 455: 971–974. [DOI] [PubMed] [Google Scholar]
- Chen Z, Sasaki T, Tan X, Carretero J, Shimamura T, Li D, Xu C, Wang Y, Adelmant GO, Capelletti M, et al. 2010. Inhibition of ALK, PI3K/MEK, and HSP90 in murine lung adenocarcinoma induced by EML4-ALK fusion oncogene. Cancer Res 70: 9827–9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YL, Takeuchi K, Soda M, Inamura K, Togashi Y, Hatano S, Enomoto M, Hamada T, Haruta H, Watanabe H, et al. 2008. Identification of novel isoforms of the EML4-ALK transforming gene in non–small cell lung cancer. Cancer Res 68: 4971–4976. [DOI] [PubMed] [Google Scholar]
- Choi YL, Soda M, Yamashita Y, Ueno T, Takashima J, Nakajima T, Yatabe Y, Takeuchi K, Hamada T, Haruta H, et al. 2010. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 363: 1734–1739. [DOI] [PubMed] [Google Scholar]
- Chou A, Fraser S, Toon CW, Clarkson A, Sioson L, Farzin M, Cussigh C, Aniss A, O'Neill C, Watson N, et al. 2015. A detailed clinicopathologic study of ALK-translocated papillary thyroid carcinoma. Am J Surg Pathol 39: 652–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JG, Zou HY, Arango ME, Li Q, Lee JH, McDonnell SR, Yamazaki S, Alton GR, Mroczkowski B, Los G. 2007. Cytoreductive antitumor activity of PF-2341066, a novel inhibitor of anaplastic lymphoma kinase and c-Met, in experimental models of anaplastic large-cell lymphoma. Mol Cancer Ther 6(12 Pt 1): 3314–3322. [DOI] [PubMed] [Google Scholar]
- Chun YS, Wang L, Nascimento AG, Moir CR, Rodeberg DA. 2005. Pediatric inflammatory myofibroblastic tumor: anaplastic lymphoma kinase (ALK) expression and prognosis. Pediatr Blood Cancer 45: 796–801. [DOI] [PubMed] [Google Scholar]
- Colleoni GW, Bridge JA, Garicochea B, Liu J, Filippa DA, Ladanyi M. 2000. ATIC-ALK: a novel variant ALK gene fusion in anaplastic large cell lymphoma resulting from the recurrent cryptic chromosomal inversion, inv(p23q35). Am J Pathol 156: 781–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools J, Wlodarska I, Somers R, Mentens N, Pedeutour F, Maes B, De Wolf-Peeters C, Pauwels P, Hagemeijer A, Marynen P. 2002. Identification of novel fusion partners of ALK, the anaplastic lymphoma kinase, in anaplastic large-cell lymphoma and inflammatory myofibroblastic tumor. Genes Chromosomes Cancer 34: 354–362. [DOI] [PubMed] [Google Scholar]
- Costa DB, Shaw AT, Ou SH, Solomon BJ, Riely GJ, Ahn MJ, Zhou C, Shreeve SM, Selaru P, Polli A, et al. 2015. Clinical experience with crizotinib in patients with advanced ALK-rearranged non-small-cell lung cancer and brain metastases. J Clin Oncol 33: 1881–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crino L, Ahn MJ, De Marinis F, Groen HJ, Wakelee H, Hida T, Mok T, Spigel D, Felip E, Nishio M, et al. 2016. Multicenter phase II study of whole-body and intracranial activity with ceritinib in patients with ALK-rearranged non-small-cell lung cancer previously treated with chemotherapy and crizotinib: results from ASCEND-2. J Clin Oncol 34: 2866–2873. [DOI] [PubMed] [Google Scholar]
- Crystal AS, Shaw AT, Sequist LV, Friboulet L, Niederst MJ, Lockerman EL, Frias RL, Gainor JF, Amzallag A, Greninger P, et al. 2014. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 346: 1480–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm-Welk C, Pillon M, Woessmann W, Mussolin L. 2015. Prognostic factors in paediatric anaplastic large cell lymphoma: role of ALK. Front Biosci (Schol Ed) 7: 205–216. [DOI] [PubMed] [Google Scholar]
- d'Amore ES, Visco C, Menin A, Famengo B, Bonvini P, Lazzari E. 2013. STAT3 pathway is activated in ALK-positive large B-cell lymphoma carrying SQSTM1-ALK rearrangement and provides a possible therapeutic target. Am J Surg Pathol 37: 780–786. [DOI] [PubMed] [Google Scholar]
- De Brouwer S, De Preter K, Kumps C, Zabrocki P, Porcu M, Westerhout EM, Lakeman A, Vandesompele J, Hoebeeck J, Van Maerken T, et al. 2010. Meta-analysis of neuroblastomas reveals a skewed ALK mutation spectrum in tumors with MYCN amplification. Clin Cancer Res 16: 4353–4362. [DOI] [PubMed] [Google Scholar]
- De Paepe P, Baens M, van Krieken H, Verhasselt B, Stul M, Simons A, Poppe B, Laureys G, Brons P, Vandenberghe P, et al. 2003. ALK activation by the CLTC-ALK fusion is a recurrent event in large B-cell lymphoma. Blood 102: 2638–2641. [DOI] [PubMed] [Google Scholar]
- Di Paolo D, Yang D, Pastorino F, Emionite L, Cilli M, Daga A, Destafanis E, Di Fiore A, Piaggio F, Brignole C, et al. 2015. New therapeutic strategies in neuroblastoma: combined targeting of a novel tyrosine kinase inhibitor and liposomal siRNAs against ALK. Oncotarget 6: 28774–28789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebele RC, Lu X, Sumey C, Maxson DA, Weickhardt AJ, Oton AB, Bunn PA Jr, Baron AE, Franklin WA, Aisner DL, et al. 2012a. Oncogene status predicts patterns of metastatic spread in treatment-naive nonsmall cell lung cancer. Cancer 118: 4502–4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebele RC, Pilling AB, Aisner DL, Kutateladze TG, Le AT, Weickhardt AJ, Kondo KL, Linderman DJ, Heasley LE, Franklin WA, et al. 2012b. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non–small cell lung cancer. Clin Cancer Res 18: 1472–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang DD, Zhang B, Gu Q, Lira M, Xu Q, Sun H, Qian M, Sheng W, Ozeck M, Wang Z, et al. 2014. HIP1-ALK, a novel ALK fusion variant that responds to crizotinib. J Thorac Oncol 9: 285–294. [DOI] [PubMed] [Google Scholar]
