Abstract
Knowledge of the evolution of pathogens is of great medical and biological significance to the prevention, diagnosis, and therapy of infectious diseases. In order to understand the origin and evolution of the SARS-CoV (severe acute respiratory syndrome-associated coronavirus), we collected complete genome sequences of all viruses available in GenBank, and made comparative analyses with the SARS-CoV. Genomic signature analysis demonstrates that the coronaviruses all take the TGTT as their richest tetranucleotide except the SARS-CoV. A detailed analysis of the forty-two complete SARS-CoV genome sequences revealed the existence of two distinct genotypes, and showed that these isolates could be classified into four groups. Our manual analysis of the BLASTN results demonstrates that the HE (hemagglutinin-esterase) gene exists in the SARS-CoV, and many mutations made it unfamiliar to us.
Key words: SARS, SARS-CoV, motif frequency profile, genomic signature, Chaos Game Representation, PUP
Introduction
The SARS-CoV (severe acute respiratory syndrome-associated coronavirus) has been generally accepted as the major pathogen of SARS, which has cost thousands of lives globally a few months ago 1., 2.. This coronavirus has been classified as a new member of Genus Coronavirus in Family Coronaviridae and Order Nidovirales (3).
Serologically, the seventeen species coronavirus have been classified into three groups. Groups I and II contain mammalian viruses, while Group III contains only avian viruses. Within each group, the viruses are further subclassified into distinct species by the host range, antigenic relationship, and genomic organization. Genomic analyses have revealed that the SARS-CoV has typical features of coronavirus, but it represents a novel virus that is phylogenetically distinct from any other member in the three known groups.
It has been suggested that the SARS-CoV is more closely related to the cow coronavirus and MHV (Murine Hepatitis Virus) by comparing a conserved 215-a.a. (amino acid) segment of the polymerase protein (4). However, the strength of the association is reduced if the entire genome is taken into consideration. In this paper, we present the phylogenetic comparison between SARS-CoV and other viruses, and the analysis of the mutation sites of 42 SARS-CoV isolates, as well as the possible recombination and horizontal transfer.
Results and Discussion
Comparison of the GC content and genome size distribution with other viruses
Genome sizes of viruses range from a few hundred base pairs to a few hundred thousand base pairs (Figure 1). According to their genome sizes, the 2,498 virus isolates can be classified as three groups.
Fig. 1.
Comparison of the GC content and genome size between SARS-CoV with other viruses.
The first group, also the smallest group corresponding to the low part of the figure, represents the viroids. The range of genome sizes of this group is between 120 bp (Avocado sunblotch viroid; NCBI accession number: AF404074) and 374 bp (Citrus exocortis viroids, NCBI accession number: N30870). The range of the GC content is between 35.5% (Avocado sunblotch viroid; genome size: 248 bp; NCBI accession number: AF404068) and 61.1% (Grapevine yellow speckle viroid 1; genome size: 365 bp; NCBI accession number: AF462165). Here, the two isolates of AF404068 and AF404074 demonstrate the sequence diversity of viroid genomes. Although they are only different isolates of Avocado sunblotch viroid, the difference of their genome sizes is still great.
The second group, located at the top of the figure, includes all double-strand viruses. The range of genome sizes of this group is between 102,653 bp (Lymphocystis disease virus 1; NCBI accession number: L63545) and 335,593 bp (Ectocarpus siliculosus virus; NCBI accession number: AF204951). The range of the GC content of the genomes is between 17.77% (Amsacta morei entomopoxvirus; genome size: 2,322,392 bp; NCBI accession number: AF250284) and 72.4% (Bovine herpesvirus type 1.1; genome size: 135,301 bp; NCBI accession number: AJ004801).
The third group, also the largest group corresponding to the middle part of the figure, is the most complicated one that includes all other kinds of viruses. The SARS-CoV belongs to this group. From the figure, it is easy to see that the GC content and genome size of the SARS-CoV are normal.
Sequence comparison with other viruses
We performed a genomic sequence comparison between Isolate BJ01 of SARS-CoV and other viruses. As a contrast, we divided the viruses into coronaviruses and non-coronaviruses. BLASTN (default parameters) was used to compare the SARS-CoV genomic sequence with the dataset of non-coronaviruses, and sequences more than 20 nt in length and identity greater than 70% were extracted to create the conservative map (Figure 2).
Fig. 2.
Conservative map created by comparing the SARS-CoV genome sequence against a database of non-coronaviruse sequence. Only the comparison result with segment length greater than 20 nt was extracted.
