Abstract
Although the capacity for electroactive bacteria to convert environmental metallic minerals and organic pollutants is well known, the role of the redox properties of microbial extracellular polymeric substances (EPS) in this process is poorly understood. In this work, the redox properties of EPS from two widely present electroactive bacterial strains (Shewanella oneidensis and Pseudomonas putida) were explored. Electrochemical analysis demonstrates that the EPS extracted from the two strains exhibited redox properties. Spectroelectrochemical and protein electrophoresis analyses indicate that the extracted EPS from S. oneidensis and P. putida contained heme-binding proteins, which were identified as the possible redox components in the EPS. The results of heme-mediated behavior of EPS may provide an insight into the important roles of EPS in electroactive bacteria to maximize their redox capability for biogeochemical cycling, environmental bioremediation and wastewater treatment.
Extracellular polymeric substances (EPS) are a complex high-molecular-weight polymer mixture composed mainly of proteins1, polysaccharides2, humic substances, etc., that are secreted by microorganisms3. The presence of EPS in pure and mixed cultures has been reported4,5. Because of their important roles in the biogeochemical cycling of pollutants (e.g., biosorption, biomineralization, and biocorrosion6), microbial EPS have attracted extensive attention. While few studies have been performed on the redox features of EPS because their complex and heterogeneous compositions and characteristics make this topic of research difficult to investigate. It has been suggested that the redox properties of EPS may arise from bacterial refractory polymers, such as proteins and, possibly, humic substances, which could serve as electron donors or acceptors in bacterial biofilms7.
Electroactive bacteria, which can transport electrons over biological membranes to or from their extracellular environment, have been successfully used to generate electricity from waste materials8,9. The redox properties of EPS in electroactive bacteria may be important to their ability to biochemically modify metals and organic pollutants. EPS extracted from a model electroactive bacterium, Shewanella, has been shown to be involved in U(VI) immobilization through sorption and reduction of U(VI)4,10. Additionally, EPS extracted from Shewanella may enhance the biotransformation of organic pollutants11,12,13. Recently, approximately 20 redox proteins in EPS from Shewanella sp. were identified14. Though there is limited research into the redox properties of Pseudomonas putida EPS, some studies have reported that P. putida can reduce arsenate15 and biotransform dibenzothiophene16. These results suggest that the redox properties of EPS play important roles in the migration and transformation of redox-sensitive contaminants by electroctive bacteria. It has also been reported that EPS extracted from Escherichia coli could reduce positively charged silver ions to silver nanoparticles17. However, the redox properties and the possible redox components of EPS from various electroactive bacteria are not well documented.
In this study, the redox properties of EPS from two widely present electroactive bacterial strains (S. oneidensis18 and P. putida19), and the relationships between the compositions of these EPS and their redox properties were investigated. The results of this work provide an insight into the relationship between microbial extracellular electron transfer and the redox properties of EPS from electroactive bacteria.
Results and Discussion
EPS components and structure analysis
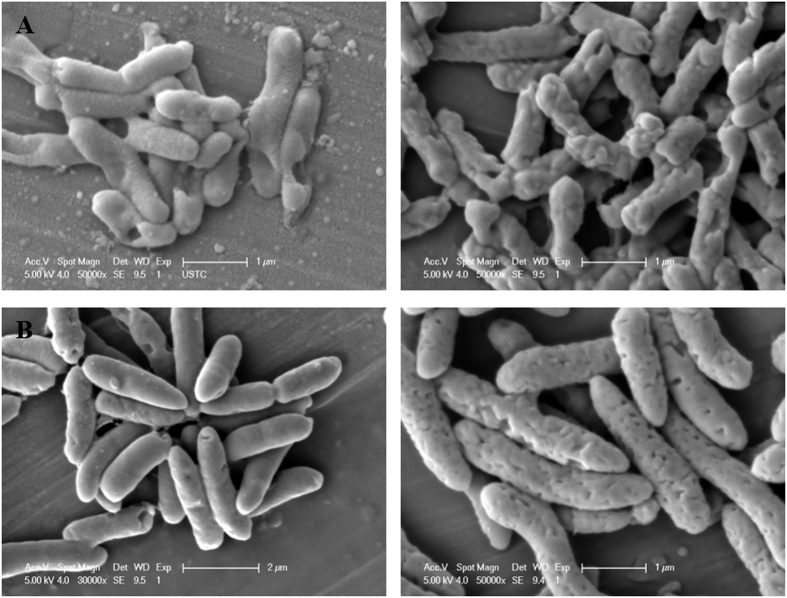
EPS of two electroactive bacteria were extracted by EDTA method and the feasibility of the extraction method was confirmed by SEM photographing before and after EPS extraction (Fig. 1). Proteins and polysaccharides were found to be the main components of the two electroactive bacterial EPS (Table 1). The EPS of S. oneidensis and P. putida contained more proteins, including heme-binding proteins. Uronic acids was the main component in the polysaccharides of the two EPS samples, and the average content of uronic acids in polysaccharides of each EPS was about 42%.
