Abstract
Differential proteome profiles of human lung squamous carcinoma tissue compared to paired tumor-adjacent normal bronchial epithelial tissue were established and analyzed by means of immobilized pH gradient-based two-dimensional polyacrylamide gel electrophoresis (2-D PAGE) and matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS). The results showed that well-resolved, reproducible 2-DE patterns of human lung squamous carcinoma and adjacent normal bronchial epithelial tissues were obtained under the condition of 0.75-mg protein-load. The average deviation of spot position was 0.733±0.101 mm in IEF direction, and 0.925±0.207 mm in SDS-PAGE direction. For tumor tissue, a total of 1241±88 spots were detected, 987±65 spots were matched with an average matching rate of 79.5%. For control, a total of 1190±72 spots were detected, and 875±48 spots were matched with an average matching rate of 73.5%. A total of 864±34 spots were matched between tumors and controls. Forty-three differential proteins were characterized: some proteins were related to oncogenes, and others involved in the regulation of cell cycle and signal transduction. It is suggested that the differential proteomic approach is valuable for mass identification of differentially expressed proteins involved in lung carcinogenesis. These data will be used to establish human lung cancer proteome database to further study human lung squamous carcinoma.
Key words: human lung squamous carcinoma tissue, normal bronchial epithelial tissue, 2-D PAGE, MALDI-TOF-MS, proteome
Introduction
Lung cancer is a malignant tumor with a very high incidence. According to histology diagnosis, lung cancer is divided into four types: squamous carcinoma, adenocarcinoma, large-cell carcinoma and small-cell carcinoma. Squamous carcinoma is the commonest type with a ratio of 30-50% of all lung cancer patients. A clinical statistical study showed that in urban China, the number of squamous carcinoma patients was rapidly increasing, and constituted 50-70% of all lung cancer patients (1). Some molecular pathogenesis studies on human lung cancer have been undertaken successfully at the gene (DNA) and transcription (mRNA) levels (2). However, since the functions of gene are determined by proteins, the proteomic study of lung cancer may directly and comprehensively explore the carcinogenesic mechanism of lung cancer. With the development of proteomic technologies and their application in lung cancer research, some tumor-related markers have been identified, and applied in clinic diagnosis. For example, TAO1 and TAO2 were the effective markers that distinguish original adenocarcinoma from transferred adenocarcinoma 3., 4.. Some studies also indicated that the proteins, antioxidant enzyme (AOE372) and ATP synthase subunit (ATP5D) were significantly over-expressed in lung adenocarcinoma, which will make them potential diagnostic markers of lung adenocarcinoma (5). However, these proteomic studies focus on lung adenocarcinoma, and there is a lack of the data on proteomic studies of squamous carcinoma. Therefore, it is necessary to develop proteomic study on lung squamous carcinoma with a high incidence.
In this study, the total proteins from human lung squamous carcinoma and control were separated with 2-D PAGE. The 2-DE gel image analysis was performed with 2-DE gel image analysis software. Some differentially expressed proteins (up- and down-regulated) between tumors versus normal tissues were identified with MALDI-TOF-MS and database analysis. The objective is to obtain some clues for further study of the basic molecular mechanisms, diagnosis, treatment and new drug development for lung squamous carcinoma.
Results
2-DE pattern of human lung squamous carcinoma tissues and of adjacent normal bronchial epithelial tissues
2-DE was carried out, separately, under the condition of 1.5-mg, 1-mg, 0.75-mg, and 0.5-mg protein-loads. The well-resolved 2-DE profiles were obtained under the condition of a 0.75-mg protein-load. In order to measure the reproducibility, 2-DE was repeated for three times under the condition of 0.75-mg protein-load, respectively, for the tumor- and normal-tissues from each patient. Three silver-stained 2-DE profiles of each sample were very similar. The image analysis showed these 2-DE maps were reproducible. For human lung squamous carcinoma tissues, a total of 1241±88 spots were detected; 987±65 spots were matched with an average matching rate of 79.5%. For adjacent normal bronchial epithelial tissues, a total of 1190±72 spots were detected, and 875±48 spots were matched with an average matching rate of 73.5%. A total of 100 well-resolved and matched spots among three tumor-gels were chosen randomly to calculate the deviation of the spot position. The spot positional deviation was 0.733±0.101 mm in the IEF direction, and 0.925±0.207 mm in the SDS-PAGE direction. The well-resolved and reproducible 2-DE patterns of human lung squamous carcinoma tissues and of adjacent normal bronchial epithelial tissues were attained (Fig. 1, Fig. 2). The tumor-2DE maps were compared with the control-2DE maps, and a total of 864±34 spots were matched.