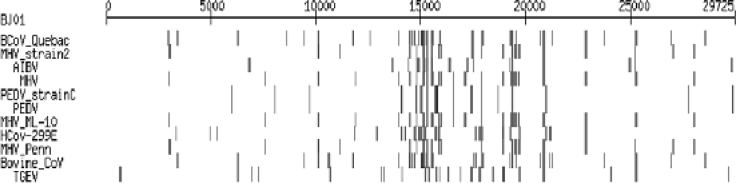
The coronaviruses are phylogenetically close to the SARS-CoV. To demonstrate their respective similar regions to the SARS-CoV, we mapped the comparison results to BJ01 sequence to see the distribution. As shown in Figure 3, the SARS-CoV genomic fragments are plotted along the horizontal axis in the order they appeared in the genome, and other coronaviruses are placed vertically. The darkness of a pixel corresponds to the strength of the match between a SARS-CoV fragment and a coronavirus genome, and the width of the rectangle corresponds to the length of the matched sequence. The length of the longest matched segment is 138 nt (Codons 14,914-15,051) with the identity of about 81.16% (112/138). This segment lies in the gene coding region of the R (replicase) protein of SARS-CoV.
Fig. 3.
Homology comparison of BJ01 with other coronaviruses. The darkness of a pixel corresponds to the strength (identities value) of the match between a SARS-CoV fragment and a coronavirus genome, and the width of rectangle corresponds to the length of the match.
Motif frequency profile comparison with other viruses
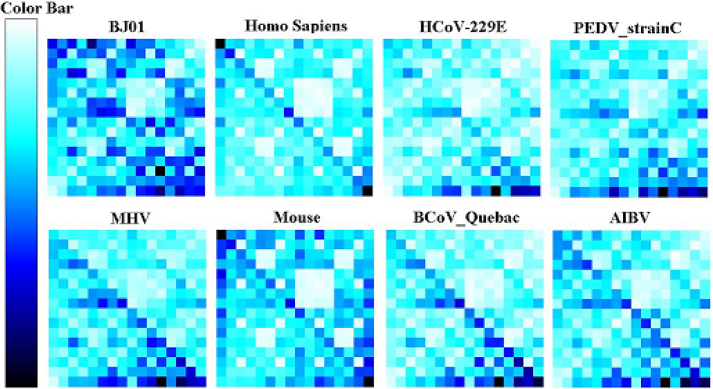
We computed the average absolute distance of the motif frequency profile (MFP) between BJ01 and other viruses. The analysis results with the motif length from 2 to 8 nt all demonstrate that the SARS-CoV is most adjacent to coronaviruses. Here we only show the comparison result with coronaviruses since the distances with non-coronavirus are very large. To present the similarity of the MFP between the SARS-CoV and other coronaviruses more clearly, we use the chaos game representation (CGR) to demonstrate the results. Only one isolate is selected to represent its species because the MFP of different isolates within the same species is very similar. To find whether there is a relationship between the viruses and their hosts, we also demonstrate the MFP of Homo Sapiens (host of the SARS-CoV) and Mus Musculus (host of the MHV).
Figure 4 presents the dinucleotide frequency profile. It is obvious that the frequency of dinucletide TT is rich in all the eight organisms. BJ01, MHV and PEDV (Porcine Epidemic Diarrhea Virus) have another similar characteristic of the high frequency of TG. The obvious difference between BJ01 and other coronaviruses is that the AA frequency of BJ01 is higher. This AA-rich characteristic is the same to Homo Sapiens, so probably in SARS-CoV it is influenced by its host—Homo Sapiens. The result that the MFPs of Homo Sapiens and mouse are very similar to each other is in accordance with the phylogenetic analysis. The lower frequency of CG in all the images could be easily explained by the reason that there is a relatively high chance of a methyl-C mutation into a T. Theoretically, the C-T methyl mutation should induce the frequency increase of TG, while the result shows that the frequency of TG is not high in about six organisms out of the eight. So there should be some other mechanisms that lead to the decrease of TG in these organisms.
Fig. 4.
Dinucleotide motif frequency profile. The darkness of a pixel corresponds to the frequency. The darker the pixel shows, the greater the frequency is.