Figure 1.
SEM images of cells of two electroactive bacteria before (left) and after (right) EPS extraction: (A) S. oneidensis and (B) P. putida.
Table 1. The main compositions of three bacterial EPS.
| Bacterium | Proteins (mg/L) | Polysaccharides (mg/L) | Uronic acids (mg/L) | Heme-binding proteins (mg/L) | DNA (mg/L) |
|---|---|---|---|---|---|
| S. oneidensis | 9.14 | 4.52 | 2.45 | 1.84 | 0.40 |
| P. putida | 36.66 | 4.69 | 2.40 | 1.38 | 0.38 |
| E. coli | 7.25 | 4.34 | 2.35 | 0.45 | 0.29 |
The structure and functional groups of EPS were analyzed using FTIR spectroscopy (Figure S1). Table S1 listed the detailed peak assignments of FTIR spectra of EPS, which were referred to the previous reports20,21. The bands of amide I (1700–1600 cm−1), amide II (1600–1500 cm−1) and amide III (1350–1250 cm−1) were attributed to proteins, while C-O (1200–1000 cm−1) was associated with polysaccharides. The amide I band at 1636 cm−1 was generally ascribed to C=O stretching vibrations of β–sheet of proteins. The band at 1259 cm−1 was the representative amide III band, which is attributed to N-H bending and C-N stretching in proteins. The broad peak at 3427 cm−1 was attributed to O-H stretching. The bands at 2925 cm−1 and 2853 cm−1 were assigned to C-H stretching of methylene group in fatty acids. The band at 1408 cm−1 was ascribed to symmetric deformation of CH3 and CH2 of proteins and symmetric C-O stretching of carboxylic groups. The bands at 1086 cm−1 and 1043 cm−1 was attributed to the C-O bond, the C-O-C bond and the C-H bond from polysaccharides. The band at 1086 cm−1 was also assigned to the asymmetric and symmetric P=O stretching of the phosphodiester backbone of nucleic acids.
Redox Properties of EPS
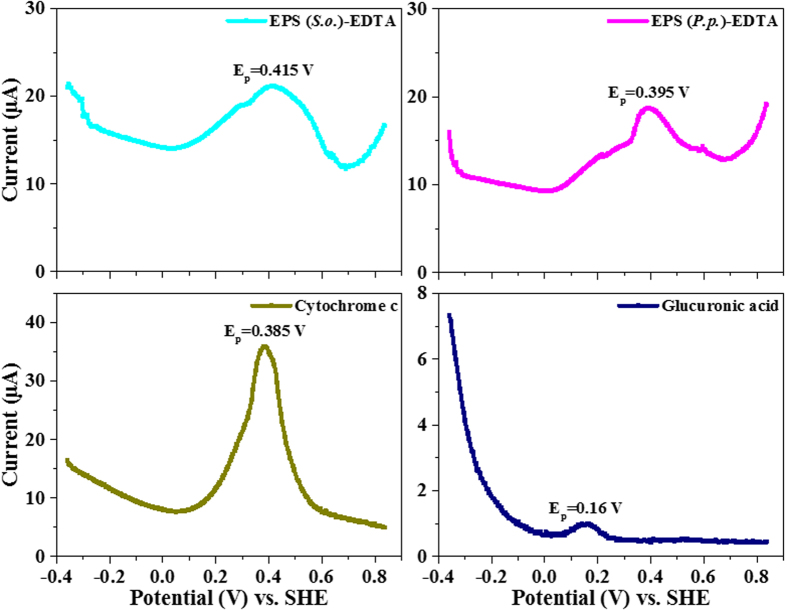
Differential pulse voltammetry (DPV) was applied to identify the redox properties of the two electroactive bacterial EPS. Simultaneously, the electrochemical activities of bovine heart cytochrome c and glucuronic acid were measured as references. Figure 2 shows that the voltammograms of two EPS solution all exist an obvious oxidation peak. The DPV peak potentials of bovine heart cytochrome c, S. oneidensis EPS and P. putida EPS are 0.385 V, 0.415 V and 0.395 V (vs. standard hydrogen electrode, SHE), respectively. It shows that the DPV peak potential of bovine heart cytochrome c was similar to those measured in EPS samples. However, glucuronic acid only exhibited a weak redox peak. These results imply that two electroactive bacterial EPS all existed redox matters, and cytochrome c might be the main redox matter in EPS.