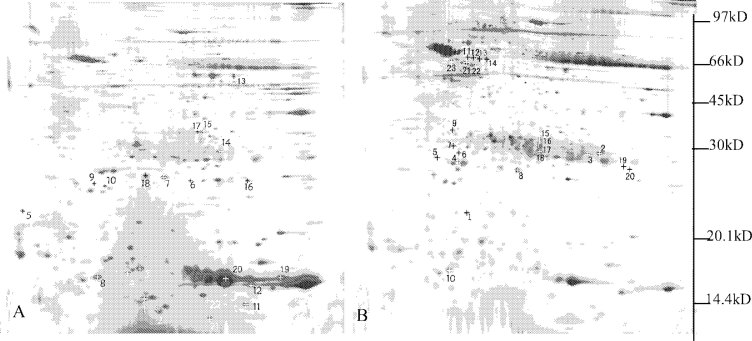
Fig. 1.
Two-dimensional electrophoretic map of human lung squamous carcinoma tissues and paired tumor-adjacent normal bronchial epithelial tissues. (A paired tumor-adjacent normal bronchial epithelial tissues B. human lung squamous carcinoma tissues.)
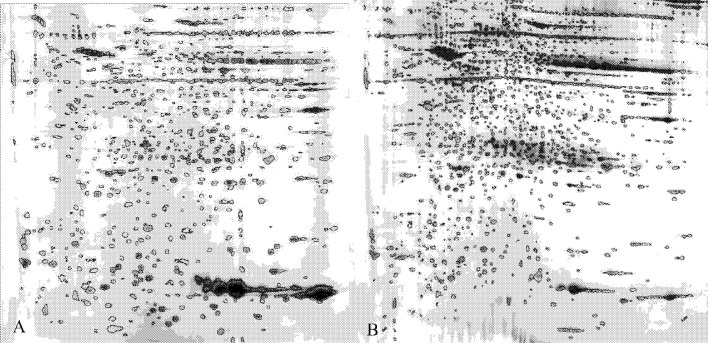
Fig. 2.
Spot detection of two-dimensional electrophoretic map of human lung squamous carcinoma tissues and paired tumor-adjacent normal bronchial epithelial tissues. (A paired tumor-adjacent normal bronchial epithelial tissues B. human lung squamous carcinoma tissues.)
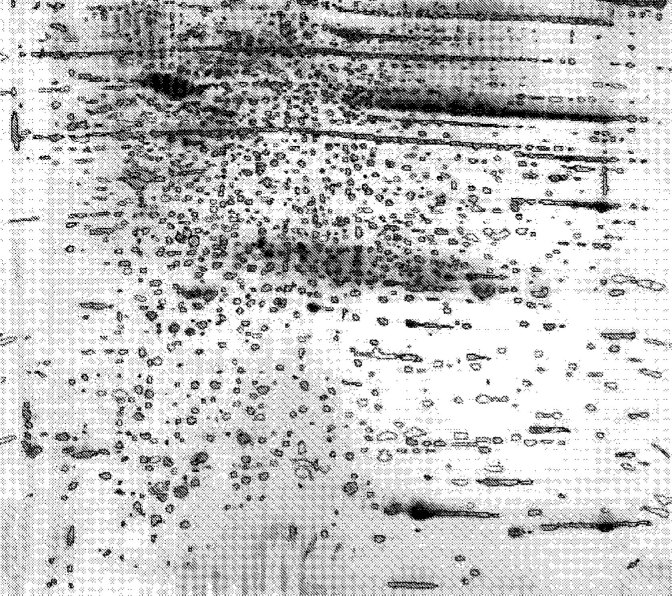
The 2-DE profiles were very similar among 8 tumor-tissues. An average electrophoresis map of human lung squamous carcinoma tissues was constructed by comparisons of the 2-DE maps from 8 tumor-tissues with the ImageMaster 2-D gel analysis software. The average electrophoresis map identified 1814 protein-spots (Fig. 3). Similarly, an average electrophoresis map of eight normal bronchial epithelial tissues was also established with 1796 protein-spots. These average electrophoresis maps were used to perform the differential expression analysis.
