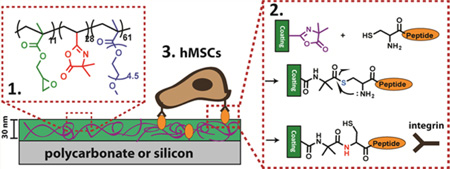
Abstract
Conjugation of biomolecules for stable presentation is an essential step toward reliable chemically defined platforms for cell culture studies. In this work, we describe the formation of a stable and site-specific amide bond via the coupling of a cysteine terminated peptide at low concentration to an azlactone containing copolymer coating. A copolymer of polyethylene glycol methyl ether methacrylate-ran-vinyl azlactone-ran-glycidyl methacrylate P(PEGMEMA-r-VDM-r-GMA) was used to form a thin coating (20–30 nm) on silicon and polycarbonate substrates. The formation and stability of coating-peptide bonds for peptides containing free thiols and amines were quantified by X-ray photoelectron spectroscopy (XPS) after exposure to cell culture conditions. Peptides containing a thiol as the only nucleophile coupled via a thioester bond; however, the bond was labile under cell culture conditions and almost all the bound peptides were displaced from the surface over a period of 2 days. Coupling with N-terminal primary amine peptides resulted in the formation of an amide bond with low efficiency (<20%). In contrast, peptides containing an N-terminal cysteine, which contain both nucleophiles (free thiol and amine) in close proximity, bound with 67% efficiency under neutral pH, and were stable under the same conditions for 2 weeks. Control studies confirm that the stable amide formation was a result of an intramolecular rearrangement through a N-acyl intermediate that resembles native chemical ligation. Through a combination of XPS and cell culture studies, we show that the cysteine terminated peptides undergo a native chemical ligation process at low peptide concentration in aqueous media, short reaction time, and at room temperature resulting in the stable presentation of peptides beyond 2 weeks for cell culture studies.
Graphical Abstract
INTRODUCTION
For basic understanding of cell-material interactions, it is necessary to develop suitable surfaces for two-dimensional in vitro cell culture studies. In particular, these surfaces are useful for cell types such as adult and embryonic stem cells whose behavior (i.e., adhesion, proliferation, and differentiation) may be directed by the cell culture surface.1,2 There is increasing evidence that these cell types can undergo changes influenced by peptide density, peptide identity, and stiffness of the surface.3,4 To systematically study these effects a chemically defined surface is required. The chemically defined surfaces that have been used to influence cell-material interactions include polymer films, hydrogels, polymer brushes and self-assembled monolayers.5–9 Often, surfaces are engineered with characteristics such as resistance to nonspecific protein adsorption via covalent incorporation of low-fouling materials like polyethylene glycol (PEG).10,11 Additionally, peptides such as Arg-Gly-Asp (RGD) can provide sites for receptor-mediated cell adhesion.12 These peptides can be coupled to a low-fouling surface through many types of bonds including amide, carbamate, alkyl sulfide, thioester, triazole, ester, thioether, and disulfide bonds.13 Previous studies have compared the relative stability of thioester, amide, and carbamate linkers mainly by using cell-based assays.14,15 In general, these studies have found that, under cell culture conditions the thioester bond is labile, whereas the amide bond is stable. However, these studies have been largely qualitative in nature, and quantitative long-term studies under cell culture conditions are lacking. As certain stem cell behaviors, such as differentiation or expansion, can be longer-term processes, quantitative understanding of peptide-material bond stability is needed to inform the design of chemically defined surfaces.
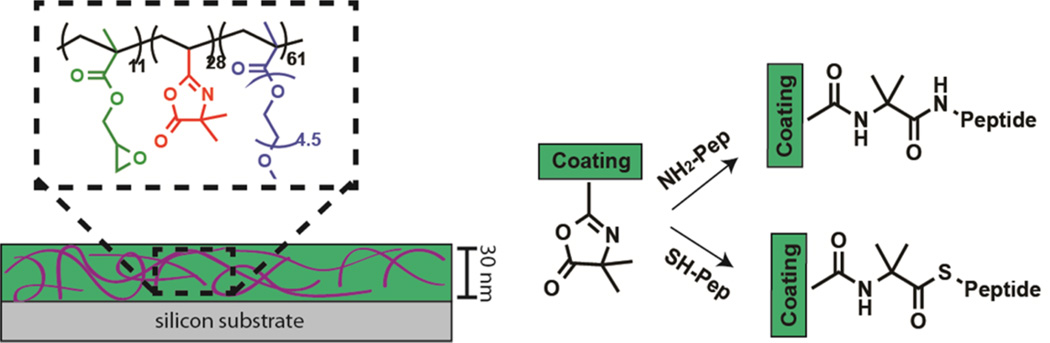
Predominantly, the chemistry used for attaching peptides involves an activation step to form an ester or an amide bond, which often results in poor efficiency in aqueous conditions. Ring opening reaction of oxazolines is an attractive alternative. They have traditionally been shown to react with either a thiol (–SH) or amine (–NH2) nucleophile, resulting in either a thioester or amide bond (Figure 1). The development of vinyl dimethylazlactone (VDM), which incorporates the oxazoline ring in a polymerizable monomeric structure has made the development of VDM copolymers possible.16,17 In a previous report, we developed a new VDM-containing copolymer coating consisting of a tercopolymer polyethylene glycol methyl ether methacrylate-ran-vinyl azlactone-ran-glycidyl methacrylate, P(PEGMEMA-r-VDM-r-GMA).14 This copolymer can be thermally cross-linked using the GMA groups into a stable coating prior to peptide attachment to the VDM units. The thermal cross-linking of GMA is independent of the substrate, and its applicability to glass, polycarbonate, gold, and silicon substrates has been demonstrated.14,18,19 Herein, we use principles from native chemical ligation (NCL) to couple linear peptides through a stable amide bond to the azlactones containing thin films of P(PEGMEMA-r-VDM-r-GMA) with high efficiency under neutral conditions. Cyclic peptides containing a thiol group were coupled via a thioester bond with high efficiency (>80%), whereas those containing primary amine group coupled with low efficiency (<20%) at neutral pH in aqueous medium. NCL was first reported for peptide synthesis using trans-thioesterification between two unprotected amino acids.20 Typically, NCL occurs in pH 7–8 buffered conditions, where an N-terminal-cysteine forms an S-acyl covalent intermediate that spontaneously undergoes an S- to N-acyl migration to form an amide bond through a five-member ring intermediate.21–23 We hypothesized that the reaction of an azlactone ring with a cysteine can also undergo NCL, because of the similar thioester intermediate formed. To investigate, two peptides, both with the sequence Cys-Arg-Gly- Asp-Ser (CRGDS), were coupled to azlactone groups on a cross-linked coating. The key difference between the two peptides was one had an unmodified N-terminus (am-CRGDS), while the other had an acetylated N-terminus (ac-CRGDS). Therefore, at pH 7.4, both am-CRGDS and ac-CRGDS were expected to form a thioester bond through the thiol group. We provide evidence for intramolecular rearrangement with am-CRGDS using X-ray photoelectron spectroscopy (XPS) as well as evidence for the instability of thioester bonds under cell culture conditions.