Figure 2. Differential pulse voltammetry of EPS extracted from S. oneidensis and P. putida by using EDTA method, bovine heart cytochrome c, and glucuronic acid.
In order to investigate the effects of various EPS extraction methods on the redox properties of electroactive bacterial EPS, three methods (i.e., heating, cation exchange resin and EDTA methods) were used, and the redox properties of the extracted EPS were compared (Figs 2 and S2). Results show that two electroactive bacterial EPS extracted by the heating and EDTA methods had similar DPV peak potentials with high peak currents, while the cation exchange resin extraction method had a great change on the DPV peak potentials of two electroactive bacterial EPS with small peak currents. This implied that the EPS extracted from the three methods were all electrochemical-activated, and extraction methods influenced the redox properties of the bacterial EPS.
Redox Components in EPS
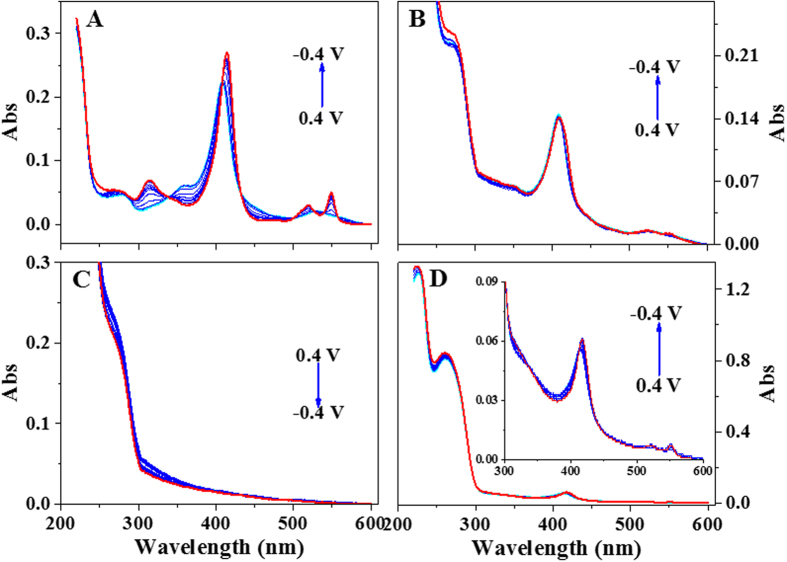
EPS were reduced at an applied negative potential, and the spectra changes of the EPS after reduction could be monitored using a spectroelectrochemical method. The UV-Vis spectra of the EPS solutions after electrochemical reduction at various negative potentials are shown in Fig. 3, the EPS of E. coli was used as a negative control with 10 g/L. Figure 3A shows that bovine heart cytochrome c at its native oxidation state showed an oxidation peak at 529 nm, and after negative potentials were applied, two reduction peaks appeared at 519 nm and 549 nm. The absorbance ratio at the two peaks (A549/A519) was related to the purity ratio of the reduced cytochrome c22, which was 1.7 at an applied potential of −0.4 V (vs. SHE). The spectra of EPS from S. oneidensis and P. putida (Fig. 3B and D) were similar to the spectrum of cytochrome c, as the same reduction peaks could be observed. The A547/A524 cytochrome purity ratio for the reduced S. oneidensis EPS was 0.81, and the A551/A519 ratio for the reduced P. putida EPS was 1. However, there were no absorbance peaks could be observed in the spectra of the EPS from a non-electroactive bacterium, E. coli (Fig. 3C).