Fig. 3.
The average electrophoretic map of human lung squamous carcinoma tissues
MALDI-TOF-MS PMF analysis of the differential protein-spots
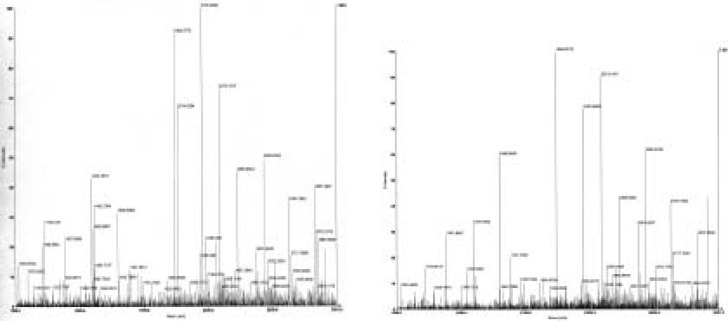
The differential protein-spots between the human lung squamous carcinoma tissues and the adjacent normal bronchial epithelial tissues were detected by 2-D gel image analysis software. To determine the accuracy of the matched-result, two matched-spots (n7 and t8) were identified with MALDI-TOF-MS. The results showed that n7 and t8 are the same protein (AC: Q8TCL7, Hypothetical protein) (Table 1). Forty-three differential protein-spots (11 spots were only expressed in tumor-tissues, 10 spots only in control-tissues, the remaining spots expressed between tumor and control tissues with at least five-fold discrepancy) were detected between at least five paired tumor- and control- tissues. These differential protein-spots were excised from the silver-stained gel, and digested in-gel with trypsin. The peptide mass fingerprinting (PMF) maps were obtained by MALDI-TOF-MS, and calibrated with TPCK-Trypsin auto-degraded peak (m/z=1993.9772 Da). Fig. 4 shows the PMF maps of the protein-spots n8 and t5. The PMF data were used to search the SWISS-PROT and TrEMBL databases with PeptIdent software. The resulting protein was identified by comprehensively considering the corresponding experimental pI, Mr, the number of matched-peptides, and the sequence coverage (Table. 1, Table. 2)
Table. 1.
Protein Spots Searched by PeptIdent Software in Database
| Spot number | #peptide matches | AC | Description | pI | Mw | coverage | |
|---|---|---|---|---|---|---|---|
| n1 | 4/13 | P82980 | Retinol-binding protein III, cellular (CRBP-III) (HRBPiso). - Homo sapiens (Human). | 6.13 | 15800.05 | 38.1% | |
| n2 | 8/50 | Q9HC90 | Calpain 10 (EC 3.4.22.17) (Calcium-activated neutral proteinase 10) (CANP 10). - Homo sapiens (Human). | 6.54 | 15582.85 | 57.1% | |
| n5 | 6/22 | Q9BQN3 | DJ601O1.1 (Novel protein with kunitz/bovine pancreatic trypsin inhibitor domain) (Fragment). - Homo sapiens (Human). | undefined | undefined | 44.2% | |
| n6 | 10/61 | P36980 (Isoform) | SPLICE TRUNCATED Isoform of Complement factor H-related protein 2 precursor (FHR-2) (H factor-like protein 2) (H factor-like 3) (DDESK59). - Homo sapiens (Human). | 6.52 | 27896.58 | 31.7% | A |
| n9 | 14/171 | O43609 | Sprouty homolog 1 (Spry-1) (Fragment). - Homo sapiens (Human). | undefined | undefined | 61.2% | |
| n10 | 5/38 | P20340 | Ras-related protein Rab-6A. - Homo sapiens (Human). | 5.42 | 23592.78 | 35.1% | |
| n12 | 14/166 | Q9UNK4 | CHAIN 1: GROUP IID SECRETORY PHOSPHOLIPASE A2. - Homo sapiens (Human). | 8.75 | 14536.64 | 81.6% | |