- Franco R, Rocco G, Marino FZ, Pirozzi G, Normanno N, Morabito A, Sperlongano P, Stiuso P, Luce A, Botti G, et al. 2013. Anaplastic lymphoma kinase: a glimmer of hope in lung cancer treatment? Expert Rev Anticancer Ther 13: 407–420. [DOI] [PubMed] [Google Scholar]
- Friboulet L, Li N, Katayama R, Lee CC, Gainor JF, Crystal AS, Michellys PY, Awad MM, Yanagitani N, Kim S, et al. 2014. The ALK inhibitor ceritinib overcomes crizotinib resistance in non–small cell lung cancer. Cancer Discov 4: 662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto J, Shiota M, Iwahara T, Seki N, Satoh H, Mori S, Yamamoto T. 1996. Characterization of the transforming activity of p80, a hyperphosphorylated protein in a Ki-1 lymphoma cell line with chromosomal translocation t(2;5). Proc Natl Acad Sci 93: 4181–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadgeel SM, Gandhi L, Riely GJ, Chiappori AA, West HL, Azada MC, Morcos PN, Lee RM, Garcia L, Yu L, et al. 2014. Safety and activity of alectinib against systemic disease and brain metastases in patients with crizotinib-resistant ALK-rearranged non-small-cell lung cancer (AF-002JG): results from the dose-finding portion of a phase 1/2 study. Lancet Oncol 15: 1119–1128. [DOI] [PubMed] [Google Scholar]
- Gainor JF, Dardaei L, Yoda S, Friboulet L, Leshchiner I, Katayama R, Dagogo-Jack I, Gadgeel S, Schultz K, Singh M, et al. 2016. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancer. Cancer Discov 6: 1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George RE, Sanda T, Hanna M, Frohling S, Luther W II, Zhang J, Ahn Y, Zhou W, London WB, McGrady P, et al. 2008. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature 455: 975–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George SK, Vishwamitra D, Manshouri R, Shi P, Amin HM. 2014. The ALK inhibitor ASP3026 eradicates NPM-ALK+ T-cell anaplastic large-cell lymphoma in vitro and in a systemic xenograft lymphoma model. Oncotarget 5: 5750–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godbert Y, Henriques de Figueiredo B, Bonichon F, Chibon F, Hostein I, Perot G, Dupin C, Daubech A, Belleannee G, Gros A, et al. 2015. Remarkable response to crizotinib in woman with anaplastic lymphoma kinase-rearranged anaplastic thyroid carcinoma. J Clin Oncol 33: e84–e87. [DOI] [PubMed] [Google Scholar]
- Griffin CA, Hawkins AL, Dvorak C, Henkle C, Ellingham T, Perlman EJ. 1999. Recurrent involvement of 2p23 in inflammatory myofibroblastic tumors. Cancer Res 59: 2776–2780. [PubMed] [Google Scholar]
- Gustafson S, Medeiros LJ, Kalhor N, Bueso-Ramos CE. 2009. Anaplastic large cell lymphoma: another entity in the differential diagnosis of small round blue cell tumors. Ann Diagn Pathol 13: 413–427. [DOI] [PubMed] [Google Scholar]
- Hallberg B, Palmer RH. 2013. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer 13: 685–700. [DOI] [PubMed] [Google Scholar]
- Hanna MG, Najfeld V, Irie HY, Tripodi J, Nayak A. 2015. Analysis of ALK gene in 133 patients with breast cancer revealed polysomy of chromosome 2 and no ALK amplification. Springerplus 4: 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez L, Pinyol M, Hernandez S, Bea S, Pulford K, Rosenwald A, Lamant L, Falini B, Ott G, Mason DY, et al. 1999. TRK-fused gene (TFG) is a new partner of ALK in anaplastic large cell lymphoma producing two structurally different TFG-ALK translocations. Blood 94: 3265–3268. [PubMed] [Google Scholar]
- Hernandez L, Bea S, Bellosillo B, Pinyol M, Falini B, Carbone A, Ott G, Rosenwald A, Fernandez A, Pulford K, et al. 2002. Diversity of genomic breakpoints in TFG-ALK translocations in anaplastic large cell lymphomas: identification of a new TFG-ALKXL chimeric gene with transforming activity. Am J Pathol 160: 1487–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuckmann JM, Holzel M, Sos ML, Heynck S, Balke-Want H, Koker M, Peifer M, Weiss J, Lovly CM, Grutter C, et al. 2011. ALK mutations conferring differential resistance to structurally diverse ALK inhibitors. Clin Cancer Res 17: 7394–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Kim RN, Song JY, Choi SJ, Oh E, Lira ME, Mao M, Takeuchi K, Han J, Kim J, et al. 2014. HIP1-ALK, a novel fusion protein identified in lung adenocarcinoma. J Thorac Oncol 9: 419–422. [DOI] [PubMed] [Google Scholar]