Figure 3. UV-Vis spectroelectrochemical analysis of EPS of the three bacterial strains and bovine heart cytochrome c.
(A) bovine heart cytochrome c, (B) EPS of S. oneidensis, (C) EPS of E. coli, and (D) EPS of P. putida. The cyan curve and the red curve at each figure represented the origin sample and the reduced sample at reduction voltage of −0.4 V.
Cytochrome c may exist in the EPS from electroactive bacteria and contribute to its redox properties. To validate this hypothesis, a heme-binding-protein LDS-PAGE electrophoresis was conducted to determine if heme-binding proteins were present in the EPS of two electroactive bacteria, the EPS of E. coli was used as a negative control with 6 g/L. As shown in Fig. 4, the LDS-PAGE of the three EPS with heme staining confirms that the EPS from S. oneidensis and P. putida contained heme-binding proteins. Furthermore, the EPS of S. oneidensis contained more types of heme-binding proteins than P. putida. The molecular weights of most of the electrophoretic bands of the S. oneidensis EPS ranged from 55 to 100 kDa, and those of P. putida EPS were mainly <35 kDa. However, there were no detected heme-binding proteins in the non-electroactive bacterium E. coli EPS. The cytochrome c species MtrC and OmcA in S. oneidensis outer membrane mediate extracellular redox reactions, and their molecular weights are 71.3 kDa and 78.7 kDa, respectively14. These molecular weights are in the range of 55 to 100 kDa shown in the electrophoresis image of S. oneidensis EPS (Fig. 4), suggesting that these two types of cytochrome c may exist in S. oneidensis EPS. This finding is in agreement with previous research, in which the two c-type cytochromes were also identified in EPS extracted from Shewanella sp. HRCR-1 by proteomics analysis14. The molecular weights of the electrophoretic bands of P. putida EPS (<35 kDa) were similar to the bands of membrane fractions of spheroplasts from Pseudomonas stutzeri23. This band interval of 25.6–19.7 kDa may be related to the di-heme protein cytochrome c422, a membrane-bound bacterial electron transfer cytochrome found in Pseudomonas aeruginosa24, P. stutzeri25 and Shewanella baltica26 that is involved in the electron-transport systems associated with both aerobic and anaerobic respiration. It has also been reported that the A551/A522 ratio of cytochromes c4 from P. aeruginosa and P. stutzeri were 1.36 and 1.21, respectively27, which were similar to the ratio of 1 found in P. putida EPS. Heme-binding-protein electrophoresis illustrates that heme-binding proteins of various molecular weights and species are present in the EPS from S. oneidensis and P. putida.
Figure 4. LDS-PAGE of EPS of the three bacterial strains and bovine heart cytochrome c with heme staining.
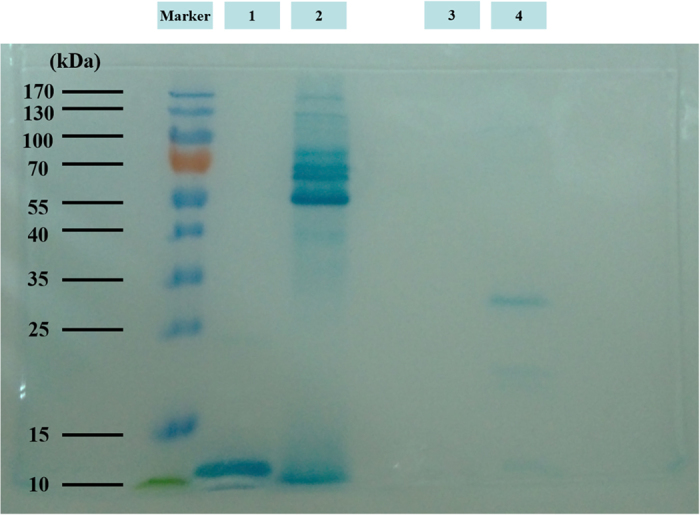
1: bovine heart cytochrome c, 2: EPS of S. oneidensis, 3: EPS of E. coli, and 4: EPS of P. putida.