| n13 | 9/44 | O60942 (Isoform) | SPLICE Isoform 3 of mRNA capping enzyme (HCE) (HCAP1) [Includes: Polynucleotide 5’- triphosphatase (EC 3.1.3.33) (mRNA 5’-triphosphatase) (TPase); mRNA guanylyltransferase (EC 2.7.7.50) (GTP-RNA guanylyltransferase) (GTase)]. - Homo sapiens (Human). | 8.3 | 52533.29 | 22.1% | |
| n21 | 4/47 | P10073 | Krueppel-related zinc finger protein 2 (HKR2 protein) (Fragment). - Homo sapiens (Human). | undefined | undefined | 34% | |
| n22 | 9/76 | Q01469 | Fatty acid-binding protein, epidermal (E-FABP) (Psoriasis-associated fatty acid-binding protein homolog) (PA-FABP). - Homo sapiens (Human). | 6.6 | 15164.43 | 48.1% | |
| n3 | 6/32 | P30085 | UMP-CMP kinase (EC 2.7.4.14) (Cytidylate kinase) (Deoxycytidylate kinase) (Cytidine monophosphate kinase). - Homo sapiens (Human). | 5.44 | 22222.34 | 60.2% | |
| n4 | 10/28 | P08833 | CHAIN 1: INSULIN-LIKE GROWTH FACTOR BINDING - Homo sapiens (Human). | 5.09 | 25270.44 | 43.6% | |
| n8 | 9/53 | P08709 | CHAIN 1: FACTOR VII LIGHT CHAIN. - Homo sapiens (Human). | 5.03 | 17025.93 | 65.1% | |
| n11 | 7/108 | P01229 | CHAIN 1: LUTROPIN BETA CHAIN. - Homo sapiens (Human). | 8.3 | 13202.57 | 36.4% | |
| n14 | 8/113 | O14753 | Putative transcription factor Ovo-like 1 (hOvol) (Fragment). - Homo sapiens (Human). | undefined | undefined | 36.2% | |
| n15 | 7/28 | Q99871 | X-linked protein STS1769. - Homo sapiens (Human). | 7.22 | 33582.38 | 32.2% | |
| n16 | 17/132 | P26842 | CHAIN 1: TUMOR NECROSIS FACTOR RECEPTOR - Homo sapiens (Human). | 7.8 | 26966.68 | 50.8% | B |
| n17 | 5/18 | P09329 | Ribose-phosphate pyrophosphokinase I (EC 2.7.6.1) (Phosphoribosyl pyrophosphate synthetase I) (PPRibP) (PRS-I). - Homo sapiens (Human), and Rattus norvegicus (Rat). | 6.56 | 34703.04 | 29.3% | |
| n18 | 14164 | Q14964 | Ras-related protein Rab-39. - Homo sapiens (Human). | 6.9 | 24869.51 | 44.2% | |
| n19 | 20/260 | Q9NYY1 | CHAIN 1: INTERLEUKIN-20. - Homo sapiens (Human). | 8.77 | 17513.32 | 63.8% | |
| n20 | 6/49 | P02023 | Hemoglobin beta chain. - Homo sapiens (Human), Pan troglodytes (Chimpanzee), and Pan paniscus (Pygmy chimpanzee) (Bonobo). | 6.81 | 15867.2 | 30.8% | |
| t1 | 9/91 | Q07812 | Apoptosis regulator BAX, membrane isoform alpha. - Homo sapiens (Human). | 5.08 | 21184.41 | 57.3% | |
| t2 | 7/32 | Q8TDH6 | CLL-associated antigen KW-1 splice variant 1 (Fragment). - Homo sapiens (Human). | undefined | undefined | 28.9% | |
| t3 | 12/31 | Q8WW21 | Similar to activator of CREM in testis. - Homo sapiens (Human). | 7.57 | 32809.7 | 62.3% | |
| t4 | 10/45 | P52744 | Zinc finger protein 138 (Fragment). - Homo sapiens (Human). | undefined | undefined | 37.2% | |
| t5 | 11/50 | Q9HAV5 | Tumor necrosis factor receptor superfamily member XEDAR (X-linked ectodysplasin-A2 receptor) (EDA-A2 receptor). - Homo sapiens (Human). | 4.91 | 32728.66 | 33.3% | C |
| t6 | 9/41 | Q9NXX7 | Hypothetical protein FLJ20001. - Homo sapiens (Human). | 5.41 | 33906.7 | 45.9% | |
