Introduction
Japanese cedar (JC) pollen is one of the most frequently inhaled outdoor allergens in Japan and a potent inducer of seasonal allergic asthma and rhinitis. Epidemiological studies of JC pollinosis have reported a more than 2.5-fold increase in JC allergy over the last 3 decades1. In recent years, different surveys conducted to estimate the prevalence of JC pollinosis have reported a prevalence ranging from 13 to 36% in the Japanese population, making it a major national health problem in Japan2, 3. This striking increase is believed to be associated with environmental factors such as reforestation and climate change. The first studies describing JC pollinosis were published in the1960s4, 5, and disease prevalence has been increasing ever since. Retrospective studies have associated this trend with the expansive afforestation era in Japan conducted from the 1950s to 1970s. During this period, billions of Japanese cedar trees (Cryptomeria japonica) were planted as a future source of construction material. However, decreasing wood tariffs and imported wood diminished the need for JC as construction material resulting in massive amounts of land covered by forests made up entirely of Japanese cedar trees at an optimum age for pollen production6, 7. In addition, climate change is promoting flower bud development and increasing pollen production7, 8.
To date, two major allergens in Japanese cedar pollen have been described; Cry j 1 and Cry j 29, 10. Both allergens are highly dominant, eliciting IgE reactivity in > 90% of JC-allergic patients tested11. In addition to Cry j 1 and 2, a number of reports have been published on other JC-derived allergenic proteins, including Cry j 3, a theumatin-like protein12, 13, Cry j 4, a calcium-biding protein14, Cry j isoflavone reductase (IFR), which is related to the minor birch allergen Bet v 6 (62 % sequence identity)15, Cry j chitinase, a class IV chitinase16, 17, Cry j Asp, an aspartic protease18, Cry j LTP, a lipid transfer protein19 and Cry j CPA9, a serine protease20. Despite this relative abundance of allergenic proteins in Japanese cedar, only Cry j 1 and 2 are considered major allergens and as a result have been the focus of more detailed studies. The T cell response to both Cry j 1 and 2 has been thoroughly characterized in Japanese patients and several immune-dominant regions have been identified21–23.
Although considerable efforts have been made to improve our understanding of the immunological processes involved in JC pollinosis, the vast majority of these studies have been performed in cohorts of JC pollen allergic, Japanese patients. However, given the degree of potential cross-reactivity of JC pollen with other tree pollens such as Italian cypress24, studies in non-Japanese subjects who have IgE reactivity against JC pollen are of great interest. Furthermore, studies on differences between immune recognition and responses of non-sensitized and sensitized donors in the JC system are extremely rare. Several reports suggest that non-sensitized individuals mount an immune response distinct from those of allergic individuals and are not oblivious to antigen stimulation25–27. Thus defining the features and targets of healthy individuals is important and understudied, and could provide insights for the development of safer and more efficacious immunotherapeutic approaches.
In this report, we aim to evaluate the effect of JC pollen exposure on the T cell response specificity and magnitude in allergic and non-allergic individuals. Studies measuring Japanese cedar pollen in Japan over a span of 10 years or more have reported detectable levels from November until April, with levels varying from 1–20 pollen grains/cm2 per months, accumulating 500–12000 pollen grains/cm2 over the entire season28, 29. In contrast, Japanese cedar and other related and cross-reactive species are not indigenous in Southern California and are hence not considered a major allergen. However, grasses (e.g. Cynodon dactylon, Lolium perenne, Phleum pratense) and ragweed (Ambroisa artemisiifolia) pollen do grow in Southern California and levels ranging from 4–50 pollen grains/cm2 per months can be detected throughout the respective season (levels determined by Erik and Ese Banck Clinical Research Center, San Diego, as reported by the American Academy of Allergy, Asthma and Immunology).
Here, we study the T cell response to three JC allergens in allergic and non-allergic Japanese patients, presumed to be exposed, versus Southern Californian patients who have never been to Japan to evaluate how exposure and sensitization influence the response magnitude and epitope-specificity of allergen-specific T cells, focusing the Cry j 1, 2 and IFR as a representative minor allergen.
Methods
Study population
Blood donations were collected (450 ml) from Japanese cedar-allergic and non-allergic patients. Each donor was recruited following institutional review board approval (IRB number VD-112). Written informed consent was for study participation was obtained. At the time of donation, information regarding whether prospective donors had lived in Japan for at least 1 year was also collected and donors were classified as “lived in Japan” (LIJ or “not lived in Japan” (NLJ). Serum samples were tested for JC pollen (Cry j)-specific IgE by the ImmunoCAP using natural JC pollen extract (Phadia, Upsala, Sweden), and donors were classified as JC-sensitized or not (Cry j IgE+/ Cry j IgE−) based on a positivity threshold of >0.35 kUA/L. Based on the criteria described above the study population was divided in the following three sub-cohorts (Table 1): 1) LIJ IgE+ (n=10); 2) NLJ IgE+ (n=24) and 3) LIJ IgE− (n=20).