- Hong S, Chen N, Fang W, Zhan J, Liu Q, Kang S, He X, Liu L, Zhou T, Huang J, et al. 2016. Upregulation of PD-L1 by EML4-ALK fusion protein mediates the immune escape in ALK positive NSCLC: implication for optional anti-PD-1/PD-L1 immune therapy for ALK-TKIs sensitive and resistant NSCLC patients. Oncoimmunology 5: e1094598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn L, Pao W. 2009. EML4-ALK: honing in on a new target in non-small-cell lung cancer. J Clin Oncol 27: 4232–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrustanovic G, Olivas V, Pazarentzos E, Tulpule A, Asthana S, Blakely CM, Okimoto RA, Lin L, Neel DS, Sabnis A, et al. 2015. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat Med 21: 1038–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Ouyang X. 2013. Predictive biochemical-markers for the development of brain metastases from lung cancer: clinical evidence and future directions. Cancer Epidemiol 37: 703–707. [DOI] [PubMed] [Google Scholar]
- Huang WS, Liu S, Zou D, Thomas M, Wang Y, Zhou T, Romero J, Kohlmann A, Li F, Qi J, et al. 2016. Discovery of brigatinib (AP26113), a phosphine oxide-containing, potent, orally active inhibitor of anaplastic lymphoma kinase. J Med Chem 59: 4948–4964. [DOI] [PubMed] [Google Scholar]
- Infarinato NR, Park JH, Krytska K, Ryles HT, Sano R, Szigety KM, Li Y, Zou HY, Lee NV, Smeal T, et al. 2015. The ALK/ROS1 inhibitor PF-06463922 overcomes primary resistance to crizotinib in ALK-driven neuroblastoma. Cancer Discov 6: 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahara T, Fujimoto J, Wen D, Cupples R, Bucay N, Arakawa T, Mori S, Ratzkin B, Yamamoto T. 1997. Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene 14: 439–449. [DOI] [PubMed] [Google Scholar]
- Janoueix-Lerosey I, Lequin D, Brugieres L, Ribeiro A, de Pontual L, Combaret V, Raynal V, Puisieux A, Schleiermacher G, Pierron G, et al. 2008. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 455: 967–970. [DOI] [PubMed] [Google Scholar]
- Ji C, Zhang L, Cheng Y, Patel R, Wu H, Zhang Y, Wang M, Ji S, Belani CP, Yang JM, et al. 2014. Induction of autophagy contributes to crizotinib resistance in ALK-positive lung cancer. Cancer Biol Ther 15: 570–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji JH, Oh YL, Hong M, Yun JW, Lee HW, Kim D, Ji Y, Kim DH, Park WY, Shin HT, et al. 2015. Identification of driving ALK fusion genes and genomic landscape of medullary thyroid cancer. PLoS Genet 11: e1005467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TW, Richardson PF, Bailey S, Brooun A, Burke BJ, Collins MR, Cui JJ, Deal JG, Deng YL, Dinh D, et al. 2014. Discovery of (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]-benzoxadiazacyclotetradecine-3-carbonitrile (PF-06463922), a macrocyclic inhibitor of anaplastic lymphoma kinase (ALK) and c-ros oncogene 1 (ROS1) with preclinical brain exposure and broad-spectrum potency against ALK-resistant mutations. J Med Chem 57: 4720–4744. [DOI] [PubMed] [Google Scholar]
- Katayama R, Khan TM, Benes C, Lifshits E, Ebi H, Rivera VM, Shakespeare WC, Iafrate AJ, Engelman JA, Shaw AT. 2011. Therapeutic strategies to overcome crizotinib resistance in non–small cell lung cancers harboring the fusion oncogene EML4-ALK. Proc Natl Acad Sci 108: 7535–7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama R, Shaw AT, Khan TM, Mino-Kenudson M, Solomon BJ, Halmos B, Jessop NA, Wain JC, Yeo AT, Benes C, et al. 2012. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung cancers. Sci Transl Med 4: 120ra117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama R, Friboulet L, Koike S, Lockerman EL, Khan TM, Gainor JF, Iafrate AJ, Takeuchi K, Taiji M, Okuno Y, et al. 2014. Two novel ALK mutations mediate acquired resistance to the next-generation ALK inhibitor alectinib. Clin Cancer Res 20: 5686–5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher FC, McDermott R. 2010. The emerging pathogenic and therapeutic importance of the anaplastic lymphoma kinase gene. Eur J Cancer 46: 2357–2368. [DOI] [PubMed] [Google Scholar]
- Kelly LM, Barila G, Liu P, Evdokimova VN, Trivedi S, Panebianco F, Gandhi M, Carty SE, Hodak SP, Luo J, et al. 2014. Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer. Proc Natl Acad Sci 111: 4233–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstein D, Gettinger S, Gold K, Langer CJ, Shaw AT, Bazhenova LA, Salgia R, Dorer DJ, Conlan MG, Camidge DR. 2015. Evaluation of anaplastic lymphoma kinase (ALK) inhibitor brigatinib [AP26113] in patients (pts) with ALK+ non–small cell lung cancer (NSCLC) and brain metastases. Ann Oncol 26(Suppl 1): i60–i61. [Google Scholar]
- Khadija K, Auger N, Lueza B, Commo F, Valent A, Rousseau V, Dorvault N, Friboulet L, Olaussen K, Rajpar L, et al. 2012. ALK amplification and crizotinib sensitivity in non-small cell lung cancer cell lines and patients report. ASCO Meeting Abstr 30: 10556. [Google Scholar]