| t9 | 10/37 | Q969U2 | Carboxy terminus of HSP70-interacting protein (Hypothetical protein) (CLL-associated antigen KW-8). - Homo sapiens (Human). | 5.61 | 34856.2 | 45.6% | |
| t11 | 32/89 | O14985 | Mucin 5B precursor (Mucin 5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). - Homo sapiens (Human). | undefined | undefined | 24.3% | |
| t12 | 10/40 | P49023 | Paxillin. - Homo sapiens (Human). | 5.92 | 60936.55 | 35% | |
| t19 | 14/148 | P17032 | Zinc finger protein 37A (Zinc finger protein KOX21) (Fragment). - Homo sapiens (Human). | undefined | undefined | 68.2% | |
| t23 | 18/145 | O95376 | Ariadne-2 protein homolog (ARI-2) (Triadl protein) (HT005). - Homo sapiens (Human). | 5.4 | 57818.86 | 34.5% | |
| t7 | 8/47 | P30279 | Gl/S-specific cyclin D2. - Homo sapiens (Human). | 5.06 | 33067.23 | 47.4% | |
| t10 | 7/51 | P08709 | CHAIN 1: FACTOR VII LIGHT CHAIN. - Homo sapiens (Human). | 5.03 | 17025.93 | 50.2% | |
| t13 | 15/102 | P02748 | CHAIN 1: COMPLEMENT COMPONENT C9. - Homo sapiens (Human). | 5.42 | 60978.68 | 24.7% | |
| t14 | 23/149 | P29320 (Isoform) | SPLICE Isoform 2 of Ephrin type-A receptor 3 precursor (EC 2.7.1.112) (Tyrosine-protein kinase receptor ETK1) (HEK) (HEK4). - Homo sapiens (Human). | 5.57 | 60945.99 | 37.1% | D |
| t16 | 26/200 | Q14192 | Skeletal muscle LIM-protein 3 (SLIM 3) (LIM-domain protein DRAL) (Four and a half LIM domains protein 2) (FHL-2). - Homo sapiens (Human). | 7.59 | 69.9% | ||
| t18 | 10/94 | Q10588 | CHAIN 1: ADP-RIBOSYL CYCLASE 2. - Homo sapiens (Human). | 6.72 | 30900.93 | 45.6% | |
| t20 | 27/238 | Q13145 | CHAIN 1: PUTATIVE TRANSMEMBRANE PROTEIN NMA. - Homo sapiens (Human). | 8.21 | 25956.82 | 71.2% | |
| t21 | 6/27 | P48643 | T-complex protein 1, epsilon subunit (TCP-l-epsilon) (CCT-epsilon). - Homo sapiens (Human). | 5.45 | 59671.02 | 25.7% | |
| t22 | 24/119 | Q13422 (Isoform) | SPLICE Isoform IK7 of DNA-binding protein Ikaros (Lymphoid transcription factor LyF-1). - Homo sapiens (Human). | 5.86 | 52705.82 | 33.8% | |
| n7 | 13/47 | Q8TCL7 | Hypothetical protein (Fragment). - Homo sapiens (Human). | undefined | undefined | 58.1% | E |
| t8 | 8/36 | Q8TCL7 | Hypothetical protein (Fragment). - Homo sapiens (Human). | undefined | undefined | 42.3% | |
*The spots in A were expressed only in normal tissue; the spots in B were expressed in normal tissue which is five times higher than in tumor tissue. The spots in C were expressed only in tumor tissue; the spots in D were expressed in tumor tissue which is five times higher than in normal tissue and the spots in E (n7 and t8) are two matched-spots.
Fig. 4.
Peptide mass fingerprinting of protein spot n8 in paired tumor-adjacent normal bronchial epithelial tissues 2-DE map and of protein spot t5 in human lung squamous carcinoma tissues 2-DE map.
Table. 2.