Table 1.
A summary of epidemiological and clinical donor characteristics.
| Donor | Cohort | Age | Gender | Cry j IgE (kU/L) |
Time lived in Japan |
Year | Location |
|---|---|---|---|---|---|---|---|
| 1569 | LIJ IgE+ | 40 | F | 2.71 | 9 years | n.d. | Yamanashi, Kofu |
| 2098 | LIJ IgE+ | 20 | F | 8.76 | 7 years | 2002 | Nagoya, Archi Preetune |
| 2102 | LIJ IgE+ | 24 | F | 14.08 | 17.5 years | 2009 | Tokyo |
| 2144 | LIJ IgE+ | 45 | F | 0.73 | 40 years | 2015 | Kobay |
| 2188 | LIJ IgE+ | 40 | F | 5.45 | 2.5 years | 2003 | sasebo |
| 2199 | LIJ IgE+ | 25 | F | 4.99 | 15 years | 2005 | Tokyo |
| 2212 | LIJ IgE+ | 32 | M | 17.62 | 13 years | 2015 | Kamamoto, Tokyo |
| 2220 | LIJ IgE+ | 21 | M | 13.42 | 22 years | 2013 | Kanagawa, Tokyo |
| 2254 | LIJ IgE+ | 31 | F | 6.45 | 28 years | 2014 | Inago?, Tokyo |
| 2258 | LIJ IgE+ | 25 | F | 8.46 | 24 years | 2014 | Tokyo |
| 1005 | NLJ IgE+ |
31 | M | 0.59 | N/A | N/A | N/A |
| 1006 | NLJ IgE+ |
39 | M | 0.67 | N/A | N/A | N/A |
| 1014 | NLJ IgE+ |
58 | M | 3.64 | N/A | N/A | N/A |
| 1020 | NLJ IgE+ |
32 | F | 0.61 | N/A | N/A | N/A |
| 1174 | NLJ IgE+ |
50 | M | 0.55 | N/A | N/A | N/A |
| 1175 | NLJ IgE+ |
38 | F | 3.39 | N/A | N/A | N/A |
| 1191 | NLJ IgE+ |
41 | F | 0.6 | N/A | N/A | N/A |
| 1198 | NLJ IgE+ |
23 | M | 3.49 | N/A | N/A | N/A |
| 1363 | NLJ IgE+ |
37 | M | 0.89 | N/A | N/A | N/A |
| 1381 | NLJ IgE+ |
56 | M | 2.2 | N/A | N/A | N/A |
| 1435 | NLJ IgE+ |
36 | M | 0.69 | N/A | N/A | N/A |
| 1437 | NLJ IgE+ |
34 | F | 1.89 | N/A | N/A | N/A |
| 1440 | NLJ IgE+ |
25 | M | 0.49 | N/A | N/A | N/A |
| 1441 | NLJ IgE+ |
43 | M | 0.41 | N/A | N/A | N/A |
| 1453 | NLJ IgE+ |
20 | M | 0.98 | N/A | N/A | N/A |
| 1460 | NLJ IgE+ |
30 | M | 1.69 | N/A | N/A | N/A |
| 1652 | IgE+ NLJ |
56 | F | 1.31 | N/A | N/A | N/A |
| 1864 | IgE+ NLJ |
34 | F | 9.01 | N/A | N/A | N/A |
| 2017 | IgE+ NLJ |
21 | M | 1.28 | N/A | N/A | N/A |
| 2021 | IgE+ NLJ |
18 | F | 1.93 | N/A | N/A | N/A |
| 2036 | IgE+ NLJ |
31 | M | 1.38 | N/A | N/A | N/A |
| 2201 | IgE+ NLJ |
20 | M | 0.59 | N/A | N/A | N/A |
| 2240 | IgE+ | 27 | F | 1.84 | N/A | N/A | N/A |
| 1553 | LIJ IgE− | 23 | F | 0 | 7 years | 1999 | n.d. |
| 1578 | LIJ IgE− | 56 | F | 0.03 | n.d. | n.d. | n.d. |
| 1591 | LIJ IgE− | 21 | F | 0.28 | 6 years | n.d. | Hiroshima |
| 1616 | LIJ IgE− | 41 | M | 0 | 5 years | 1996 | yokasuka, Atsugi |
| 1617 | LIJ IgE− | 30 | F | 0.02 | 4.5 years | 2014 | Kyoto |
| 2095 | LIJ IgE− | 26 | F | 0 | 3 years | 2014 | Yogi |
| 2145 | LIJ IgE− | 52 | M | 0.13 | 3 years | 2014 | Kabay |
| 2186 | LIJ IgE− | 51 | F | 0.01 | 28 years | 1993 | Tokyo |
| 2187 | LIJ IgE− | 45 | M | 0 | 2 years | 1992 | Tokyo, Yokohima |
| 2221 | LIJ IgE− | 56 | M | 0 | 1.5 years | 1982 | Okanawa |
| 1599 | LIJ IgE− | 33 | M | 0.05 | 32 years | 2013 | Hyogo, Tokyo |
| 1626 | LIJ IgE− | 23 | M | 0.03 | 5.5 years | 2007 | Okanawa |
| 1632 | LIJ IgE− | 54 | M | 0.07 | 9 years | n.d. | Hiroshima |
| 1635 | LIJ IgE− | 41 | F | 0.06 | 26 years | 1999 | Hiroshima |
| 1694 | LIJ IgE− | 32 | F | 0 | N/A | n.d. | N/A |
| 2185 | LIJ IgE− | 31 | F | 0.01 | 5 years | 1989 | Okanela |
| 2198 | LIJ IgE− | 29 | F | 0 | 23 years | 2009 | Tokyo |