- Kim DW, Mehra R, Tan DSW, Felip E, Chow LQM, Camidge DR, Vansteenkiste JF, Sharma S, De Pas T, Riely GJ, et al. 2014. Ceritinib in advanced anaplastic lymphoma kinase (ALK)-rearranged (ALK+) non–small cell lung cancer (NSCLC): results of the ASCEND-1 trial. ASCO Meeting Abstracts 32(15 suppl): 8003. [Google Scholar]
- Kim MH, Lee S, Koo JS, Jung KH, Park IH, Jeong J, Kim SI, Park S, Park HS, Park BW, et al. 2015. Anaplastic lymphoma kinase gene copy number gain in inflammatory breast cancer (IBC): prevalence, clinicopathologic features and prognostic implication. PLoS One 10: e0120320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Mehra R, Tan DS, Felip E, Chow LQ, Camidge DR, Vansteenkiste J, Sharma S, De Pas T, Riely GJ, et al. 2016. Activity and safety of ceritinib in patients with ALK-rearranged non-small-cell lung cancer (ASCEND-1): updated results from the multicentre, open-label, phase 1 trial. Lancet Oncol 17: 452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama T, Tsukaguchi T, Yoshida M, Kondoh O, Sakamoto H. 2014. Selective ALK inhibitor alectinib with potent antitumor activity in models of crizotinib resistance. Cancer Lett 351: 215–221. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S, Woodward W, Yang W, Reuben JM, Tepperberg J, Ogura D, Niwa S, Huo L, Gong Y, El-Zein R, et al. 2013. Status of the anaplastic lymphoma kinase (ALK) gene in inflammatory breast carcinoma. Springerplus 2: 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo AH, Stoica GE, Riegel AT, Wellstein A. 2007. Recruitment of insulin receptor substrate-1 and activation of NF-κB essential for midkine growth signaling through anaplastic lymphoma kinase. Oncogene 26: 859–869. [DOI] [PubMed] [Google Scholar]
- Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, et al. 2010. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363: 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamant L, Dastugue N, Pulford K, Delsol G, Mariame B. 1999. A new fusion gene TPM3-ALK in anaplastic large cell lymphoma created by a (1;2)(q25;p23) translocation. Blood 93: 3088–3095. [PubMed] [Google Scholar]
- Lawrence B, Perez-Atayde A, Hibbard MK, Rubin BP, Dal Cin P, Pinkus JL, Pinkus GS, Xiao S, Yi ES, Fletcher CD, et al. 2000. TPM3-ALK and TPM4-ALK oncogenes in inflammatory myofibroblastic tumors. Am J Pathol 157: 377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Lim SM, Rha SY, Roh JK, Cho YJ, Shin KH, Yang WI, Kim SH, Kim HS. 2014. Prognostic implications of anaplastic lymphoma kinase gene aberrations in rhabdomyosarcoma; an immunohistochemical and fluorescence in situ hybridisation study. J Clin Pathol 67: 33–39. [DOI] [PubMed] [Google Scholar]
- Lee J, Kim HC, Hong JY, Wang K, Kim SY, Jang J, Kim ST, Park JO, Lim HY, Kang WK, et al. 2015. Detection of novel and potentially actionable anaplastic lymphoma kinase (ALK) rearrangement in colorectal adenocarcinoma by immunohistochemistry screening. Oncotarget 6: 24320–24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ye X, Liu J, Zha J, Pei L. 2011. Evaluation of EML4-ALK fusion proteins in non-small cell lung cancer using small molecule inhibitors. Neoplasia 13: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao EH, Hung W, Abrams B, Zhen M. 2004. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature 430: 345–350. [DOI] [PubMed] [Google Scholar]
- Lin E, Li L, Guan Y, Soriano R, Rivers CS, Mohan S, Pandita A, Tang J, Modrusan Z. 2009. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non–small cell lung cancers. Mol Cancer Res 7: 1466–1476. [DOI] [PubMed] [Google Scholar]
- Lopes SS, Yang X, Muller J, Carney TJ, McAdow AR, Rauch GJ, Jacoby AS, Hurst LD, Delfino-Machin M, Haffter P, et al. 2008. Leukocyte tyrosine kinase functions in pigment cell development. PLoS Genet 4: e1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loren CE, Englund C, Grabbe C, Hallberg B, Hunter T, Palmer RH. 2003. A crucial role for the anaplastic lymphoma kinase receptor tyrosine kinase in gut development in Drosophila melanogaster. EMBO Rep 4: 781–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovly CM, Heuckmann JM, de Stanchina E, Chen H, Thomas RK, Liang C, Pao W. 2011. Insights into ALK-driven cancers revealed through development of novel ALK tyrosine kinase inhibitors. Cancer Res 71: 4920–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovly CM, McDonald NT, Chen H, Ortiz-Cuaran S, Heukamp LC, Yan Y, Florin A, Ozretic L, Lim D, Wang L, et al. 2014. Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer. Nat Med 20: 1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KV, Jong KA, Kim GY, Singh J, Dia EQ, Yoshimoto K, Wang MY, Cloughesy TF, Nelson SF, Mischel PS. 2005. Differential induction of glioblastoma migration and growth by two forms of pleiotrophin. J Biol Chem 280: 26953–26964. [DOI] [PubMed] [Google Scholar]
- Ma Z, Hill DA, Collins MH, Morris SW, Sumegi J, Zhou M, Zuppan C, Bridge JA. 2003. Fusion of ALK to the Ran-binding protein 2 (RANBP2) gene in inflammatory myofibroblastic tumor. Genes Chromosomes Cancer 37: 98–105. [DOI] [PubMed] [Google Scholar]