Figure 1.
Schematic representation of the copolymer coating and the ring opening of azlactone groups with two nucleophiles. Reaction with thiols (ac-CRGDS) and primary amines (am-GRGDSP) are expected to result in thioester and amide bonds, respectively.
EXPERIMENTAL SECTION
Materials
Poly(ethylene glycol) methyl ether methacrylate (PEGMEMA, Mn ~ 300 g/mol), glycidyl methacrylate (GMA), 2-cyano-2-propyl benzodithioate, 2,2′-azobis(2-methylpropionitrile), anisole, ethanolamine, sulfuric acid (H2SO4), hydrogen peroxide 30% in water (H2O2), were purchased from Sigma-Aldrich Co. 2-Vinyl-4,4-dimethyl azlactone (VDM) was a gift from Dr. Steve Heilmann from the 3 M Corporation (Milwaukee, WI). Silicon wafers (〈100〉, p-type) were purchased from University Wafer (Boston, MA). Minimum essential medium, alpha (1x; αMEM) was from CellGro (Mannassas, VA). Penicillin/streptomycin was from Hyclone (Logan, UT). VDM and GMA were purified by vacuum distillation. Peptides am-CRGDS, ac-CRGDS, and am-GRGDSP were purchased from GenScript and had purities over 90%. All other materials were used as received.
Polymer Synthesis
Polymer synthesis of P(PEGMEMA-r-GMA-r-VDM) was reported in a prior publication.14 Using reversible addition–fragmentation chain transfer (RAFT), PEGMEMA (7.2 mmol, 2.16 g), GMA (0.7 mmol, 99.5 mg), and VDM (2.1 mmol, 292 mg)\ were added to a 25 mL Schlenk flask. The solvent anisole (13.0 mL) was added with a chain transfer agent (CTA), 2-cyano-2-propyl benzodithioate (0.01 mmol, 2.2 mg). Lastly the initiator 2,2′-azobis(2-methylpropionitrile), (0.01 mmol, 1.6 mg) was added at an initiator to CTA ratio of 1:1. The mixture was degassed with three freeze–pump–thaw cycles. Polymerization was allowed to proceed at 70 °C for 15 h, and then the mixture was precipitated in n-hexanes and then dissolved in tetrahydrofuran (THF, Fisher Scientific) and reprecipated in hexane three times. The resulting light pink copolymer was stored in THF at −20 °C. P(PEGMEMA-r-GMA-r-VDM) was analyzed using gel permeation chromatography (GPC) giving a Mn = 39 500 and dispersity of 1.9. Proton nuclear magnetic resonance spectroscopy (1H NMR), data gave 61.1% PEGMEMA, 28% VDM, and 10.9% GMA.
Coating Formation
Silicon wafers were treated with piranha solution (3:1 H2SO4:H2O2) at 95 °C for 30 min, and washed with deionized (DI) water and ethanol, and used within 24 h of cleaning. Caution! Piranha reacts violently in contact with organic matter. Polycarbonate coverslips (GraceBio, HybriSlip 22 mm × 22 mm) were rinsed twice with 100% ethanol (Decon Laboratories) followed by DI water and used directly. Copolymers of P(PEGMEMA-GMA-VDM) were diluted in 100% ethanol and spin coated onto the prepared substrates using a CEE 100 spin coater (Brewer Science). A concentration of the copolymer of 12 mg/mL was used to fabricate a 30 nm thick (4000 rpm for 60 s) film by spin coating. Films were annealed at 110 °C for 3 h under vacuum to cross-link. Thickness was measured by ellipsometry using a Rudolph Auto EL null ellipsometer. Measurements were made at wavelengths 633, 546, and 405 nm with an angle of incidence of 70°. Thickness was determined using FilmEllipse software version 1.1 (Scientific Company Intl.). For cell culture experiments, polycarbonate substrates were rinsed with DI water and ethanol and used directly with the same polymer solution, spin speeds, and cross-linking temperatures.
Peptide Coupling
On the benchtop at room temperature, a 1 mM solution of the peptide (ac-CRGDS, am-CRGDS, or am-GRGDSP) in phosphate buffered saline (PBS, Fisher Scientific) at pH 7.4 was pipetted on to the coating and reacted for 1 h. Coupling was followed by rinses with deionized water and ethanol. The cell culture samples were then soaked in PBS solution for 1 h and in 70% ethanol for 20 min to sterilize.
Displacement Assays
Coatings coupled with either am-CRGDS or ac-CRGDS peptides were placed into one of the following solutions: (1) minimum essential media alpha (αMEM, Cellgrow, Manassas, VA) with 1% penicillin/streptomycin (Hycolne, Logan UT) and 10% fetal bovine serum (Invitrogen, Carlsbad CA), (2) DI water, (3) PBS at pH 7.2–7.4, (4) 0.1 M HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffer at pH 7.2, (5) 10 mM of 4,4,4-trifluorobutylamine concentration in DI water pH 7.2, or (6) 0.2 M sodium bicarbonate solution adjusted to pH 9.0–9.5 with sodium hydroxide (NaOH). The samples were then kept covered in an incubator at 37 °C for 1, 2, 7, or 14 days. Samples were not placed back into solution after XPS measurement. All experiments were repeated independently twice.
Maleimide Tagging
The maleimide molecule, N-(4,4,5,5,6,6,7,7,8,8,9,9,10,10,11,11,11-heptadecafluoroundecyl)-maleimide (Sigma-Aldrich) was dissolved at 2 mM in dimethylformamide (DMF, Fisher Scientific). The DMF solution was then mixed with PBS pH 6.9–7.4 at a 1:1 ratio. This solution was then placed onto fresh coatings coupled with am-CRGDS for 1 h. After reaction, the coating was soaked in DMF for 1 h with agitation to remove excess maleimide, and rinsed with DI water followed by ethanol. DMF was required to solubilize the maleimide.
Ninhydrin Assay
A 4 M sodium acetate buffer was prepared by dissolving anhydrous sodium acetate in 10% glacial acetic acid (1 mL glacial acetic acid, 9 mL water), and titrated to pH 5.5 with glacial acetic acid. One hundred fifty millimolar ninhydrin stock solution was prepared in ethylene glycol, to which 0.33 mL of sodium acetate buffer were added. Stannous chloride solution was prepared by dissolving Tin(II) chloride at 527 mM in ethylene glycol. Immediately prior to running the assay, a working solution of ninhydrin reagent was prepared by adding 125 µL of stannous chloride solution to 5 mL ninhydrin stock solution. Ethanolamine standards were prepared by serial dilution in phosphate-buffered saline. Five microliter volume samples or standards were added to wells of an assay plate and heated at 105 °C for 15 min. The plate was allowed to cool for 15 min before measuring absorbance at 570 nm on a microplate reader.