Matching of Protein Spot-n8 Peptide Mass Fingerprint Data with Protein P08709 in Database
| User mass | Matching mass | Mass (Da) | #MC | Modification | Positon | Peptide |
|---|---|---|---|---|---|---|
| 1707.7813 | 1708.0164 | 0.2351 | 2 | 198-212 | IPILEKRNASKPQGR | |
| 2134.0172 | 2134.4144 | 0.3972 | 1 | 1xCys_PAM | 61-78 | ANAFLEELRPGSLER ECK |
| 2150.0881 | 2150.3391 | 0.2510 | 2 | 1xGGLU | 76-92 | ECKEEQCSPEEAREI FK |
| 2194.0103 | 2194.3489 | 0.3386 | 2 | 2xGGLU | 76-92 | ECKEEQCSPEEAREI FK |
| 2251.0155 | 2251.4009 | 0.3854 | 2 | 1xCys_CAM, 2xGGLU | 76-92 | ECKEEQCSFEEAREI FK |
| 2295.1210 | 2295.4107 | 0.2897 | 2 | 1xCys_CAM, 3xGGLU | 76-92 | ECKEEQCSFEEAREI FK |
| 2305.0513 | 2305.3909 | 0.3396 | 2 | 2xGGLU | 79-96 | EEQCSFEEAREIFKD AER |
| 2588.1566 | 2587.8964 | −0.2602 | 0 | 1xCys_CAM | 174-197 | CHEGYSLLADGVSCT PTVEYPCGK |
| 2612.1725 | 2611.8344 | −0.3381 | 0 | 1xCys_CAM | 99-122 | LFWISYSDGDQCASS PCQNGGSCK |
65.1% of sequence covered:
| 61 | 71 | 81 | 91 | 101 | 111 | ||
| | | | | | | | | | | | | ||
| 61 | ANAFLEELRP | GSLERECKEE | QCSFEEAREI | FKDAERtkLF | WISYSDGDQC | ASSPCQNGGS | 120 |
| 121 | CKdqlqsyic | fclpafegrn | cethkddqli | cvnenggceq | ycsdhtgtkr | scrCHEGYSL | 180 |
| 181 | LADGVSCTPT | VEYPCGKIPI | LEKRNASKPQ | GR |
Discussion
The topics of proteomics research at present mainly include the establishment of 2-DE reference maps and database from organs, tissues, and cells, and the characterization of special functional proteins by proteomic comparisons between healthy and diseased states. In the future, the special functional proteins will be used to develop diagnostic markers for clarifying the molecular mechanisms of diseases, and to detect the novel drug targets.
Our research group has previously studied the protein expression maps of a lung cancer cell line (6). The protein expression profiles are not stable due to the inconsistent time and environment of in vitro cultured-cells. In order to establish the protein expression profiles of lung cancer and characterize the tumor-related proteins, the human lung squamous carcinoma tissues and the adjacent normal bronchial epithelial tissues were analyzed by 2-DE and MS in this study. The extraction method of the total protein from tissue and the amount of protein loaded onto a gel were optimized. The samples were precipitated in TCA in order to get rid of high-molecular weight polysaccharides that were abundant in cartilage and which were not removed from bronchial epithelial tissues completely. The well-resolved, reproducible 2-DE profiles from the human lung squamous carcinoma tissues and from the paired tumor-adjacent normal bronchial epithelial tissues were established. Images with higher quality were obtained with Labscan software–Imagescanner image system–compared to the previous Gel Doc2000 gel image system. Compared with Bio-Rad PDQuest 2-D gel analysis software, the Pharmacia ImageMaster 2D Elite software (4.01 version) demonstrated higher sensitivity. However, both the two types of 2-D gel analysis software were time-consuming and labor-intensive. It is necessary to improve the degree of automation of their analysis. The average gel of lung squamous carcinoma tissues was established by ImageMaster 2D analysis software. The average gel is a statistically synthetic gel of a batch of gels, representing a set of gels from the homogeneity sample, but containing more information, which may improve the accuracy of the analysis results (7).
The comparative proteomics study was performed between the human lung squamous carcinoma tissues and the control tissues. Forty-three differential protein-spots were selected to perform in-gel Trypsin digestion and MALDI-TOF-MS-based peptide mass fingerprinting (PMF) analysis. Forty-one protein-spots were identified by searching the SWISS-PROT and TrEMBL databases. Two protein-spots were not identified, which may be due to obtaining an insufficient tryptic peptide fragment.