The spectroelectrochemical results and heme-binding-protein electrophoresis images suggest that membrane-bound heme-binding proteins may exist in the EPS isolated from S. oneidensis and P. putida and are likely the redox-active components accepting electrons through electrochemical reduction. These data suggest that heme-binding proteins are important redox components mediating electron transfer in electroactive bacteria and other environmental microorganisms, as cytochrome c that contain heme is an important class of enzymes that participate in the respiratory chain. In natural environments, electroactive bacteria can use iron or manganese oxides and hydroxides as terminal electron acceptors to generate energy for biosynthesis and cell maintenance. EPS can facilitate the attachment of microorganisms to the mineral surface, and the EPS layer comprises several redox species to enhance attacking the mineral surface through an electrochemical dissolution6. Thus, electroactive bacterial EPS mediate their direct contact with electron acceptors to accelerate extracellular electron transfer for mineral biotransformation28.
Conclusions
In summary, heme-binding proteins were identified as the key redox species in the EPS of S. oneidensis and P. putida for electroactive bacterial extracellular electron transfer. Heme-binding proteins in EPS of electroactive bacteria are usually able to transfer electrons between membrane-bound reduction systems and terminal oxidases. Thus, exploring the redox properties of electroactive bacterial EPS could provide in-depth understanding of the interactions between electroactive bacteria and external electron acceptors, such as metals, organic contaminants, and insoluble minerals, which is important for understanding microbial extracellular electron transfer. So, establishing the relationships between electroactive bacterial EPS and the process of extracellular electron transfer and developing the methods to discover other potential redox species in electroactive bacterial EPS continue to pose challenges for the field.
Methods
Bacterial Strains
Wild-type S. oneidensis MR-1 (ATCC® number 700550TM) was kindly provided by Prof. K. H. Nealson from the University of Southern California29. P. putida (CCTCC® number AB2015303TM) was isolated through a photometric high-throughput method30 for electroactive bacteria screening from the lab-scale sequencing batch reactor that treated municipal wastewater.
EPS Extraction
S. oneidensis and P. putida were cultivated at 30 °C for 22 h in Luria-Bertani medium. The dispersed biomass was harvested by centrifugation at 5000 g for 5 min. EPS were isolated from these bacterial cells by an EDTA method, as previously described14. The dispersed cells were resuspended in 0.9% NaCl. An equal volume of 2% Na2-EDTA in 0.9% NaCl (pH 7.0) was mixed with the cell suspension and the mixture was incubated at 4 °C for 3 h. The cells were pelleted by centrifugation with 5000 g at 4 °C for 20 min, then the supernatant was filtered through 0.22 μm membrane filter. EPS were purified by ultrafiltration using a cellulose membrane (Millipore Inc., USA) with a molecular weight cut-off of 1 kDa. The EPS solution was then lyophilized to obtain pristine EPS and stored at −20 °C before use. The morphology of cell surfaces was imaged using a scanning electronic microscope (SEM, SIRION200, FEI Co., Netherlands) to investigate the microbial cell integrity before and after EPS isolation.
In order to compare the redox property of EPS extrated from different methods, heating and cation exchange resin (CER) methods were also used. EPS extraction by heating referred a previous report31. The dispersed biomass was harvested by centrifugation at 5000 g for 5 min. The bacterial pellet was re-suspended into 0.9% NaCl solution, and then was heated to 60 °C in a water bath for 30 min. After that, the solution was centrifuged at 5000 g for 15 min and then was filtrated. EPS extraction by CER was conducted according to a reported method32. The bacterial suspension was transferred to an extraction beaker and the CER (Dowex, 20–50 mesh in the sodium form, Sigma, USA) was added. The suspension was stirred at 300 rpm for 3 h at 4 °C. After that, the CER was removed and the suspension was centrifuged for 15 min at 5000 g and 4 °C in order to remove remaining bacterial cells. The supernatant was filtrated through 0.22 μm membrane and used as EPS solution.