XPS Analysis
XPS measurements were performed using a Thermo Scientific Model K-Alpha XPS instrument with monochromatic Al Kα radiation (1486.7 eV). Survey spectra and high-resolution spectra were acquired using analyzer pass energies of 200 and 50 eV, respectively, with an X-ray spot size of 400 µm for single point analysis. Data was analyzed using Avantage XPS software package and peak fitting with Gaussian/ Lorentzian peak shapes and a Shirley/Smart type baseline. At least three points were taken per sample, and the averages and standard deviations were reported. Depth profiling was done using large Argon clusters with 4000 eV for etching, and data were collected every 30 s using the snapshot function. Actual atomic percent was calculated using eq 1 by the Avantage software, where N is nitrogen, S is sulfur, O is oxygen, C is carbon and A is the area under the XPS peak, and s is the sensitivity factor of each element.
| (1) |
Theoretical atomic percent of all elements after complete reaction with a peptide was calculated by counting the number of N, C, O, or S elements (n), multiplying by the 1H NMR fraction for each component of the copolymer, as shown in eq 2 for N. Here we assume all azlactone units have reacted with peptide.
| (2) |
The theoretical atomic percent of each element was then calculated using eq 3 and the values derived from eq 2. Lastly, the extent of reaction was determined by the measured N/C ratio divided by the theoretical N/C ratio. Further details are given in Supporting Information Example S2.
| (3) |
Cell Culture
hMSCs (Lonza, Cat PT2501) were expanded at low density on tissue culture treated polystyrene plates. Passage 6 cells were harvested using 0.05% trypsin solution and suspended in 1 mL of minimum essential medium alpha (αMEM) (Cellgrow, Manassas, VA). hMSCs were then seeded at 10 000 cells/cm2 onto polycarbonate substrates coated with the copolymer and patterned with peptide spots in 4 mL of αMEM containing 1% penicillin/streptomycin (Hyclone, Logan, UT) and 10% MSC qualified fetal bovine serum (FBS) (Invitrogen, Carslbad, CA) and were added to a 6-well plate (Thermofisher Scientific/Nunc, Rochester, NY). The plate was rocked to ensure even distribution of the cells. Samples were incubated at 37 °C and 5% CO2 to promote cell attachment. At 4 h postseeding, samples were placed in an incubation chamber and live-imaged every hour for 12 h; timepoints at 5 and 12 h were reported.
Statistical Analysis
Values were reported as the mean with the standard deviation. Error propagation of the standard deviation was done using Supporting Equations S1 and S2.
RESULTS AND DISCUSSION
Initial Coupling Efficiency
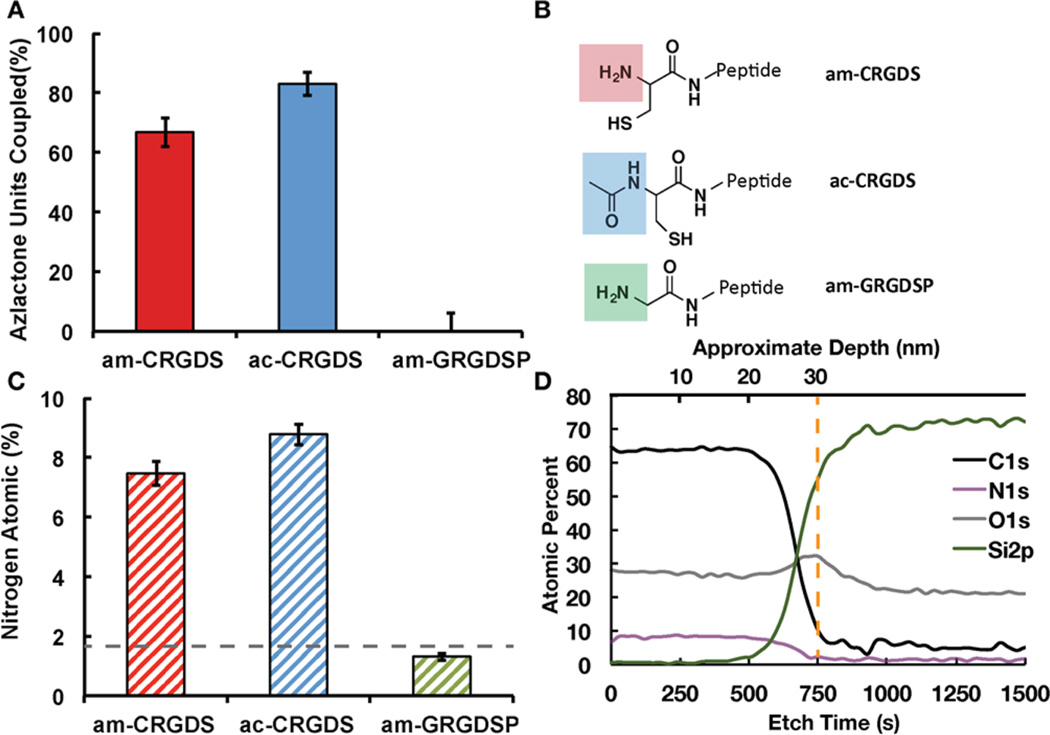
The reaction efficiency of the peptides am-CRGDS, ac-CRGDS and a control with a free N-terminus but without a cysteine, NH2-Gly-Arg-Gly-Asp-Ser-Pro (am-GRGDSP) was evaluated using XPS. All reactions were conducted with 1 mM peptide concentration in phosphate buffered saline (PBS) at pH 7.4 on the coating at room temperature for 1 h. These conditions were chosen as low concentrations of peptides are desirable for cost effectiveness and aqueous based conditions are desirable to broaden the applicability of the coating to plastic substrates (e.g., polycarbonate, polystyrene). ac-CRGDS reacted with a higher percent of azlactone groups (83.1% ± 3.9%) than am-CRGDS (66.8% ± 4.9%) (Figure 2a). The differences in reactivity may correspond to the end terminus functionality shown in Figure 2b. Figure 2c shows the percentage of nitrogen atoms on the surface, which was used to calculate the percentage of the azlactone units that had been ring opened using eqs 2 and 3. Further optimization of the concentration of ac-CRGDS and am-CRGDS and the incubation time resulted in a 9% and 7% increase in the total percent of bound peptides, respectively (Supporting Figure S1). Differences in reaction efficiency between ac-CRGDS (83%) and literature reported cyclic Arg-Gly-Asp-D-Phe-Cys (cRGDfC)14 (100%), which has a free thiol, may be due to the distinct conformations the peptides adapt in water.24,25 Lastly, the distribution of the ac-CRGDS and am-CRGDS peptides throughout the thickness of the coating was evaluated using an ion-etch gun on the XPS (Figure 2d and Supporting Figure S2). Both ac-CRGDS and am-CRGDS reacted evenly throughout the entire thickness (30 nm), as evident by the consistent nitrogen (N) atomic percent as a function of depth.