Some of the identified proteins with differently expressed levels are the oncogene-encoded products, and others are the regulation proteins of cell cycle and signal transduction. For examples, Cyclin D2, XEDAR (X-linked ectodysplasin-A2 receptor), HSP70-interacting protein (CLL-associated antigen KW-8), Retinol-binding protein III, Ras-related protein Rab6A and Ras-related protein Rab-39 are differently expressed in both tissues. Niklinski’s study showed that Cyclin D is related with carcinogenesis of lung cancer, which is possibly a prognostic marker for non-small cell lung cancer (8). XEDAR (X-linked ectodysplasin-A2 receptor) is a recently isolated member of the tumor necrosis factor receptor family, ligand of which is ectodysplasin-A2 (EDA-A2). XEDAR activates the NF-kappaB and JNK pathways in an EDA-A2-dependent fashion and plays a major role in the process of tumor development and progress (9). Heat shock proteins participate in the intracellular dislocation and assembly of many proteins and are associated with cacinogenesis. Retinol-binding protein III (CRBP) inhibits activity of the PI3-K/Akt survival pathway and suppresses anchorage-independent growth of tumor cell, the expression of which is down-regulated in a subset of human breast cancer cell lines (10). Our results showed that Cyclin D2, XEDAR and HSP70-interacting proteins were up-regulated or expressed only in lung squamous carcinomas, while Retinol-binding protein III was down-regulated in lung squamous carcinoma tissues compared with controls. These data suggest that these four proteins may play roles in the carcinogenesis of lung squamous carcinoma. Ras-related protein Rab6A and Ras-related protein Rab-39 were down-regulated in lung squamous carcinomas in controlled comparisons, the mechanism of which awaits further study (11). We have found the comparative proteomics to be an effective platform for studying the human lung squamous carcinoma.
Materials and Methods
Tissue specimens and preparation
Eight human lung squamous carcinoma tissues and paired tumor-adjacent normal tissues were obtained from the First and the Second Xiangya Hospital. All samples were from male patients and were diagnosed by histopathology. Patients’ medical records were reviewed. Fresh tumor tissues and paired tumor-adjacent normal bronchial tissues that were at least 5 cm away from tumorous tissues were obtained immediately after surgery, and stored in liquid nitrogen. Epithelial was exfoliated from bronchial tissues. Specimens were washed repeatly with NS in asepsic condition in order to remove blood and get rid of excrescent tissues, the processed specimens were immediately used to extract the total proteins, or were stored at −80°C before use. A total of 30-80 mg tissues was ground into powder in liquid nitrogen; dissolved in 400 μL lysis buffer consisting of 7 M urea, 2 M thiourea, 2% NP-40, 1% Triton X-100, 100 mM DTT, 5 mM PMSF, 4% CHAPS, 0.5 mM EDTA, 40 mM Tris, 2% pharmalyte, 1 mg/mL DNase I, and 0.25 mg/mL RNase A; votexed, and incubated (37°C, 1 h). The mixture was centrifuged (15,000 r/min, 30 min, 4°C) (12). The supernatant was the total protein solution, which was precipitated by using TCA sediment method. The concentration of the total proteins was determined with the BCA protein Assay Kit (Pierce).
IPG-2D PAGE
IPG-2D PAGE were consisted of IEF that was performed using IPGstrip (pH 3-10L, 180 mm×3 mm×0.5 mm) on IPGphor isoelectric focusing cell (Amersham Pharmacia Biotech) and Second-dimension SDS-PAGE (Bio-Rad) as described by the manufacturer and Gorg (13). After separation, the protein spots were visualized by silver-based staining technique with the protein silver stain kit (Amersham Pharmacia Biotech).
Image analysis
The stained 2-DE gels were scanned with LabScan software on Imagescanner (Amersham Pharmacia Biotech). The spot-intensity calibration, spot detection, background abstraction, matching, 1-D calibration, and the establishment of average-gel were performed with ImageMaster 2D Elite 4.01analysis software (Amersham Pharmacia Biotech). The reproducibility of spot position was calculated according to Gorbett’s method (14). Statistical analysis was carried out with SPSS for Windows 10.0 and Excel.
MALDI-TOF-MS
Forty-three differentiated spots were excised from preparative gels using biopsy punches and transferred to a 1.5ml siliconized Eppendorf tube. One protein-free gel piece was treated in parallel as a negative control. Proteins were digested in-gel as previously described (15). The samples were analyzed with Applied Biosystems Voyager System 4307 MALDI-TOF Mass Spectrometer (ABI). The parameters were set up as following: positive ion-reflector mode, accelerating voltage 20 kV, grid voltage 64.5%, mirror voltage ratio 1.12, N2 laser wavelength 337 nm, pulse width 3 ns, the number of laser shots 50, acquisition mass range 1000-3000 Da, and delay 100 nsec, and vacuum degree 4×10−7Torr. A trypsin-fragment peak served as an internal standard for mass calibration. A list of the corrected mass peaks was the peptide mass fingerprinting (PMF). Proteins were identified with PMF data by searching software PeptIdent (http://www.expasy.pku.edu.cn).