Chemical Analysis
The protein contents of the EPS were measured using a bicinchoninic acid (BCA) procedure33 with bovine serum albumin as the standard. Polysaccharides were determined using the anthrone method33 with glucose as the standard. Uronic acids were measured using the m-hydroxydiphenyl sulfuric acid method34 with glucuronic acid as the standard. Heme was quantified in a fluorescence assay35 with bovine heart cytochrome c (95%, Aladdin Co., USA) as the standard. DNA was detected by the diphenylamine colorimetric method32 with calf thymus DNA as the standard. The infrared spectra (IR) of EPS samples were obtained with a VERTEX 70 FTIR spectrometer (Bruker Co., Germany) to determine the functional groups in the EPS.
The electrochemical properties of the EPS were analyzed by DPV using an electrochemical workstation (660 C, Chenhua Instruments, Inc., China). The DPV measurements were performed with a 10 mL three-electrode cell with an Ag/AgCl reference electrode, a Pt counter electrode and an indium tin oxide working electrode. The parameters for DPV were: pulse increments 5 mV, pulse amplitude 50 mV, pulse width 300 ms, pulse period 5 s. The scanning range was from −0.4 V to 0.8 V vs. SHE and the scanning rate was 1 mV/s. Before measurements were taken, 10 mL blank solution (0.9% NaCl) were added to the three-electrode cell, and then the bovine heart cytochrome c (0.2 g/L), glucuronic acid (2 g/L) or the EPS solution was added to obtain a final concentration of 800 mg/L. The cell was purged with O2-free N2 for 15 min before electrochemical measurements. The curves of samples were deducted by the blank solution as background.
The UV-Vis spectra of the EPS at various potentials were measured using spectroelectrochemical analysis, which was performed using a thin-layer spectroelectrochemical cell with a Pt gauze working electrode, an Ag/AgCl reference electrode, and a Pt wire counter electrode36. The optical length of the spectroelectrochemical cell was 1 mm. To obtain a high resolution, a high concentration of EPS (10 g/L) was used for the spectroelectrochemical measurement. Bovine heart cytochrome c at 0.2 g/L was used as a reference. The sample volume in the spectroelectrochemical cell was 0.8 mL. The potential ranged from 0.4 V to −0.4 V vs. SHE, and the equilibration time at each potential was set to 15 min. The UV-Vis spectra of the EPS samples at each potential were then measured using a spectrophotometer (UV-2450, Shimadzu, Japan). The cell was purged with O2-free N2 for 15 min before electrochemical measurements and continued through the entire experiment to avoid the interference of oxygen.
Electrophoresis of EPS Heme-Binding Proteins
Heme-binding protein electrophoresis was conducted to identify the presence of this redox component in the EPS, which was confirmed by lithium dodecyl sulfate (LDS)-PAGE and staining with tetramethylbenzidine (TMBZ, a chromogen that specifically binds to proteins with heme-associated peroxidase activity)37,38. Before analysis, the EPS samples were dissolved in deionized water, and the final concentration was adjusted to 6 g/L. Bovine heart cytochrome c (0.04 g/L) was used as control.
Additional Information
How to cite this article: Li, S.-W. et al. Redox properties of extracellular polymeric substances (EPS) from electroactive bacteria. Sci. Rep. 6, 39098; doi: 10.1038/srep39098 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors wish to thank the NSFC (51322802 and 21377123), the Fundamental Research Funds for the Central Universities (WK3530000002 and WK2060190040), and the Key Research Program of Frontier Sciences, CAS (QYZDB-SSW-QDC020) for the partial support of this study.
Footnotes
Author Contributions S.W.L. conceived the study and manuscript design, performed research, analyzed results and wrote the manuscript; G.P.S. developed the research project, conceived the study, analyzed results and revised the manuscript. Y.Y.C. conceived the study and revised the manuscript; H.Q.Y. developed the research project and revised the manuscript.