Figure 2.
(A) XPS data showing percent of azlactone groups in the copolymer coating coupled to peptides am-CRGDS, ac-CRGDS, or am-GRGDSP. “100%” represents the theoretical maximum. Standard deviation from the mean is shown from five samples. (B) Structure of the unmodified N-terminus on am-CRGDS, the acetylated N-terminus on ac-CRGDS, and the unmodified primary amine on am-GRGDSP with no neighboring cysteine amino acid. (C) The Nitrogen (N) atomic percent detected on each substrate from the XPS data. Dotted line shows the average detected in a blank (unreacted) coating. (D) Depth profile of the ac-CRGDS coupled surface, showing uniform reaction of the peptide throughout the coating thickness (30 nm). The dotted orange line shows the approximate polymer–substrate interface.
am-GRGDSP was included as a control to assess the reactivity of primary N-terminal amines that were not directly on an N-terminal cysteine. The dotted line shows the average nitrogen (N) atomic percent detected in a blank sample (Figure 2c), which primarily comes from the azlactone in the coating. Hence am-GRGDSP did not react with the copolymer under these conditions; however, the efficiency of am-GRGDSP coupling can be increased marginally to 12 ± 1.8% in aqueous media at a much higher pH 9.5 in the presence of 1.5 M sodium sulfate.
Soaking in Cell Culture Media
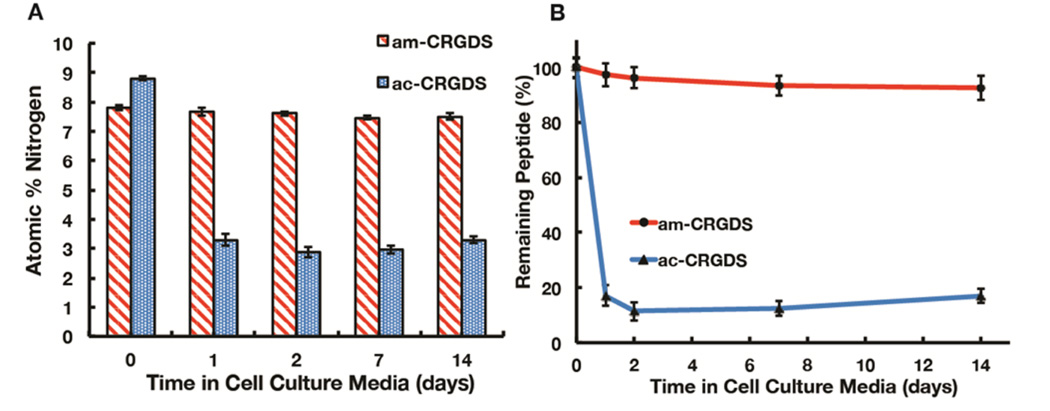
The P(PEGMEMA-r-GMA-r-VDM) coating was designed to have extended stability in cell culture conditions.14 While the stability of the coating has been shown in a previous study for over a month in various media, the stability of the peptide linkage to the coating itself and the conjugation efficiency was not optimized. Therefore, to assess the stability of the polymer–peptide bond, both am-CRGDS and ac-CRGDS peptides were coupled to the coating and soaked in cell culture media (alpha minimum essential medium, αMEM) with 10% fetal bovine serum (FBS) for 1, 2, 7, or 14 days. FBS is a common additive to cell cultures and contains many proteins and other compounds.26 It should be noted that the atomic percent of nitrogen from the peptide and nitrogen from any nonspecifically adsorbed proteins was not distinguishable in the XPS. To evaluate the peptide concentration on the coating over time, the ratio of the carbon (C) to nitrogen (N) peak was quantified (see supplemental example S2). XPS analysis revealed a significant decrease in concentration of the ac-CRGDS peptide over a period of 2 days before leveling off (Figure 3a,b). Approximately 12% of the peptide remained on the surface at day 2. By contrast, the immobilized am-CRGDS was stable over the 14-day period. Differences in the presence of am-CRGDS and ac-CRGDS on the surface most likely result from differences in the polymer–peptide bond, namely thioester versus amide. Possible reasons for thioester lability include hydrolysis, displacement by primary amines in the cell culture media, or protease-mediated degradation. The reported half-life of a thioester bond in aqueous media is 2 h at pH 7.5, hence this bond was expected to be labile under cell culture conditions.27
Figure 3.
(A) Detected N atomic percent on coatings with either am-CRGDS or ac-CRGDS after soaking in cell culture media with 10% FBS. The mean and standard deviation are shown. (B) The calculated remaining peptides on the surface from the ratio of detected atomic percent of N shown in panel a and the atomic percent of carbon (C).
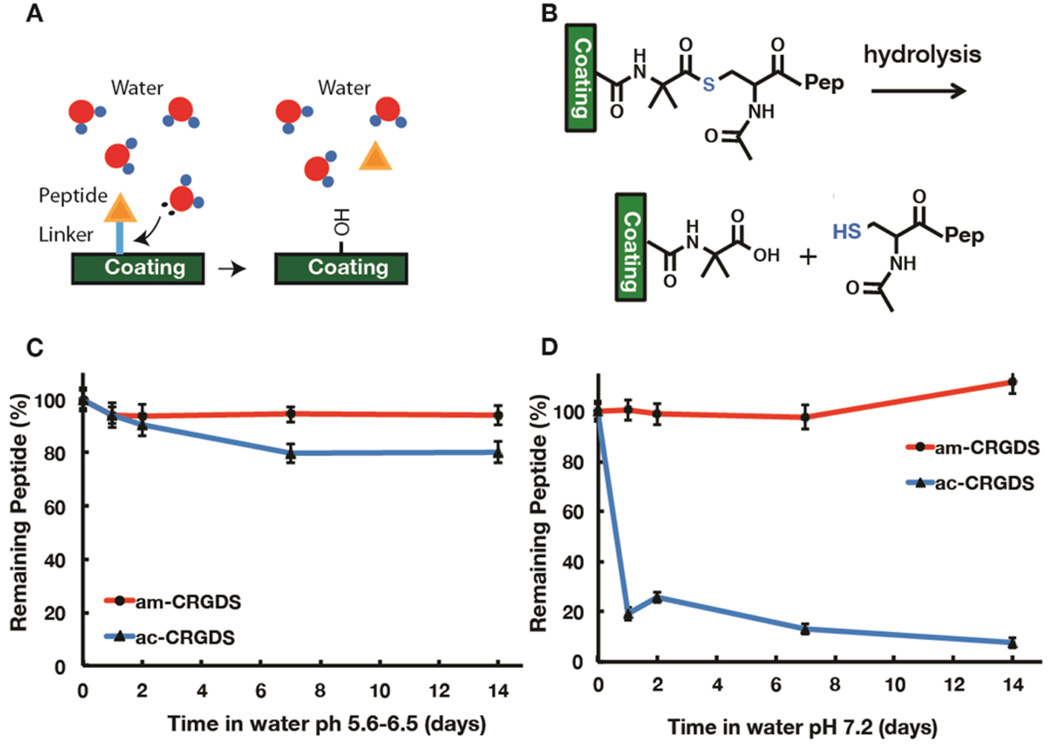
Hydrolysis assay
To probe whether hydrolysis (Figure 4a,b) was causing the lability of ac-CRGDS peptide, coatings were soaked under acidic conditions in water at pH 5.6–6.5 or under slightly basic conditions in water at pH 7.2 or in 0.1 M HEPES buffer at pH 7.2 for 2 weeks. The basic conditions mimic the pH in cell culture medium, typically pH 7.2–7.4. The atomic percent of N at various time points was determined by XPS to quantify the bound peptide. At lower pH (5.6–6.5) in DI water, hydrolysis played a minor role in the thioester linkage lability, as only 15% of the peptide was lost over 2 weeks (Figure 4c). However, upon raising the pH (Figure 4d) or using a HEPES buffer adjusted to pH 7.2 (Figure S3), loss of peptide was substantial (93%) after 2 weeks. Under all hydrolysis conditions tested, the am-CRGDS peptide–polymer linkage was stable over the entire two-week period.