- Martinsson T, Eriksson T, Abrahamsson J, Caren H, Hansson M, Kogner P, Kamaraj S, Schonherr C, Weinmar J, Ruuth K, et al. 2011. Appearance of the novel activating F1174S ALK mutation in neuroblastoma correlates with aggressive tumor progression and unresponsiveness to therapy. Cancer Res 71: 98–105. [DOI] [PubMed] [Google Scholar]
- Mathivet T, Mazot P, Vigny M. 2007. In contrast to agonist monoclonal antibodies, both C-terminal truncated form and full length form of Pleiotrophin failed to activate vertebrate ALK (anaplastic lymphoma kinase)? Cell Signal 19: 2434–2443. [DOI] [PubMed] [Google Scholar]
- McManus DT, Catherwood MA, Carey PD, Cuthbert RJ, Alexander HD. 2004. ALK-positive diffuse large B-cell lymphoma of the stomach associated with a clathrin-ALK rearrangement. Hum Pathol 35: 1285–1288. [DOI] [PubMed] [Google Scholar]
- Medeiros LJ, Elenitoba-Johnson KS. 2007. Anaplastic large cell lymphoma. Am J Clin Pathol 127: 707–722. [DOI] [PubMed] [Google Scholar]
- Meech SJ, McGavran L, Odom LF, Liang X, Meltesen L, Gump J, Wei Q, Carlsen S, Hunger SP. 2001. Unusual childhood extramedullary hematologic malignancy with natural killer cell properties that contains tropomyosin 4–anaplastic lymphoma kinase gene fusion. Blood 98: 1209–1216. [DOI] [PubMed] [Google Scholar]
- Megiorni F, McDowell HP, Camero S, Mannarino O, Ceccarelli S, Paiano M, Losty PD, Pizer B, Shukla R, Pizzuti A, et al. 2015. Crizotinib-induced antitumour activity in human alveolar rhabdomyosarcoma cells is not solely dependent on ALK and MET inhibition. J Exp Clin Cancer Res 34: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengoli MC, Barbieri F, Bertolini F, Tiseo M, Rossi G. 2016. K-RAS mutations indicating primary resistance to crizotinib in ALK-rearranged adenocarcinomas of the lung: report of two cases and review of the literature. Lung Cancer 93: 55–58. [DOI] [PubMed] [Google Scholar]
- Meric-Bernstam F, Johnson A, Holla V, Bailey AM, Brusco L, Chen K, Routbort M, Patel KP, Zeng J, Kopetz S, et al. 2015. A decision support framework for genomically informed investigational cancer therapy. J Natl Cancer Inst 107: djv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake I, Hakomori Y, Shinohara A, Gamou T, Saito M, Iwamatsu A, Sakai R. 2002. Activation of anaplastic lymphoma kinase is responsible for hyperphosphorylation of ShcC in neuroblastoma cell lines. Oncogene 21: 5823–5834. [DOI] [PubMed] [Google Scholar]
- Mologni L, Ceccon M, Pirola A, Chiriano G, Piazza R, Scapozza L, Gambacorti-Passerini C. 2015. NPM/ALK mutants resistant to ASP3026 display variable sensitivity to alternative ALK inhibitors but succumb to the novel compound PF-06463922. Oncotarget 6: 5720–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moog-Lutz C, Degoutin J, Gouzi JY, Frobert Y, Brunet-de Carvalho N, Bureau J, Creminon C, Vigny M. 2005. Activation and inhibition of anaplastic lymphoma kinase receptor tyrosine kinase by monoclonal antibodies and absence of agonist activity of pleiotrophin. J Biol Chem 280: 26039–26048. [DOI] [PubMed] [Google Scholar]
- Mori M, Ueno Y, Konagai S, Fushiki H, Shimada I, Kondoh Y, Saito R, Mori K, Shindou N, Soga T, et al. 2014. The selective anaplastic lymphoma receptor tyrosine kinase inhibitor ASP3026 induces tumor regression and prolongs survival in non-small cell lung cancer model mice. Mol Cancer Ther 13: 329–340. [DOI] [PubMed] [Google Scholar]
- Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT. 1994. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science 263: 1281–1284. [DOI] [PubMed] [Google Scholar]
- Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, Laquaglia MJ, Sennett R, Lynch JE, Perri P, et al. 2008. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 455: 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosse YP, Lim MS, Voss SD, Wilner K, Ruffner K, Laliberte J, Rolland D, Balis FM, Maris JM, Weigel BJ, et al. 2013. Safety and activity of crizotinib for paediatric patients with refractory solid tumours or anaplastic large-cell lymphoma: a Children's Oncology Group phase 1 consortium study. Lancet Oncol 14: 472–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PB, Lax I, Reshetnyak A, Ligon GF, Lillquist JS, Natoli EJ Jr, Shi X, Folta-Stogniew E, Gunel M, Alvarado D, et al. 2015. Heparin is an activating ligand of the orphan receptor tyrosine kinase ALK. Sci Signal 8: ra6. [DOI] [PubMed] [Google Scholar]
- Murugan AK, Xing M. 2011. Anaplastic thyroid cancers harbor novel oncogenic mutations of the ALK gene. Cancer Res 71: 4403–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio M, Murakami H, Horiike A, Takahashi T, Hirai F, Suenaga N, Tajima T, Tokushige K, Ishii M, Boral A, et al. 2015. Phase I study of ceritinib (LDK378) in Japanese patients with advanced, anaplastic lymphoma kinase-rearranged non-small-cell lung cancer or other tumors. J Thorac Oncol 10: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normant E, Paez G, West KA, Lim AR, Slocum KL, Tunkey C, McDougall J, Wylie AA, Robison K, Caliri K, et al. 2011. The Hsp90 inhibitor IPI-504 rapidly lowers EML4-ALK levels and induces tumor regression in ALK-driven NSCLC models. Oncogene 30: 2581–2586. [DOI] [PubMed] [Google Scholar]