References
- Zhang P. et al. Extracellular protein analysis of activated sludge and their functions in wastewater treatment plant by shotgun proteomics. Sci Rep 5, 12041 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gil G., Thomas L., Emwas A. H., Lens P. N. & Saikaly P. E. NMR and MALDI-TOF MS based characterization of exopolysaccharides in anaerobic microbial aggregates from full-scale reactors. Sci Rep 5, 14316 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J. et al. Composition and aggregation of extracellular polymeric substances (EPS) in hyperhaline and municipal wastewater treatment plants. Sci Rep 6, 26721 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B. et al. Contribution of extracellular polymeric substances from Shewanella sp. HRCR-1 biofilms to U(VI) immobilization. Environ Sci Technol 45, 5483–5490 (2011). [DOI] [PubMed] [Google Scholar]
- Yu G. H., Tang Z., Xu Y. C. & Shen Q. R. Multiple fluorescence labeling and two dimensional FTIR-C-13 NMR heterospectral correlation spectroscopy to characterize extracellular polymeric substances in biofilms produced during composting. Environ Sci Technol 45, 9224–9231 (2011). [DOI] [PubMed] [Google Scholar]
- Tourney J. & Ngwenya B. T. The role of bacterial extracellular polymeric substances in geomicrobiology. Chem Geol 386, 115–132 (2014). [Google Scholar]
- Flemming H.-C., Neu T. R. & Wozniak D. J. The EPS matrix: the “house of biofilm cells”. J Bacteriol 189, 7945–7947 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan B. E. Exoelectrogenic bacteria that power microbial fuel cells. Nat Rev Microbiol 7, 375–381 (2009). [DOI] [PubMed] [Google Scholar]
- Logan B. E. & Rabaey K. Conversion of wastes into bioelectricity and chemicals by using microbial electrochemical technologies. Science 337, 686–690 (2012). [DOI] [PubMed] [Google Scholar]
- Marshall M. J. et al. c-Type cytochrome-dependent formation of U(IV) nanoparticles by Shewanella oneidensis. Plos Biol 4, 1324–1333 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. Enhanced biotransformation of nitrobenzene by the synergies of Shewanella species and mediator-functionalized polyurethane foam. J Hazard Mater 252–253, 227–232 (2013). [DOI] [PubMed] [Google Scholar]
- Gao D. W., Wang X. L. & Xing M. Dynamic variation of microbial metabolites and community involved in membrane fouling in A/O-MBR. J Membrane Sci 458, 157–163 (2014). [Google Scholar]
- Wang J. et al. Mediators-assisted reductive biotransformation of tetrabromobisphenol-A by Shewanella sp. XB. Bioresour Technol 142, 192–197 (2013). [DOI] [PubMed] [Google Scholar]
- Cao B. et al. Extracellular polymeric substances from Shewanella sp. HRCR-1 biofilms: characterization by infrared spectroscopy and proteomics. Environ Microbiol 13, 1018–1031 (2011). [DOI] [PubMed] [Google Scholar]
- Freikowski D., Winter J. & Gallert C. Hydrogen formation by an arsenate-reducing Pseudomonas putida, isolated from arsenic-contaminated groundwater in West Bengal, India. Appl Microbiol Biotechnol 88, 1363–1371 (2010). [DOI] [PubMed] [Google Scholar]
- Alcon A., Santos V. E., Martin A. B., Yustos P. & Garcia-Ochoa F. Biodesulfurisation of DBT with Pseudomonas putida CECT5279 by resting cells: influence of cell growth time on reducing equivalent concentration and HpaC activity. Biochem Eng J 26, 168–175 (2005). [Google Scholar]
- Kang F., Alvarez P. J. & Zhu D. Microbial extracellular polymeric substances reduce Ag+ to silver nanoparticles and antagonize bactericidal activity. Environ Sci Technol 48, 316–322 (2014). [DOI] [PubMed] [Google Scholar]
- Uria N., Munoz Berbel X., Sanchez O., Munoz F. X. & Mas J. Transient storage of electrical charge in biofilms of Shewanella oneidensis MR-1 growing in a microbial fuel cell. Environ Sci Technol 45, 10250–10256 (2011). [DOI] [PubMed] [Google Scholar]
- Majumder D. et al. Electricity generation and wastewater treatment of oil refinery in microbial fuel cells using Pseudomonas putida. Int J Mol Sci 15, 16772–16786 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C. et al. Chemical composition and relative hydrophobicity of microbial exopolymeric substances (EPS) isolated by anion exchange chromatography and their actinide-binding affinities. Mar Chem 126, 27–36 (2011). [Google Scholar]