Figure 4.
Schematic showing (A) the hydrolysis of peptide from the coating and (B) the chemistry of hydrolysis at the surface. (C) The amount of peptide bound to the surface after soaking in (C) DI water and (D) water at pH 7.2 as determined by XPS. Percentages were calculated from the N and C atomic percent detected on the coating for up to 14 days. One standard deviation is shown on each plot.
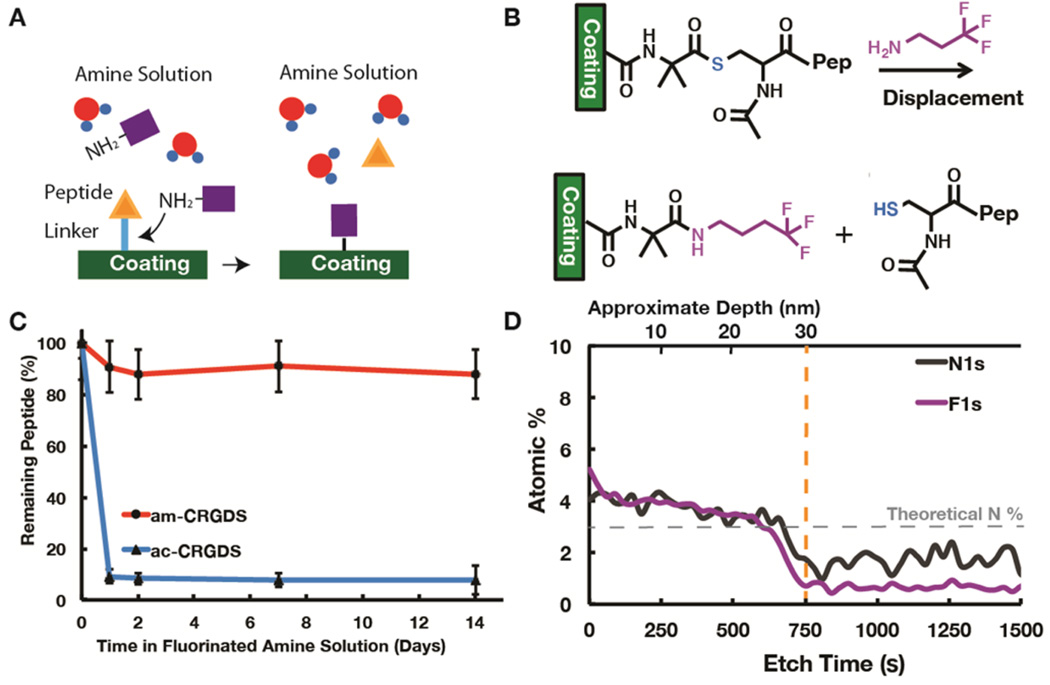
Amine Displacement
Displacement of the thioester bond by a primary amine was also a potential cause of peptide lability (Figure 5a). To recreate a similar amine concentration to cell culture media, but in a defined manner, the free amine content was first measured using a ninhydrin assay to give a primary amine concentration of 22.2 ± 4.1 mM. This method used an oxidative deamination reaction to change the yellow ninhydrin reagent to a purple color in the presence of primary amines (Figure S2).28 This assay takes into account proteins that contain many free amines per molecule, all of which cannot bind the coating at once, and may have a hydrodynamic radius too large to penetrate the cross-linked network. The αMEM contains many amino acids: in total 1.252 mg/mL or ~9.63 mM (assuming an average molecular weight of 130 Da for amino acids). The total protein contribution from FBS, at 10% concentration, was 3.2–3.7 mg/mL.26,29 We chose a conservative concentration of 10 mM small molecule amine 4,4,4-trifluorobutylamine (Fl-NH2) in water at pH 7.2 to mimic the cell culture media. Upon displacement of the thioester bond by F1-NH2, the fluorine (F) signal can be detected by XPS (Figure 5b). The F signal on the amine allows us to distinguish between hydrolysis and displacement by the amine using XPS. The Fl-NH2 caused a significant loss of the ac-CRGDS peptide within the first day of incubation, very similar to the loss in cell culture media (Figure 5c), as the N atomic percent decreased (from 7.57% to 3.48%) and the F atomic percent increased (from 0% to 5.05%) (Figure S4). Almost all of the peptide lost from the coating was replaced by the Fl-NH2, as evident by the increase in the F/N ratio to 1.47, which is close to the theoretical estimate of 1.51. This theoretical F/N ratio is the calculated ratio if all peptides are replaced by Fl-NH2 (calculated by eqs 2 and 3 in the Experimental Section). XPS depth profile (Figure 5d), confirms the displacement of ac-CRGDS through the entire thickness of the coating, although the surface reaction was more quantitative. Results indicate that only 7.9% of the bound ac-CRGDS remained on the surface of the coating after 14 days. From Figure 5 and Figure 4 we conclude that the lability of the thioester bond between the coating and ac-CRGDS in cell culture medium was due to amine displacement reaction by the amino acids in the medium.
Figure 5.
Schematic showing (A) the displacement of the ac-CRGDS peptide by a molecule with a free primary amine and (B) the displacement chemistry using the Fl-NH2 molecule. XPS data showing (C) percent of bound peptide remaining calculated using the N to C ratio in XPS and (D) depth profile on ac-CRGDS after soaking for 1 day in Fl-NH2 amine solution. The orange line shows the polymer–substrate interface, and the gray dotted line shows the theoretical N atomic percent that should be detected if all of the peptide was displaced from the surface. Error bars represent one standard deviation about the mean, where present.