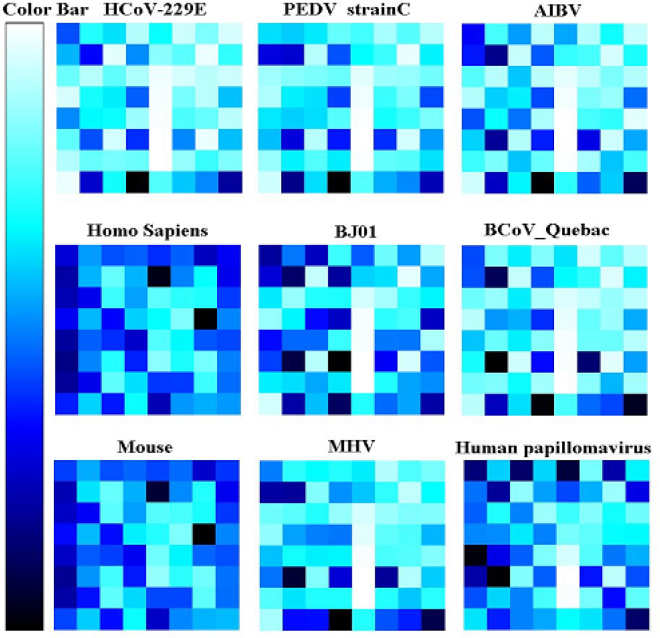
To see what contributes to the high content of TT, we also did the MFP analysis with the motif length from 3 to 8 nt. We found out that at motif length 4, almost all the tetranucleotide of coronaviruses with the highest frequency are TGTT except those in SARS-CoV (Figure 5). The richest tetranucleotide of the SARS-CoV is TGCT, while TGTT is the second. This fact demonstrates that the SARS-CoV is phylogenetically far from other coronaviruses, and this conclusion is the same as the serological analysis result. To see whether this characteristic is unique to coronaviruses, we searched 2,497 virus genomes, and found only two viruses, Beet soil-borne mosaic virus (genome size: 4,616 bp; NCBI accession number: AF061869) and Soil-borne cereal mosaic virus (genome size: 3,683 bp; NCBI accession number: AJ132577), have this characteristic. Both these two exceptions are ssRNA positively stranded viruses and belong to two different subspecies: Furovirus and Benyvirus. We computed the TGTT content of coronavirus genomes and the result is shown in Table 1. As a comparison, the contents of other viruses have also been computed, but the results are not listed here. The lowest content is 0.13% appeared in TT-like mini virus (NCBI accession number: AF291073), and the highest content is 7.33% in HCoV-229. The difference of TGTT frequency between different viruses is great. Nothing significant is found at other motif lengths. Thus the richest tetranucleotide, TGTT, could be a common characteristic of coronaviruses.
Fig. 5.
Tetranucleotide motif frequency profile.
Table 1.
The Frequency and Content of the First Two Richest Tetranucleotides
| Primary | Secondary | Viruses |
|---|---|---|
| TGTT(67, 1.45%) | TGGT(62, 1.34%) | BMV1 |
| TGTT(42, 1.14%) | GTTA(39, 1.06%) | CMV2 |
| TGCT(318,1.07%) | TGTT(274,0.92%) | BJ01 |
| TGTT(353, 1.23%) | TTGT(329, 1.15%) | TGEV |
| TGTT(387, 1.40%) | TTGT(354, 1.28%) | AIBV |
| TGTT(397, 1.42%) | TTTT(337, 1.20%) | PEDV |
| TGTT(397, 1.42%) | TTTT(337, 1.20%) | PEDV_strainC |
| TGTT(413, 1.32%) | TTGT(355, 1.14%) | MHV_strain2 |
| TGTT(417, 1.33%) | TTGT(369, 1.18%) | MHV |
| TGTT(418, 1.34%) | TTGT(371, 1.19%) | MHV_ML-10 |
| TGTT(426, 1.37%) | TTGT(365, 1.17%) | MHV_Penn |
| TGTT(499, 1.61%) | TTTT(456, 1.47%) | Bovine_CoV |
| TGTT(501, 1.83%) | TTGT(402, 1.47%) | HCoV-229E |
| TGTT(502, 1.61%) | TTTT(466, 1.50%) | BCoV_Quebac |
Codon usage bias comparison with other viruses
DNA sequence data from various organisms have clearly shown that synonymous codons for each amino acid are not used with equal frequency, even though choices among the codons should be equivalent in terms of protein structures. The statistical data suggest that the choices among synonymous codons are consistently similar for all genes within a certain genome 5., 6., 7.. Generally speaking, if two organisms were adjacent in evolution, their codon usage bias should be similar (7).
We made statistics of the codon usage frequency of all viruses and computed the standard deviation of their codon usage frequency relative to BJ01. When we computed the codon usage frequency of the SARS-CoV, the five function-known gene-coding regions for the R, S (spike), E (envelope), M (membrane), N (nucleocapsid) and five putative uncharacterized proteins (PUP) were all analyzed (8). Results show that all other coronaviruses have a small difference of codon usage frequency with the SARS-CoV. Astonishingly, some viruses that do not belong to coronavirus also have the same difference with the SARS-CoV, such as human papillomavirus (NCBI accession number: U85660). This fact suggests that the codon usage bias is not valid at all conditions.