- Osajima-Hakomori Y, Miyake I, Ohira M, Nakagawara A, Nakagawa A, Sakai R. 2005. Biological role of anaplastic lymphoma kinase in neuroblastoma. Am J Pathol 167: 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota K, Azuma K, Kawahara A, Hattori S, Iwama E, Tanizaki J, Harada T, Matsumoto K, Takayama K, Takamori S, et al. 2015. Induction of PD-L1 expression by the EML4-ALK oncoprotein and downstream signaling pathways in non-small cell lung cancer. Clin Cancer Res 21: 4014–4021. [DOI] [PubMed] [Google Scholar]
- Ou SH, Klempner SJ, Greenbowe JR, Azada M, Schrock AB, Ali SM, Ross JS, Stephens PJ, Miller VA. 2014. Identification of a novel HIP1-ALK fusion variant in non-small-cell lung cancer (NSCLC) and discovery of ALK I1171 (I1171N/S) mutations in two ALK-rearranged NSCLC patients with resistance to alectinib. J Thorac Oncol 9: 1821–1825. [DOI] [PubMed] [Google Scholar]
- Ou SH, Greenbowe J, Khan ZU, Azada MC, Ross JS, Stevens PJ, Ali SM, Miller VA, Gitlitz B. 2015. I1171 missense mutation (particularly I1171N) is a common resistance mutation in ALK-positive NSCLC patients who have progressive disease while on alectinib and is sensitive to ceritinib. Lung Cancer 88: 231–234. [DOI] [PubMed] [Google Scholar]
- Ouchi K, Miyachi M, Tsuma Y, Tsuchiya K, Iehara T, Konishi E, Yanagisawa A, Hosoi H. 2015. FN1: a novel fusion partner of ALK in an inflammatory myofibroblastic tumor. Pediatr Blood Cancer 62: 909–911. [DOI] [PubMed] [Google Scholar]
- Palmer RH, Vernersson E, Grabbe C, Hallberg B. 2009. Anaplastic lymphoma kinase: signalling in development and disease. Biochem J 420: 345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Zhang W, Chang Y, Vega JA, Deuel TF. 2007. Anaplastic lymphoma kinase is activated through the pleiotrophin/receptor protein-tyrosine phosphatase β/ζ signaling pathway: an alternative mechanism of receptor tyrosine kinase activation. J Biol Chem 282: 28683–28690. [DOI] [PubMed] [Google Scholar]
- Perner S, Wagner PL, Demichelis F, Mehra R, Lafargue CJ, Moss BJ, Arbogast S, Soltermann A, Weder W, Giordano TJ, et al. 2008. EML4-ALK fusion lung cancer: a rare acquired event. Neoplasia 10: 298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrantonio F, Maggi C, Di Bartolomeo M, Facciorusso MG, Perrone F, Testi A, Iacovelli R, Miceli R, Bossi I, Leone G, et al. 2014. Gain of ALK gene copy number may predict lack of benefit from anti-EGFR treatment in patients with advanced colorectal cancer and RAS-RAF-PI3KCA wild-type status. PLoS One 9: e92147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers C, Aigner A, Stoica GE, McDonnell K, Wellstein A. 2002. Pleiotrophin signaling through anaplastic lymphoma kinase is rate-limiting for glioblastoma growth. J Biol Chem 277: 14153–14158. [DOI] [PubMed] [Google Scholar]
- Pulford K, Morris SW, Turturro F. 2004. Anaplastic lymphoma kinase proteins in growth control and cancer. J Cell Physiol 199: 330–358. [DOI] [PubMed] [Google Scholar]
- Rangachari D, Yamaguchi N, VanderLaan PA, Folch E, Mahadevan A, Floyd SR, Uhlmann EJ, Wong ET, Dahlberg SE, Huberman MS, et al. 2015. Brain metastases in patients with EGFR-mutated or ALK-rearranged non-small-cell lung cancers. Lung Cancer 88: 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner DJ, Ailion M, Thomas JH, Meyer BJ. 2008. C. elegans anaplastic lymphoma kinase ortholog SCD-2 controls dauer formation by modulating TGF-β signaling. Curr Biol 18: 1101–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Tan ZP, Zhu X, Crosby K, Haack H, Ren JM, Beausoleil S, Moritz A, Innocenti G, Rush J, et al. 2012. Identification of anaplastic lymphoma kinase as a potential therapeutic target in ovarian cancer. Cancer Res 72: 3312–3323. [DOI] [PubMed] [Google Scholar]
- Rohrbough J, Broadie K. 2010. Anterograde Jelly belly ligand to Alk receptor signaling at developing synapses is regulated by Mind the gap. Development 137: 3523–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R Jr. 2013. Anaplastic lymphoma kinase (ALK): structure, oncogenic activation, and pharmacological inhibition. Pharmacol Res 68: 68–94. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Tsukaguchi T, Hiroshima S, Kodama T, Kobayashi T, Fukami TA, Oikawa N, Tsukuda T, Ishii N, Aoki Y. 2011. CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell 19: 679–690. [DOI] [PubMed] [Google Scholar]