Understanding CRGDS–Polymer Bond Stability
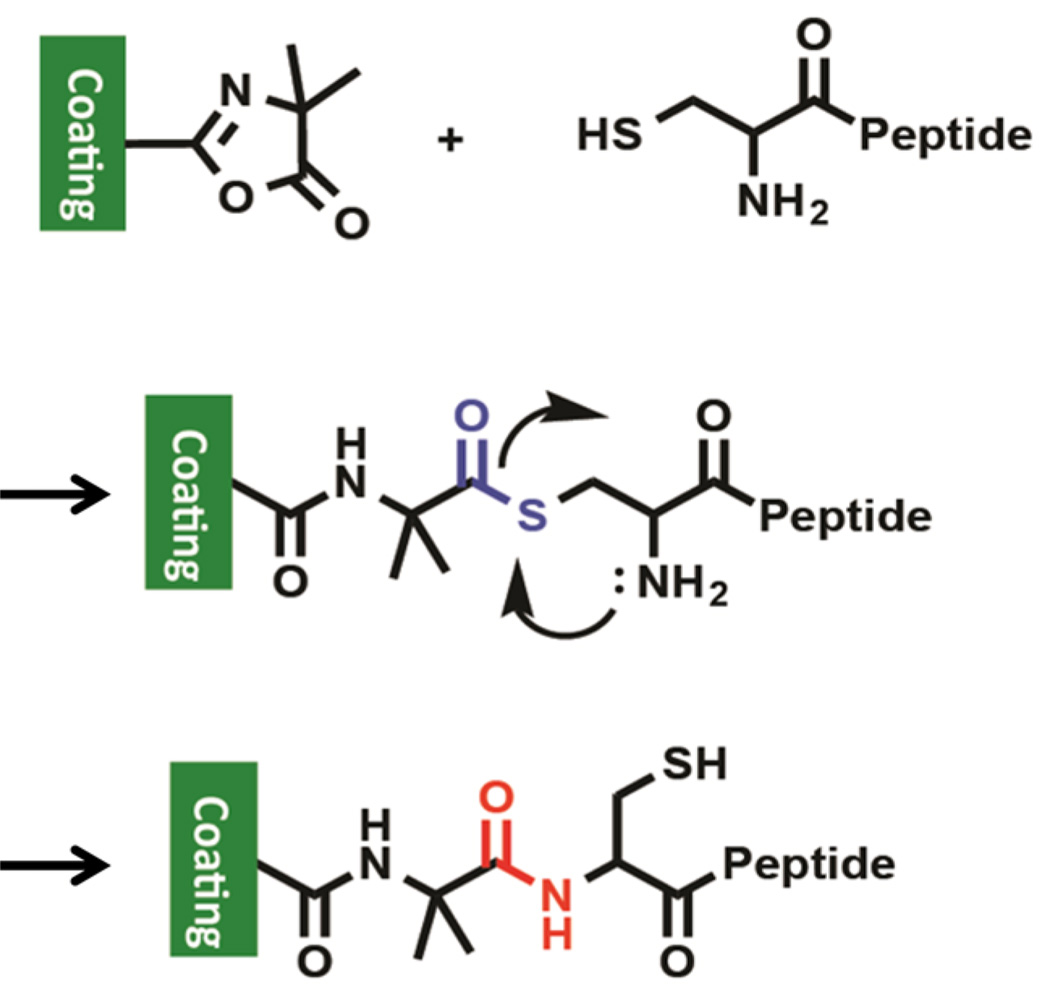
From these studies it is clear that under all test conditions, including cell culture medium, the am-CRGDS bond with the substrate was stable. Theoretically, the am-CRGDS peptide can react with the VDM in the coating through either the free –NH2 or –SH group on the terminal cysteine. The data show that ring opening of azlactone by –NH2 in the terminal cysteine was unlikely, as similar peptides (am-GRGDSP) with no free thiol do not react under these reaction conditions. Amine reactions were only quantitative in aqueous solution at high amine concentrations (1 M and above).30 At low concentrations (10 mM or less) using alkaline pH and 1.5 M sodium sulfate, less than 20% of azlactone groups reacted through the primary amine, depending on peptide sequence. Our studies show that it is likely that the N-terminal cysteine first forms an S-acyl covalent intermediate that undergoes an intramolecular 5 membered N-acyl migration to yield a hydrolytically stable amide bond (Figure 6). Similar intermediates that are formed during native chemical ligation processes have been exploited for the synthesis of complex peptides, macromolecules and hydrogels.22,31,32 Native chemical ligation is a well-known and useful tool, although no such rearrangements have been previously reported with azlactone groups. Typically, native chemical ligation is reported to occur in aqueous conditions between pH 7 and 8 at room temperature. Specifically, hydrogels using native chemical ligation show this rearrangement to occur within a 35 min to 2 h time frame in 0.1 M PBS at pH 7.6, depending on the concentration.22 Therefore, as our reaction peptide coupling (0.01 M PBS at pH 7.4) conditions are similar to native chemical ligation, intramolecular rearrangement is probable.
Figure 6.
Mechanism of amide bond formation between cysteine terminated peptides and azlactone groups in the coating at pH 7.4 in PBS. Here the free thiol first reacts with the azlactone ring to form a thioester bond, which is subsequently displaced by the neighboring amine to form a more stable amide bond.
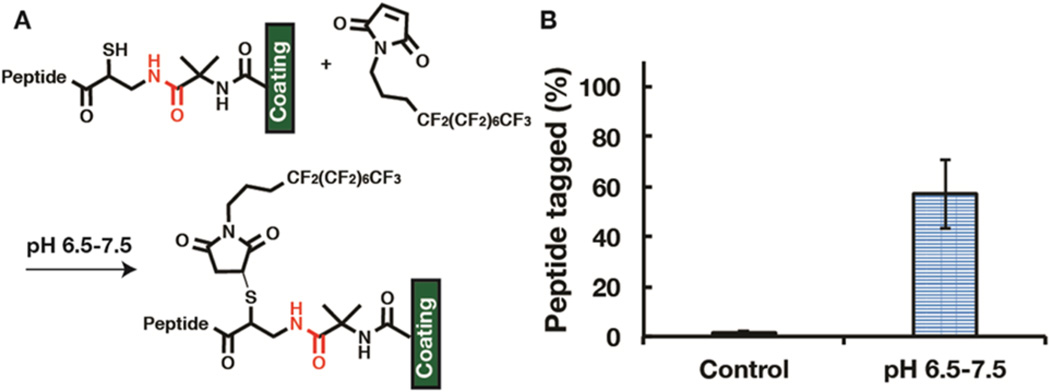
Since these reactions occur on a cross-linked polymer coating, 1H NMR cannot be used to determine whether the am-CRGDS was bound through an amide or thioester linkage to the azlactone. To provide evidence for amide bond formation, the am-CRGDS coupled surface was reacted with a heptadecafluoroundecyl maleimide (Fl-maleimide). Maleimides are highly selective to thiols between pH 6.5 and pH 7.5.13,33 If the am-CRGDS peptide was bound through an amide linkage, the peptide should have a free thiol group, and this free thiol group can react with the maleimide (Figure 7a). Figure 7b shows that 57% ± 13% of the peptide was tagged by the Fl-maleimide. This reaction was done immediately after the am-CRGDS peptide was coupled to the coating. A quantitative maleimide reaction was not achieved, potentially due to steric hindrance or suboptimal reaction conditions; however, the results showed that a large portion of the peptide was bound through amide bonds. This, in addition to am-CRGDS’s excellent stability in cell culture medium (essentially no loss), provides direct evidence for the amide bond formation.