For the very similar codon usage of different strains of the same subspecies, we selected only one of the several isolates in Figure 6.
Fig. 6.
The codon usage frequency of mouse, human, human papillomavirus and six coronaviruses.
Function-known protein comparison with other viruses
The SARS-CoV has a similar genome organization, especially in its gene orders, with other members in coronavirus. This is good evidence that the SARS-CoV is closely related with coronavirus. The five proteins of the SARS-CoV whose functions had been known were compared with the proteins of other viruses. The results demonstrated that these five proteins are similarest to the proteins of coronaviurses.
Mutations analysis
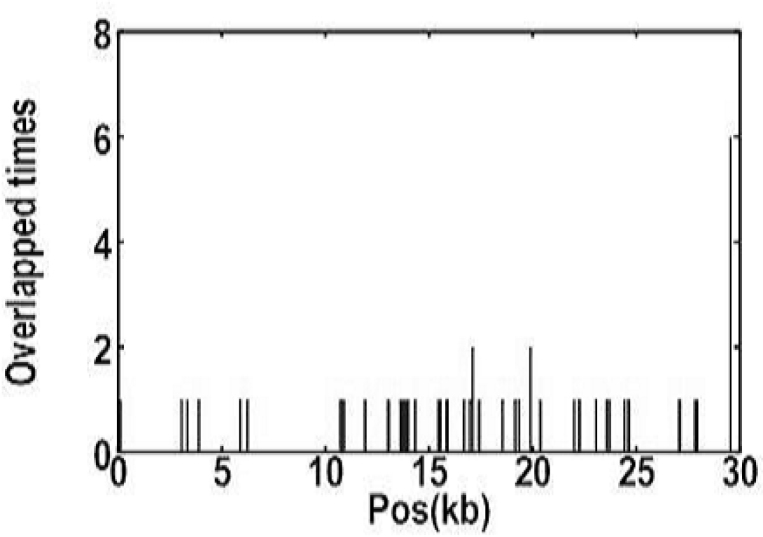
We aligned 42 complete genome sequences of the SARS-CoV by using the software, Crossmatch (version 0.990329), to look for variations. With BJ01 as the reference, we found 338 substitution sites. All the results are listed in the supplementary table.
Substitution errors replacing a purine with a purine and a pyrimidine with a pyrimidine were more easily made for steric reasons. The resulting mutations were transitions. Transversions, purine to pyrimidine changes and the reverse, are less likely made. When resulting in an amino-acid change, transversions often have a larger impact on the protein than transitions. There are four possible transition errors (A←→G, C←→T) and eight possible transversion errors (A←→C, A←→T, G←→C, G←→T). Therefore, if a mutation occurs randomly, a transversion would be more likely than a transition. However, in many organisms, transitions are two times more likely to occur than transversions (9).
Only one substitution occurred at each of the 338 substitution sites, of which 225 were transitions, including 98 AG transitions and 127 CT transitions, and 113 were transversions, including 40 AC, 35 AT, 9 GC, and 29 GT specifically. The odds ratio of transition and tranversion was about 2.0 (225/113), which just equalled the universal possibility of 2.0.
The statistics of types and codon phases of nucleotide substitution in coding region (Table 2) demonstrate that, as long as the substitution number or rate is concerned, there is no great difference among the three codon phases. In the 98 mutations that happened at the first codon phase, the number of non-synonymous substitutions is 93 and the rate is 95% (93/98), which is very close to the statistical possibility of 96%. The 112 substitutions that happened at the second codon phase were all non-synonymous substitutions. In the 119 substitutions that happened at the third codon phase, the number of non-synonymous substitutions is 18, and the rate is 15.13% (18/119), which is lower than the statistical possibility of 30%. If we take the substitution type into consideration, the first two substitution types are with 92 AG and 125 CT, which constitute about 65.96% (217/329) of the total number. The substitution number of CG is the smallest, only 9.
Table 2.
The Statistics of Type and Codon Phase of Nucleotide Substitution in Coding Region
| Codon phase | Transition |
Transversion |
Total | Percent | ||||
|---|---|---|---|---|---|---|---|---|
| AG | CT | AC | AT | GC | GT | |||
| 1 | 33 | 28(5) | 17 | 8 | 2 | 10 | 98(5) | 29.79% |
| 2 | 35 | 40 | 11 | 12 | 5 | 9 | 112 | 34.04% |
| 3 | 24(23)1 | 57(57) | 12(7) | 15(10) | 2 | 9(4) | 119(101) | 36.17% |
| Total | 92(23) | 125(63) | 40(7) | 35(10) | 9 | 28(4) | 329(106) | 100% |
| Percent | 27.96% | 37.99% | 12.16% | 10.64% | 2.74% | 8.51% | 100% | |
The number in the parentheses is synonymous number.