- Jain R. et al. Extracellular polymeric substances govern the surface charge of biogenic elemental selenium nanoparticles. Environ Sci Technol 49, 1713–1720 (2015). [DOI] [PubMed] [Google Scholar]
- Thuesen M. H., Norgaard A., Hansen A. M., Caspersen M. B. & Christensen H. E. M. Expression of recombinant Pseudomonas stutzeri di-heme cytochrome c(4) by high-cell-density fed-batch cultivation of Pseudomonas putida. Protein Expres Purif 27, 175–181 (2003). [DOI] [PubMed] [Google Scholar]
- Beller H. R. et al. Identification of c-type cytochromes involved in anaerobic, bacterial U(IV) oxidation. Biodegradation 20, 45–53 (2009). [DOI] [PubMed] [Google Scholar]
- Williams H. D., Ziosnik J. E. A. & Ryall B. Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Advances in Microb Physiol 52, 1–71 (2007). [DOI] [PubMed] [Google Scholar]
- Raffalt A. C., Schmidt L., Christensen H. E. M., Chi Q. & Ulstrup J. Electron transfer patterns of the di-heme protein cytochrome c(4) from Pseudomonas stutzeri. J Inorg Biochem 103, 717–722 (2009). [DOI] [PubMed] [Google Scholar]
- De March M., Di Rocco G., Hickey N. & Geremia S. High-resolution crystal structure of the recombinant diheme cytochrome c from Shewanella baltica (OS155). J Biomol Struct Dyn 33, 395–403 (2015). [DOI] [PubMed] [Google Scholar]
- Pettigrew G. W. & Brown K. R. Free and membrane-bound forms of bacterial cytochrome-c. Biochem J 252, 427–435 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham C. A. et al. A novel electrochemically active and Fe(III)-reducing bacterium phylogenetically related to Aeromonas hydrophila, isolated from a microbial fuel cell. Fems Microbiol Lett 223, 129–134 (2003). [DOI] [PubMed] [Google Scholar]
- Myers C. R. & Nealson K. H. Respiration-linked proton translocation coupled to anaerobic reduction of manganese(IV) and iron(III) in Shewanella putrefaciens MR-1. J Bacteriol 172, 6232–6238 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S. J. et al. A plate-based electrochromic approach for the high-throughput detection of electrochemically active bacteria. Nat Protoc 9, 112–119 (2014). [DOI] [PubMed] [Google Scholar]
- Xu J., Sheng G. P., Ma Y., Wang L. F. & Yu H. Q. Roles of extracellular polymeric substances (EPS) in the migration and removal of sulfamethazine in activated sludge system. Water Res 47, 5298–5306 (2013). [DOI] [PubMed] [Google Scholar]
- Frolund B., Palmgren R., Keiding K. & Nielsen P. H. Extraction of extracellular polymers from activated sludge using a cation exchange resin. Water Res 30, 1749–1758 (1996). [Google Scholar]
- Zhang H. L. et al. Phosphorus removal in an enhanced biological phosphorus removal process: roles of extracellular polymeric substances. Environ Sci Technol 47, 11482–11489 (2013). [DOI] [PubMed] [Google Scholar]
- Ghods S., Sims I. M., Moradali M. F. & Rehm B. H. A. Bactericidal compounds controlling growth of the plant pathogen Pseudomonas syringae pv. actinidiae, which forms biofilms composed of a novel exopolysaccharide. Appl Environ Microbiol 81, 4026–4036 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair P. R., Gorman N. & Jacobs J. M. In Current Protocols in Toxicology 8.3.1-8.3.7 John Wiley & Sons, Inc. (2001). [DOI] [PubMed] [Google Scholar]
- Tam T. K., Chen B., Lei C. & Liu J. In situ regeneration of NADH via lipoamide dehydrogenase-catalyzed electron transfer reaction evidenced by spectroelectrochemistry. Bioelectrochemistry 86, 92–96 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. P., Holt S. C. & Kolodrubetz D. Cloning and expression of two novel hemin binding protein genes from Treponema denticola. Infect Immun 69, 4465–4472 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stugard C. E., Daskaleros P. A. & Payne S. M. A 101-kilodalton heme-binding protein associated with congo red binding and virulence of Shigella flexneri and enteroinvasive Escherichia coli strains. Infect Immun 57, 3534–3539 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.