- Salido M, Pijuan L, Martinez-Aviles L, Galvan AB, Canadas I, Rovira A, Zanui M, Martinez A, Longaron R, Sole F, et al. 2011. Increased ALK gene copy number and amplification are frequent in non-small cell lung cancer. J Thorac Oncol 6: 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang J, Acquaviva J, Friedland JC, Smith DL, Sequeira M, Zhang C, Jiang Q, Xue L, Lovly CM, Jimenez JP, et al. 2013. Targeted inhibition of the molecular chaperone Hsp90 overcomes ALK inhibitor resistance in non-small cell lung cancer. Cancer Discov 3: 430–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Okuda K, Zheng W, Butrynski J, Capelletti M, Wang L, Gray NS, Wilner K, Christensen JG, Demetri G, et al. 2010. The neuroblastoma-associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK-translocated cancers. Cancer Res 70: 10038–10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonherr C, Ruuth K, Yamazaki Y, Eriksson T, Christensen J, Palmer RH, Hallberg B. 2011. Activating ALK mutations found in neuroblastoma are inhibited by crizotinib and NVP-TAE684. Biochem J 440: 405–413. [DOI] [PubMed] [Google Scholar]
- Schoppmann SF, Streubel B, Birner P. 2013. Amplification but not translocation of anaplastic lymphoma kinase is a frequent event in oesophageal cancer. Eur J Cancer 49: 1876–1881. [DOI] [PubMed] [Google Scholar]
- Sequist LV, Gettinger S, Senzer NN, Martins RG, Janne PA, Lilenbaum R, Gray JE, Iafrate AJ, Katayama R, Hafeez N, et al. 2010. Activity of IPI-504, a novel heat-shock protein 90 inhibitor, in patients with molecularly defined non-small-cell lung cancer. J Clin Oncol 28: 4953–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto T, Kiura K, Nishio M, Nakagawa K, Maemondo M, Inoue A, Hida T, Yamamoto N, Yoshioka H, Harada M, et al. 2013. CH5424802 (RO5424802) for patients with ALK-rearranged advanced non-small-cell lung cancer (AF-001JP study): a single-arm, open-label, phase 1-2 study. Lancet Oncol 14: 590–598. [DOI] [PubMed] [Google Scholar]
- Shaw AT, Engelman JA. 2013. ALK in lung cancer: past, present, and future. J Clin Oncol 31: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, Solomon B, Stubbs H, Admane S, McDermott U, et al. 2009. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 27: 4247–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AT, Yeap BY, Solomon BJ, Riely GJ, Gainor J, Engelman JA, Shapiro GI, Costa DB, Ou SH, Butaney M, et al. 2011. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol 12: 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AT, Kim DW, Nakagawa K, Seto T, Crino L, Ahn MJ, De Pas T, Besse B, Solomon BJ, Blackhall F, et al. 2013. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 368: 2385–2394. [DOI] [PubMed] [Google Scholar]
- Shaw AT, Kim DW, Mehra R, Tan DS, Felip E, Chow LQ, Camidge DR, Vansteenkiste J, Sharma S, De Pas T, et al. 2014. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med 370: 1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AT, Friboulet L, Leshchiner I, Gainor JF, Bergqvist S, Brooun A, Burke BJ, Deng YL, Liu W, Dardaei L, et al. 2016a. Resensitization to crizotinib by the lorlatinib ALK resistance mutation L1198F. N Engl J Med 374: 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AT, Gandhi L, Gadgeel S, Riely GJ, Cetnar J, West H, Camidge DR, Socinski MA, Chiappori A, Mekhail T, et al. 2016b. Alectinib in ALK-positive, crizotinib-resistant, non-small-cell lung cancer: a single-group, multicentre, phase 2 trial. Lancet Oncol 17: 234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaw JT, Wan H, Pfeifer K, Rivera VM, Guan J, Palmer RH, Hallberg B. 2016. Brigatinib, an anaplastic lymphoma kinase inhibitor, abrogates activity and growth in ALK-positive neuroblastoma cells, Drosophila and mice. Oncotarget 7: 29011–29022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoff MJ, Lally B, Slade MG, Goldberg WG, Lee P, Michaud GC, Wahidi MM, Chawla M. 2013. Symptom management in patients with lung cancer: diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 143(5 Suppl): e455S–e497S. [DOI] [PubMed] [Google Scholar]
- Siraj AK, Beg S, Jehan Z, Prabhakaran S, Ahmed M, R Hussain A, Al-Dayel F, Tulbah A, Ajarim D, Al-Kuraya KS. 2015. ALK alteration is a frequent event in aggressive breast cancers. Breast Cancer Res 17: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. 2007. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448: 561–566. [DOI] [PubMed] [Google Scholar]
- Solomon B, Varella-Garcia M, Camidge DR. 2009. ALK gene rearrangements: a new therapeutic target in a molecularly defined subset of non–small cell lung cancer. J Thorac Oncol 4: 1450–1454. [DOI] [PubMed] [Google Scholar]
- Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa K, Mekhail T, Felip E, Cappuzzo F, Paolini J, Usari T, et al. 2014. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 371: 2167–2177. [DOI] [PubMed] [Google Scholar]
- Stoica GE, Kuo A, Aigner A, Sunitha I, Souttou B, Malerczyk C, Caughey DJ, Wen D, Karavanov A, Riegel AT, et al. 2001. Identification of anaplastic lymphoma kinase as a receptor for the growth factor pleiotrophin. J Biol Chem 276: 16772–16779. [DOI] [PubMed] [Google Scholar]