To eliminate mutational noises induced by sequencing errors or other factors, we only considered the forty-nine mutations found within two or more isolates. The total transition number was 37, including 13 AG and 24 CT. The total tranversion number was 12, including 3 AC, 4 AT, and 5 GT. The odds ratio of transition and tranversion was about 3.1 (37/12), larger than the universal possibility of 2.0. This change of the odds ratio from 2.0 to 3.1 means that we omitted some real substitution sites when we only considered those that happened in at least two isolates.
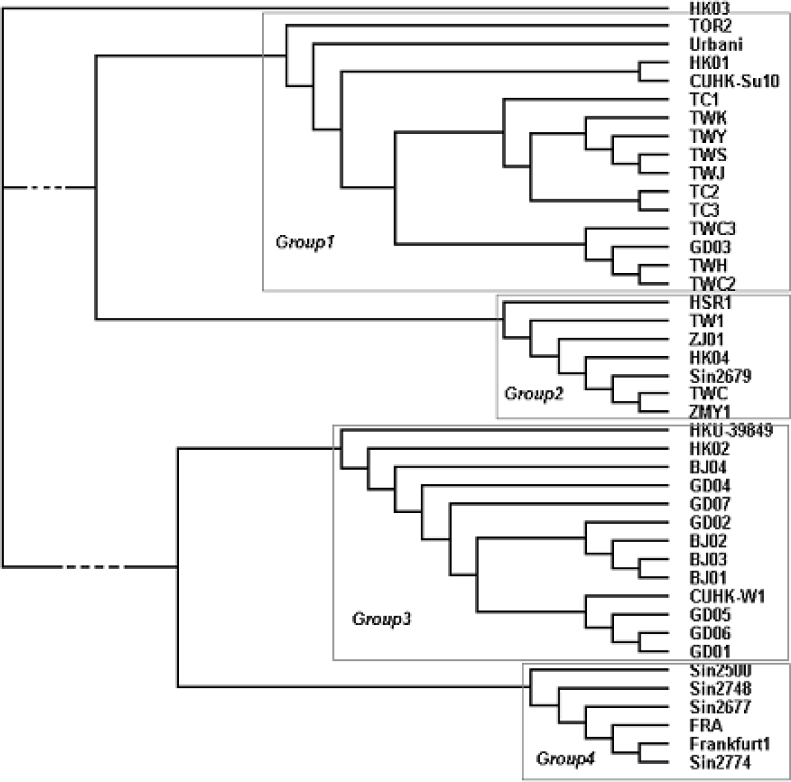
Among 42 isolates of the SARS-CoV compared with BJ01, ZYM1 has the highest substitution mutation rate. It has 86 mutation sites with 62 non-synonymous substitutions, and the whole genome mutation rate is 0.29% (86/29726). The strain that has the lowest substitution mutation rate is GD02, which has five mutation sites with four non-synonymous substitutions and the whole genome mutation rate is 0.017% (5/29726). 42 new sequences were produced by using the nucleotides at the 338 substitution sites to represent the 42 isolates of viruses. We used the software of Clustalw to analyze these 42 new sequences and produced a phylogenetic tree (Figure 7), which is almost the same as another one produced by the 49 substitution sites that occurred in at least 2 isolates. The only difference lies in the branch lengths of the tree. There are obviously four groups in the tree. Group 1, mainly constituted by Taiwan (TW) isolates (10/15), is called “TW Group”. Group 3, including 4 isolates from Beijing (BJ), 6 from Guangdong (GD), and 3 from Hong Kong (HK), is called “BJ-GD Group”. FRA, Frankfurt1, and four isolates from Singapore (SP), constitute Group 4 which is named “SP Group”. Due to the complex members in Group 2, we could not give it a specific name. The HK03 does not belong to any group because the difference between it and other isolates is maximal. This group division is different from the one that was made on geography, which suggests the complexity of virus propagation.
Fig. 7.
The phylogenetic tree of 42 SARS-CoV isolates with every sequence being constructed by the nucleotides of the 338 substitution sites.