| 2234 | LIJ IgE− | 18 | F | 0.02 | 10 months |
2000 | Nishinomiya |
| 2237 | LIJ IgE− | 20 | F | 0.01 | 20 years | 2015 | Tokyo |
| 2239 | LIJ IgE− | 29 | F | 0 | 1 year | 2013 | Kyoto |
Polysensitization determination by ImmunoCAP
In addition to Japanese cedar, plasma samples from all donors were additionally tested by ImmunoCAP (Phadia, Upsala, Sweden) for IgE titers against a panel of 13 natural extracts made from allergenic species including tree pollen (Italian cypress (Cup s), Mountain cedar (Jun s), Red cedar (Jun v) Birch (Bet v), Oak (Que e), Olive (Ole e) and White Ash (Fra a)), grasses (Timothy (Phl p), Bermuda (Cyn d), Sweet vernal (Ant o), Kentucky blue (Poa p) and Rye (Lol p)) and weed (Ragweed (Amb a)) species.
Selection of peptides from Japanese cedar allergen sequences
Sequences of all known Cry j 1 and Cry j 2 isoforms were obtained from the WHO/IUIS Allergen Nomenclature database (www.allergen.org)30. In addition, the sequence from Cry j IFR was obtained from the NCBI protein database (Accession number AAK27264.1). Sets of peptides of 16 amino acids in length, overlapping by 8 residues, were generated to cover all three allergen protein sequences. Peptides from isoforms were synthesized as well as any other peptides appearing in at least 3 (25%) isoforms. Overall, a total of 148 peptides were assembled in 15 pools of ~10 peptides each, and screened for their T cell reactivity.
Peptide synthesis
Peptides were purchased from A and A (San Diego, CA) as crude material on a small (1 mg, >70% purity) scale. Individual peptides were resuspended in DMSO at a final concentration of 40 mg/ml.
Stimulation and expansion of Japanese cedar-specific T cells
PBMCs were isolated from whole blood by density gradient centrifugation according to manufacturers’ instructions (Ficoll-Hypaque, Amersham Biosciences, Uppsala, Sweden) and cryopreserved. For in vitro expansion of JC-specific T cells, PBMCs were thawed and stimulated with JC pollen extract (ALK-Abello A/S, Horsholm, Denmark) at 2 µg/ml. Cells were cultured in RPMI 1640 supplemented with 5% human AB serum in 24 well plates (BD Biosciences, San Diego, CA) at a density of 2 × 106/ml and incubated at 37 °C. IL-2 (10 U/ml) was added every 3 days after initial stimulation. Cells were harvested on day 14 and screened for reactivity to peptide pools by ELISPOT.
Dual ELISPOT assays
The production of IFNγ and IL-5 from cultured PBMCs in response to antigenic stimulation was assessed by dual ELISPOT assays as described previously31.
Peptide pools that elicited a positive response (>100 spot forming cells/ 1× 106 PBMC) were deconvoluted to identify the individual epitopes at day 17 using cells from the same original culture.
Statistical correlation to determine hierarchy of T cell reactivity
To compare the T cell epitope repertoire in each cohort for each of the 3 Cry j allergens, average responses to each peptide from all three cohorts were compared by Spearman’s rank correlation analysis, a two-tailed, non-parametric measure of rank correlation. The R2 value was calculated from the Spearman r value. P-values <0.05 are considered significant.