- Stoica GE, Kuo A, Powers C, Bowden ET, Sale EB, Riegel AT, Wellstein A. 2002. Midkine binds to anaplastic lymphoma kinase (ALK) and acts as a growth factor for different cell types. J Biol Chem 277: 35990–35998. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Sonobe M, Kobayashi M, Yoshizawa A, Menju T, Nakayama E, Mino N, Iwakiri S, Sato K, Miyahara R, et al. 2010. Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann Surg Oncol 17: 889–897. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Choi YL, Togashi Y, Soda M, Hatano S, Inamura K, Takada S, Ueno T, Yamashita Y, Satoh Y, et al. 2009. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res 15: 3143–3149. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Soda M, Togashi Y, Ota Y, Sekiguchi Y, Hatano S, Asaka R, Noguchi M, Mano H. 2011. Identification of a novel fusion, SQSTM1-ALK, in ALK-positive large B-cell lymphoma. Haematologica 96: 464–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizaki J, Okamoto I, Takezawa K, Sakai K, Azuma K, Kuwata K, Yamaguchi H, Hatashita E, Nishio K, Janne PA, et al. 2012. Combined effect of ALK and MEK inhibitors in EML4-ALK-positive non-small-cell lung cancer cells. Br J Cancer 106: 763–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togashi Y, Soda M, Sakata S, Sugawara E, Hatano S, Asaka R, Nakajima T, Mano H, Takeuchi K. 2012. KLC1-ALK: a novel fusion in lung cancer identified using a formalin-fixed paraffin-embedded tissue only. PLoS One 7: e31323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gaal JC, Flucke UE, Roeffen MH, de Bont ES, Sleijfer S, Mavinkurve-Groothuis AM, Suurmeijer AJ, van der Graaf WT, Versleijen-Jonkers YM. 2012. Anaplastic lymphoma kinase aberrations in rhabdomyosarcoma: clinical and prognostic implications. J Clin Oncol 30: 308–315. [DOI] [PubMed] [Google Scholar]
- van Gaal JC, Roeffen MH, Flucke UE, van der Laak JA, van der Heijden G, de Bont ES, Suurmeijer AJ, Versleijen-Jonkers YM, van der Graaf WT. 2013. Simultaneous targeting of insulin-like growth factor-1 receptor and anaplastic lymphoma kinase in embryonal and alveolar rhabdomyosarcoma: a rational choice. Eur J Cancer 49: 3462–3470. [DOI] [PubMed] [Google Scholar]
- Wang YW, Tu PH, Lin KT, Lin SC, Ko JY, Jou YS. 2011. Identification of oncogenic point mutations and hyperphosphorylation of anaplastic lymphoma kinase in lung cancer. Neoplasia 13: 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Krishnan C, Nguyen EP, Meyer KJ, Oliveira JL, Yang P, Yi ES, Erickson-Johnson MR, Yaszemski MJ, Maran A, et al. 2012. Fusion of dynactin 1 to anaplastic lymphoma kinase in inflammatory myofibroblastic tumor. Hum Pathol 43: 2047–2052. [DOI] [PubMed] [Google Scholar]
- Wang M, Zhou C, Sun Q, Cai R, Li Y, Wang D, Gong L. 2013. ALK amplification and protein expression predict inferior prognosis in neuroblastomas. Exp Mol Pathol 95: 124–130. [DOI] [PubMed] [Google Scholar]
- Wang D, Li D, Qin G, Zhang W, Ouyang J, Zhang M, Xie L. 2015. The structural characterization of tumor fusion genes and proteins. Comput Math Methods Med 2015: 912742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb TR, Slavish J, George RE, Look AT, Xue L, Jiang Q, Cui X, Rentrop WB, Morris SW. 2009. Anaplastic lymphoma kinase: role in cancer pathogenesis and small-molecule inhibitor development for therapy. Expert Rev Anticancer Ther 9: 331–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner T, He J, Yelensky R, Esteve-Puig R, Botton T, Yeh I, Lipson D, Otto G, Brennan K, Murali R, et al. 2014. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat Commun 5: 3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner T, Lee W, Obenauf AC, Ran L, Murali R, Zhang QF, Wong EW, Hu W, Scott SN, Shah RH, et al. 2015. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature 526: 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Cheng M, Zhang Q, Wasik M, Kelsh R, Winkler C. 2013. Anaplastic lymphoma kinase is required for neurogenesis in the developing central nervous system of zebrafish. PLoS One 8: e63757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Shibata T, Wakai S, Ushiku T, Tsuta K, Fukayama M, Makimoto A, Furuta K, Tsuda H. 2013. Anaplastic lymphoma kinase status in rhabdomyosarcomas. Mod Pathol 26: 772–781. [DOI] [PubMed] [Google Scholar]
- Yoshimura Y, Kurasawa M, Yorozu K, Puig O, Bordogna W, Harada N. 2016. Antitumor activity of alectinib, a selective ALK inhibitor, in an ALK-positive NSCLC cell line harboring G1269A mutation: efficacy of alectinib against ALK G1269A mutated cells. Cancer Chemother Pharmacol 77: 623–628. [DOI] [PubMed] [Google Scholar]
- Zou HY, Friboulet L, Kodack DP, Engstrom LD, Li Q, West M, Tang RW, Wang H, Tsaparikos K, Wang J, et al. 2015. PF-06463922, an ALK/ROS1 inhibitor, overcomes resistance to first and second generation ALK inhibitors in preclinical models. Cancer Cell 28: 70–81. [DOI] [PMC free article] [PubMed] [Google Scholar]