We found that there are two specific genotypes after analyzing six mutation sites that happened in at least ten isolates. The first one corresponds to the positions 3,838, 11,474, and 26,458. The nucleotides of the eleven isolates (GD03, TC1, TC2, TC3, TWC2, TWC3, TWH, TWJ, TWK, TWS, TWY) at the three positions are all C, T, and G, respectively. The second one corresponds to the positions 17,545, 22,203 and 27,808, and the nucleotides of the eleven isolates (BJ01, BJ02, BJ03, BJ04, GD01, GD02, GD04, GD05, GD06, GD07, CUHK-W1) at the three positions are all G, C and C, respectively. Compared with the division result above, the eleven isolates corresponding to the first genotype all belong to “TW Group”, whereas the eleven isolates corresponding to the second genotype all belong to “BJ-GD Group”. It is very unlikely that the C:T:G and G:C:C genotypes emerge by chance. Rather, this should be the evidence for the genetic signature of strain differences in the SARS-CoV.
Except for the single nucleotide substitutions, there were some big segment insertions or deletions. The 29-nt insertion has been reported in our former paper (10). Our research has discovered the 29-nt insertion in two newly sequenced isolates (GD02, GD05), as well as a 54-nt deletion and a 386-nt deletion in GD06 and HK02, respectively (Table 3). The region near Codon 27,863 seems like a hotspot, because these five big segment insertions all occurred nearby. The details will be shown in another paper.
Table 3.
The Big Segment Insertion and Deletion of SARS-CoV
| Isolate | Genome size (nt) | Indel (ref. to BJ01) | Source |
|---|---|---|---|
| GD01 | 29,757 | 29 nt insertion (27,863–27,864) | Guangdong |
| GD02 | 29,753 | 29 nt insertion (27,863–27,864) | Guangdong |
| GD05 | 29,757 | 29 nt insertion (27,863–27,864) | Guangdong |
| GD06 | 29,675 | 54 nt deletion (27,837–27,900) | Guangdong |
| HK02 | 29,339 | 386 nt deletion (27,698–28,083) | Hong Kong |
Possible recombination and horizontal transfer
The recombination in the RNA virus genomes is a general phenomenon, and is considered to play a major role as a driving force in the virus variability and thus in virus evolution. An ever-increasing number of RNA viruses has been shown to undergo the RNA recombination, whether under natural or experimental conditions (11). Recent reports strongly suggest that RNA recombination is related to the virus replication and occurs by a copy-choice mechanism (12).
We analyzed the possible recombination between the SARS-CoV and other viruses, especially other coronaviruses by using the software of SIMPLOT (version 2.5, http://sray.med.som.jhmi.edu/RaySoft/SimPlot/), but didn’t find any hint of recombinations between the SARS-CoV and other viruses.
The HE (hemagglutinin-esterase) gene found in some of the coronaviruses in Group II is homologous to that of the influenza C virus. The HE gene, which is present between the ORF1b and the S protein in Group I and sometimes in Group III, was not found in the SARS-CoV (13). However, when we analyzed the comparison result manually, we found a region in the BJ01 genome has some similarity with the HE gene. Many specific segments existing in the HE gene sequence appear in this region, and the order of these segments is the same to that in HE gene. So we postulate that the HE gene exists in the SARS-CoV genome, and a lot of mutations happened in this region have broken it and made it unfamiliar to us, since it is not very important for the life and propagation of the SARS-CoV.
Materials and Methods
Data
The complete genome sequences of 2,497 isolates of viruses were obtained from GenBank (NCBI/GenBank/ftp.ncbi.nlm.nih.gov/Feb-14,2003). They represent 493 species or subspecies, and among them, there are 11 isolates of coronaviruses (Table 4).
Table 4.
The Complete Genome Sequences of 12 Isolates of Coronavirus
| Isolate | Accession number | Genome size(nt) | Modification date |
|---|---|---|---|
| BJ01 | AY278488 | 29,726 | 12-May-03 |
| AIBV | NC_001451.1 | 27,608 | 19-Nov-02 |
| HCoV-229E | NC_002645.1 | 27,317 | 19-Apr-03 |
| PEDV | NC_003436.1 | 28,033 | 26-Apr-03 |
| PEDV_strainC | AF353511.1 | 28,033 | 29-Nov-01 |
| TGEV | NC_002306.2 | 28,586 | 28-Apr-03 |
| Bovine_CoV | NC_003045.1 | 31,026 | 25-Apr-03 |
| BCoV_Quebac strain | AF220295.1 | 31,100 | 1-Apr-03 |
| MHV_Penn 97-1 | AF208066 | 31,112 | 11-May-00 |
| MHV_ML-10 | AF208067 | 31,233 | 3-Jan-02 |
| MHV_strain2 | AF201929.1 | 31,276 | 3-Jan-02 |
| MHV | NC_001846 | 31,357 | 7-Jan-03 |
Until August 17, 2003, 32 complete genome sequences of the SARS-CoV had been submitted to GenBank (Table 5). Besides, we have newly sequenced 10 complete SARS-CoV genomes (Table 6). All of our analyses included are referred to the SARS-CoV Isolate BJ01 except those specially mentioned.