Figure 7.
(A) Schematic showing Fl-maleimide reactivity with the free thiol under pH specific conditions. (B) XPS measurement showing the percent of am-CRGDS peptide on the surface that has been labeled by the Fl-maleimide. The control was the surface incubated in the reaction solution (1:1 dimethylformamide to PBS at pH 7.4). One standard deviation is shown.
This NCL based rearrangement is not limited to this particular copolymer studied. To show the generalizability of this reaction to any VDM containing copolymer, the two peptides were reacted with a thin coating of poly(vinyl azlactone-ran-glycidyl methacrylate) P(VDM-r-GMA), which does not contain the PEG comonomer. Similar loss of the ac-CRGDS peptide and stability of the am-CRGDS peptides was observed (Figure S5).
Use as Culture Substrate for hMSCs
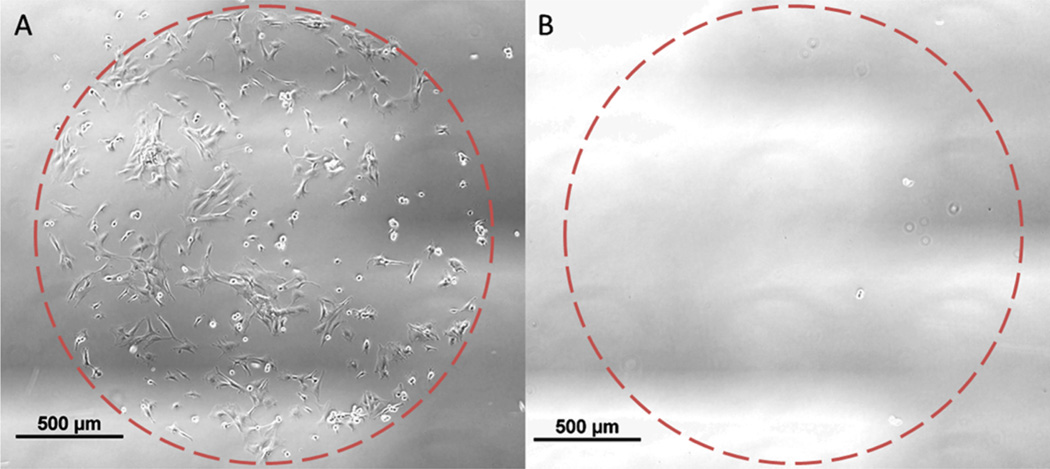
These coatings do not support hMSC attachment without an adhesive peptide on the surface, as they contain a large amount of PEG, which is bioinert. hMSC survival in vitro requires receptor-mediated adhesion to surface peptides or proteins.14 Human mesenchymal stem cells (hMSCs) were cultured on substrates coupled with either ac-CRGDS or am-CRGDS. At 5 h after seeding, attachment of hMSCs was visible on spots coupled with am-CRGDS but not on ac-CRGDS (Figure 8a,b). Cells remained attached to the am-CRGDS spots over the 12-h time-lapse period after which the experiment was stopped. Cells seeded onto coatings with ac-CRGDS spots did not attach or spread after 4 h and further incubation out to 12 h failed to result in any cell attachment (Figure S6).
Figure 8.
Representative phase images of hMSC culture on the thin film coating functionalized with 2 mm spots of (A) am-CRGDS and (B) ac-CRGDS, respectively. Images taken 5 h after hMSC seeding.
Further studies with XPS explained the lack of cell adhesion on ac-CRGDS peptide spots. For this study, coatings were soaked in cell culture media in the absence of cells and XPS time points were taken every few hours. Within the first 4 h of soaking, at least 40% of the initial peptide was removed from the coating (Figure S7). The displaced ac-CRGDS was released into the culture media, where it can act as a soluble adhesion ligand, which further impaired hMSC attachment to the coating by occupying integrin binding sites on the cell surface. This simple comparison of amide and thioester linkages underscores the importance of forming stable peptide–polymer bonds in the design of cell culture surfaces.
CONCLUSIONS
In summary, we have shown that cysteine terminated peptides (am-CRGDS and ac-CRGDS) can be reacted with azlactone groups in a cross-linked polymer coating with high efficiency through the formation of a thioester bond. However, unlike am-CRGDS, thioester linkage formed by ac-CRGDS proved to be labile in cell culture media, detaching from the polymer coating within 1 to 2 days. Further investigations revealed that hydrolysis under cell culture conditions as well as displacement by other amines were two main factors for the lability of thioester bond. The lability of the thioester bond can be advantageous for creating dynamic surfaces, which present certain peptides for short timeframes. Unexpectedly, the peptide am-CRGDS, which has a free amine and thiol group in the Cys terminal, first forms an S-acyl covalent intermediate that undergoes an intramolecular 5 membered N-acyl migration to yield a hydrolytically stable amide bond. This conclusion is supported by control experiments where under similar conditions the N-terminal of am-GRGDSP does not react with azlactone in the coating. For a primary amine to react with the azlactone, higher pH, the presence of salt, and higher concentrations of peptides or organic solvents were required. Additionally, the free –SH group on the surface bound am-CRGDS peptide was tagged with a fluorinated maleimide providing evidence for an amide linkage. From stability studies, the am-CRGDS peptide was stable under all conditions tested, unlike the ac-CRGDS peptide bound by thioester bond. Hence, with cysteine-terminated peptides, the formation of the S-acyl intermediate lowers the energetic barrier for the formation of the amide bond through the intramolecular N-acyl migration. This process is reminiscent of native chemical ligation in protein synthesis, but is utilized here to form a stable amide bond from the otherwise less reactive N-terminal of a peptide in aqueous solution at low concentrations (1 mM). Formation of stable amide bonds in aqueous solutions at low concentrations has been a challenge as common chemistries such as N-hydroxysuccinimide (NHS) esters are susceptible to hydrolysis, resulting in variable efficiencies. The work lays out an efficient pathway for the formation of a stable amide bond with the advantages of a selective reaction, high reaction efficiency, short reaction time (1 h), low peptide concentration (1 mM) and use of aqueous solvent. The high conjugation efficiency and excellent stability under cell culture conditions makes these polymer–peptide coatings potentially useful for longer-term stem cell culture experiments (e.g., proliferation, differentiation, expansion) where a stable and chemically defined surface is desired.