Acknowledgments
This work was supported by a grant from National 973 Project (2001CB5102), for Outstanding Scholars of New Era from the Ministry of Education of China (2002-48), National Natural Science Foundation of China (30000028), the key research program from Science and Technology Committee of Hunan, China (02SSY2001-1) and the key research program from Public Health Bureau of Hunan Province, China (Z02-04).
References
- 1.Lopez A.D. Counting the dead in China: measuring tobacco’s impact in the developing world. Br. Med. J. 1998;317:1399. doi: 10.1136/bmj.317.7170.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sekido Y. Progress in understanding the molecular pathogenesis of human lung cancer. Bioch. Biophy. Acta. 1998;1378:F21–F59. doi: 10.1016/s0304-419x(98)00010-9. [DOI] [PubMed] [Google Scholar]
- 3.Hlrano T. Relationship between TAO1 and TAO2 polypeptides associated with lung adenocarcinoma and histocytological features. Br. J. Cancer. 1997;75:978–985. doi: 10.1038/bjc.1997.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oh J.M.C. A database of protein expression in lung cancer. Proteomics. 2001;1:1303–1319. doi: 10.1002/1615-9861(200110)1:10<1303::AID-PROT1303>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 5.Chen G.A. Proteomic analysis of lung adenocarcinoma: Identification of a highly expressed set of proteins in tumors. Clin. Cancer Res. 2002;8:2298–2305. [PubMed] [Google Scholar]
- 6.Zhan X.Q. Differential proteomic analysis of human lung adenocarcinama cell line A-549 and normal cell line HBE. Acta. Biochim. Biophys. Sin. 2002;34:50–56. [PubMed] [Google Scholar]
- 7.Jia Y.F. The image analysis of two-dimensional gel electrophoresis. Prog. Biochem. Biophys. 2001;28:246–250. [Google Scholar]
- 8.Niklinski J. Prognostic molecular markers in non-small cell lung cancer. Lung Cancer. 2001;34(suppl 2):s53–s58. doi: 10.1016/s0169-5002(01)00345-2. [DOI] [PubMed] [Google Scholar]
- 9.Sinha S.K. Role of TRAF3 and −6 in the Activation of the NF-kappa B and JNK Pathways by X-linked Ectodermal Dysplasia Receptor. J. Biol. Chem. 2002;277:44953–44961. doi: 10.1074/jbc.M207923200. [DOI] [PubMed] [Google Scholar]
- 10.Kuppumbatti Y.S. CRBP suppresses breast cancer cell survival and anchorage-independent growth. Oncogene. 2001;20:7413–7419. doi: 10.1038/sj.onc.1204749. [DOI] [PubMed] [Google Scholar]
- 11.Pfeifer G.P. Methylation of the RASSF1A gene in human cancers. Biol. Chem. 2002;383:907–914. doi: 10.1515/BC.2002.097. [DOI] [PubMed] [Google Scholar]
- 12.Araki N. Comparative analysis of brain proteins from P53-decificient mice by two-dimensional electrophoresis. Electrophoresis. 2000;21:1880–1889. doi: 10.1002/(SICI)1522-2683(20000501)21:9<1880::AID-ELPS1880>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 13.Gorg A. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 2000;21:1037–1053. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1037::AID-ELPS1037>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 14.Gorbett J.M. Posotional reproducibility of protein spots in two -dimensional polyacrylamide gel electrophoresis using immobilized pH gradients isoelectric focusing in the first dimension: An interlaboratory comparison. Electrophoresis. 1994;15:1205–1211. doi: 10.1002/elps.11501501182. [DOI] [PubMed] [Google Scholar]
- 15.Chen P. Identification of protein spots in silver-stained two-dimensional gels by MALDI-TOF mass peptide map analysis. Acta. Biochim. Biophys. Sin. 2000;32:387–391. [PubMed] [Google Scholar]