Table 5.
The Complete Genome Sequences of 32 SARS-CoV Isolates in GenBank (17-Aug-03 update)
| Isolate | Genome size(nt) | Accession number | Modification date |
|---|---|---|---|
| BJ01 | 29,725 | AY278488.2 | 1-May-03 |
| BJ02 | 29,745 | AY278487.3 | 5-Jun-03 |
| BJ03 | 29,740 | AY278490.3 | 5-Jun-03 |
| BJ04 | 29,732 | AY279354.2 | 5-Jun-03 |
| GD01 | 29,757 | AY278489.2 | 5-Jun-03 |
| ZMY1 | 29,749 | AY351680.1 | 3-Aug-03 |
| ZJ01 | 29,715 | AY297028.1 | 19-May-03 |
| TOR2 | 29,751 | NC_004718.3 | 13-Aug-03 |
| Urbani | 29,727 | AY278741.1 | 12-Aug-03 |
| CUHK-Su10 | 29,736 | AY282752.1 | 7-May-03 |
| CUHK-W1 | 29,736 | AY278554.2 | 31-Jul-03 |
| HKU-39849 | 29,742 | AY278491.2 | 18-Apr-03 |
| Frankfurt1 | 29,727 | AY291315.1 | 11-Jun-03 |
| FRA | 29,740 | AY310120.1 | 12-Aug-03 |
| HSR1 | 29,751 | AY323977.2 | 22-Jul-03 |
| Sin2500 | 29,711 | AY283794.1 | 12-Aug-03 |
| Sin2677 | 29,705 | AY283795.1 | 12-Aug-03 |
| Sin2679 | 29,711 | AY283796.1 | 12-Aug-03 |
| Sin2748 | 29,706 | AY283797.1 | 12-Aug-03 |
| Sin2774 | 29,711 | AY283798.1 | 12-Aug-03 |
| TC1 | 29,573 | AY338174.1 | 28-Jul-03 |
| TC2 | 29,573 | AY338175.1 | 28-Jul-03 |
| TC3 | 29,573 | AY348314.1 | 29-Jul-03 |
| TW1 | 29,729 | AY291451.1 | 14-May-03 |
| TWC | 29,725 | AY321118.1 | 26-Jun-03 |
| TWC2 | 29,727 | AY362698.1 | 13-Aug-03 |
| TWC3 | 29,727 | AY362699.1 | 13-Aug-03 |
| TWH | 29,727 | AP006557.1 | 2-Aug-03 |
| TWJ | 29,725 | AP006558.1 | 2-Aug-03 |
| TWK | 29,727 | AP006559.1 | 2-Aug-03 |
| TWS | 29,727 | AP006560.1 | 2-Aug-03 |
| TWY | 29,727 | AP006561.1 | 2-Aug-03 |
Table 6.
Ten Newly Sequenced Complete SARS-CoV Genomes by Beijing Genomics Institute
| Isolate | Genome size (nt) | Source |
|---|---|---|
| HK01 | 29,720 | Hong Kong |
| HK02 | 29,339 | Hong Kong |
| HK03 | 29,721 | Hong Kong |
| HK04 | 29,723 | Hong Kong |
| GD02 | 29,753 | Guangdong |
| GD03 | 29,720 | Guangdong |
| GD04 | 29,725 | Guangdong |
| GD05 | 29,757 | Guangdong |
| GD06 | 29,675 | Guangdong |
| GD07 | 29,725 | Guangdong |
Motif frequency profile
Analyses of the GC content or amino acid composition bias have long been a standard method in biological sequence research. By extending a single nucleotide to longer words, we could reveal more and more species-specific features (14). Recent investigations have reported differences in the frequency of occurrence of many short oligonucleotides, hereafter called “motifs”. The existence of specific MFP has been reported for all motif lengths, such as dinucleotides and trinucleotides.
For the sequence a with the length L, there are (L-m+1) overlapped motifs. When the motif length is m, the total number of possible motif, N, is 4m. Computing the frequency of appearance of the N motif, and put these N values in a fixed order, we form a MFP vector A specific for the sequence a:
A=(a1, a2, a3, ···, aN)
Here, the ak, k=1 to N, is a motif with the length m.