Supplementary Material
Acknowledgments
This research was funded by the National Science Foundation (NSF DMR 1306482). The authors acknowledge support from staff and the use of equipment at the Materials Science Center at UW-Madison (NSF DMR-1121288). John D. Krutty acknowledges the Holton Fellowship from the University of Wisconsin-Madison and the Holton family. Angela W. Xie acknowledges the National Institutes of Health (Biotechnology Training Program NIGMS 5 T32-GM08349) and the National Science Foundation (DGE-1256259).
ABBREVIATIONS
- NCL
native chemical ligation
- CRGDS
Cys-Arg-Gly-Asp-Ser peptide
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Complementary data, equations, and sample calculations referred to in the text (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
REFERENCES
- 1.Lanniel M, Huq E, Allen S, Buttery L, Williams PM, Alexander MR. Soft Matter. 2011;7(14):6501–6514. [Google Scholar]
- 2.Watt FM, Huck WTS. Nat. Rev. Mol. Cell Biol. 2013;14(8):467–473. doi: 10.1038/nrm3620. [DOI] [PubMed] [Google Scholar]
- 3.Kolind K, Leong KW, Besenbacher F, Foss M. Biomaterials. 2012;33(28):6626–6633. doi: 10.1016/j.biomaterials.2012.05.070. [DOI] [PubMed] [Google Scholar]
- 4.Fossett E, Khan WS. Stem Cells Int. 2012;2012(5):465259. doi: 10.1155/2012/465259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irvine DJ, Mayes AM, Griffith LG. Biomacromolecules. 2001;2(1):85–94. doi: 10.1021/bm005584b. [DOI] [PubMed] [Google Scholar]
- 6.DeForest CA, Anseth KS. Annu. Rev. Chem. Biomol. Eng. 2012;3:421–444. doi: 10.1146/annurev-chembioeng-062011-080945. [DOI] [PubMed] [Google Scholar]
- 7.Villa-Diaz LG, Nandivada H, Ding J, Nogueira-de-Souza NC, Krebsbach PH, O’Shea KS, Lahann J, Smith GD. Nat. Biotechnol. 2010;28(6):581–583. doi: 10.1038/nbt.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koepsel JT, Murphy WL. ChemBioChem. 2012;13(12):1717–1724. doi: 10.1002/cbic.201200226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ameringer T, Fransen P, Bean P, Johnson G, Pereira S, Evans RA, Thissen H, Meagher L. J. Biomed. Mater. Res., Part A. 2012;100A(2):370–379. doi: 10.1002/jbm.a.33194. [DOI] [PubMed] [Google Scholar]
- 10.Alconcel SNS, Baas AS, Maynard HD. Polym. Chem. 2011;2(7):1442–1448. [Google Scholar]
- 11.Surman F, Riedel T, Bruns M, Kostina NY, Sedláková Z, Rodriguez-Emmenegger C. Macromol. Biosci. 2015;15:636–646. doi: 10.1002/mabi.201400470. [DOI] [PubMed] [Google Scholar]
- 12.Ruoslahti E. Annu. Rev. Cell Dev. Biol. 1996;12(1):697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 13.Hermanson GT. Bioconjugate Techniques. San Diego, CA: Academic Press; 2013. [Google Scholar]
- 14.Schmitt SK, Xie AW, Ghassemi RM, Trebatoski DJ, Murphy WL, Gopalan P. Adv. Healthcare Mater. 2015;4:1555–1564. doi: 10.1002/adhm.201500191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt SK, Murphy WL, Gopalan P. J. Mater. Chem. B. 2013;1(9):1349–1360. doi: 10.1039/c2tb00253a. [DOI] [PubMed] [Google Scholar]
- 16.Heilmann SM, Rasmussen JK, Krepski LR. J. Polym. Sci., Part A: Polym. Chem. 2001;39(21):3655–3677. [Google Scholar]
- 17.Ho HT, Levere ME, Fournier D, Montembault V, Pascual S, Fontaine L. Aust. J. Chem. 2012;65(8):970–977. [Google Scholar]
- 18.Zdyrko B, Klep V, Luzinov I. Langmuir. 2003;19(24):10179–10187. [Google Scholar]
- 19.Sweat DP, Kim M, Yu X, Gopalan P. Langmuir. 2013;29(11):3805–3812. doi: 10.1021/la305060z. [DOI] [PubMed] [Google Scholar]
- 20.Dawson P, Muir T, Clark-Lewis I, Kent S. Science. 1994;266(5186):776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 21.Malins LR, Payne RJ. Curr. Opin. Chem. Biol. 2014;22:70–78. doi: 10.1016/j.cbpa.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 22.Hu B-H, Su J, Messersmith PB. Biomacromolecules. 2009;10(8):2194–2200. doi: 10.1021/bm900366e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boere KWM, Soliman BG, Rijkers DTS, Hennink WE, Vermonden T. Macromolecules. 2014;47(7):2430–2438. [Google Scholar]
- 24.Hruby VJ. Life Sci. 1982;31(3):189–199. doi: 10.1016/0024-3205(82)90578-1. [DOI] [PubMed] [Google Scholar]
- 25.Mas-Moruno C, Rechenmacher F, Kessler H. Anti-Cancer Agents Med. Chem. 2010;10(10):753–768. doi: 10.2174/187152010794728639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gstraunthaler G. ALTEX. 2003;4:275–281. [PubMed] [Google Scholar]
- 27.Thapa P, Zhang RY, Menon V, Bingham JP. Molecules. 2014;19(9):14461. doi: 10.3390/molecules190914461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abernathy DG, Spedding G, Starcher B. J. Inst. Brew. 2009;115(2):122–127. [Google Scholar]
- 29.Price P, Gregory E. In Vitro. 1982;18(6):576–584. doi: 10.1007/BF02810081. [DOI] [PubMed] [Google Scholar]
- 30.Coleman PL, Walker MM, Milbrath DS, Stauffer DM, Rasmussen JK, Krepski LR, Heilmann SM. J. Chromatogr. A. 1990;512(0):345–363. [Google Scholar]
- 31.Byun E, Kim J, Kang SM, Lee H, Bang D, Lee H. Bioconjugate Chem. 2011;22(1):4–8. doi: 10.1021/bc100285p. [DOI] [PubMed] [Google Scholar]
- 32.Khan S, Sur S, Dankers PYW, da Silva RMP, Boekhoven J, Poor TA, Stupp SI. Bioconjugate Chem. 2014;25(4):707–717. doi: 10.1021/bc400507v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brewer CF, Riehm JP. Anal. Biochem. 1967;18(2):248–255. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.