When m is 3 and a is the complete sequence of a gene, A represents the codon usage bias of the gene.
Average absolute distance
Suppose A and B are two MFP vectors corresponding to sequence a and b respectively, A=(a1, a2, a3, ···, aN), B =(b1, b2, b3, ···, bN). A simple measure of the difference between A and B is the average absolute motif relative frequency difference.
Chaos game representation
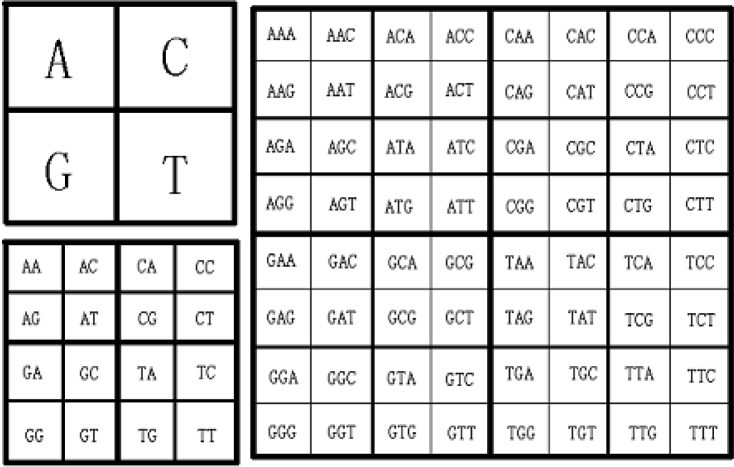
Chaos game representation (CGR) is a new tool derived from the “chaotic dynamic systems” theory. The whole set of frequencies of the motif found in a given genomic sequence can be displayed in the form of a single image in which each pixel is associated with a specific motif. Frequencies of motifs found in a sequence are displayed in a square image, with the location of a given motif being chosen according to a recursive procedure. The gray scale indicates the relative frequency per image of each motif: the darker the pixel, the greater the frequency (15). Figure 8 shows the different CGR arrangement images with motif length from 1 to 3 nt.
Fig. 8.
CGR arrangement of motif or codon.
The programs to analyze the MFP of a given genomic sequence and to draw the CGR images were written by ourselves using the perl language.
Acknowledgements
We thank the colleague of Beijing Genomics Institute in Chinese Academy of Sciences for their hard work on the SARS-CoV sequencing and data analyzing. We are grateful to collaborators and clinicians from Peking Union Medical College Hospital and National Center of Disease Control of China. We also thank National Science Foundation of China for the financial support.
Contributor Information
Jun Yu, Email: junyu@genomics.org.cn.
Huanming Yang, Email: yanghm@genomics.org.cn.
Supporting Online Material
http://www.gpbjournal.org/journal/pdf/GPB1(3)-06.pdf
Table S1
References
- 1.Ksiazek T.G. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 2.Ruan Y. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rota P.A. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.Peiris J.S.M. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grantham R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucl. Acids Res. 1981;9:43–74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grantham R. Workings of the genetic code. Trends Bioch. Sci. 1980;5:327–331. [Google Scholar]
- 7.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 8.Qin E.D. A complete sequence and comparative analysis of a SARS-associated interaction in infected cells. J. Virol. 2003;74:8127–8134. [Google Scholar]
- 9.Yang Z., Yoder A.D. Estimation of the transition/transversion rate bias and species sampling. Mol. Bio. Evol. 1999;48:274–283. doi: 10.1007/pl00006470. [DOI] [PubMed] [Google Scholar]
- 10.Qin E.D. A genome sequence of novel SARS-CoV isolates: the genotype, GD-Ins29, leads to a hypothesis of viral transmission in south China. Geno., Prot. & Bioinfo. 2003;2:101–107. doi: 10.1016/S1672-0229(03)01014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aaziz R., Tepfer M. Recombination in RNA viruses and in virus-resistant transgenic plants. J. General Virol. 1999;80:1339–1364. doi: 10.1099/0022-1317-80-6-1339. [DOI] [PubMed] [Google Scholar]
- 12.Kirkegaard K., Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986;47:433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marra M.A. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 14.Hao B.L., Qi J. Prokaryote phylogeny without sequence alignment: from avoidance signature to composition distance. Proceedings of the Computational Systems Bioinformatics. 2003 [PubMed] [Google Scholar]
- 15.Deschavanne P.J. Genomic signature: characterization and classification of species assessed by chaos game representation of sequences. Mol. Biol. Evol. 1999;16:1391–1399. doi: 10.1093/oxfordjournals.molbev.a026048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1