HLA typing and restriction
HLA typing for Class I (HLA-A; HLA-B; HLA-C) and Class II (HLA-DQA1; HLA-DQB1, HLA-DRB1,3,4,5; HLA-DPB1) was performed by an ASHI-accredited (American society for histocompatibility and immunogenetics) laboratory at Murdoch University (Western Australia) as previously described32. Potential HLA-epitope restriction odds ratios and relative frequencies were calculated using the RATE program33.
Results
Higher JC pollen-specific IgE titers and different polysensitization patterns in allergic patients who have lived in Japan (LIJ IgE+) compared to allergic patients who have not (NLJ IgE+)
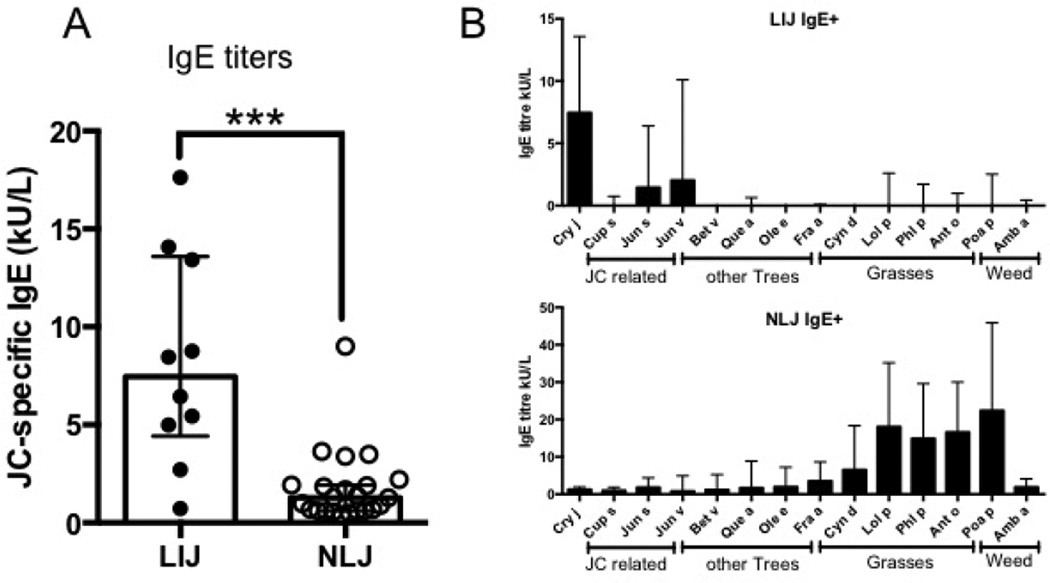
JC pollen-specific IgE titers in LIJ IgE+ and NLJ IgE+ patients were determined by ImmunoCAP (Figure 1A). As expected on the basis of the likely exposure history, titers were significantly higher in the LIJ IgE+ cohort (median 7.46, range 0.73–17.62 kU/L) compared to the NLJ IgE+ cohort (median 1.28, range 0.41–9.01 kU/L).
Figure 1.
Bar graphs depicting the median of A) Japanese cedar-specific IgE titers (kUA/L) in allergic patients who lived in Japan (LIJ) and allergic patients who did not (NLJ) and B) IgE titers to a panel of 13 pan pollen allergens in LIJ patients (upper panel) and NLJ patients (lower panel). Error bars indicate interquartile range. Statistical analysis was performed by Mann-Whitney test (non-parametric, two-tailed), ***- p <0.001. LIJ cohort: N=10; NLJ cohort: N=24
Next, we analyzed the level of polysensitzation to other tree, grass and weed pollens. For this purpose, IgE titers from the LIJ IgE+ and NLJ IgE+ to a panel of 13 allergenic species were assessed (Figure 1B). The test panel was grouped into 4 categories: 1. JC-related tree pollens; 2. other tree pollens; 3. grass pollens and 4. weed pollen. Interestingly, in the LJI IgE+ cohort, highest IgE titers were observed to Cry j (median 7.5 kU/L). Other sensitizations in this cohort were effectively limited to JC-related tree species, namely red cedar (Jun v, median titer 2.03 kU/L) and mountain cedar (Jun s, median titer 1.46 kU/L) (Figure 1B). In contrast, in the NLJ IgE+ cohort, IgE titers measured were mostly targeted towards grasses and weeds. As described above, JC pollen (Cry j) IgE titers were much lower compared to the LIJ patients (median 1.28 kU/L). Similar titers were observed for JC-related trees and other tree pollen (Figure 1B). The highest titers were observed for Kentucky blue (Poa a) and Ryegrass (Lol p) (22.4 and 18.6 kU/L, respectively). These differences in IgE titers to a panel of common pan-pollen allergens suggest fundamental differences in the exposure and origin of sensitization.
JC pollen-specific T cell responses are significantly higher in allergic patients who have lived in Japan (LIJ IgE+) compared to allergic patients who have not (NLJ IgE+)
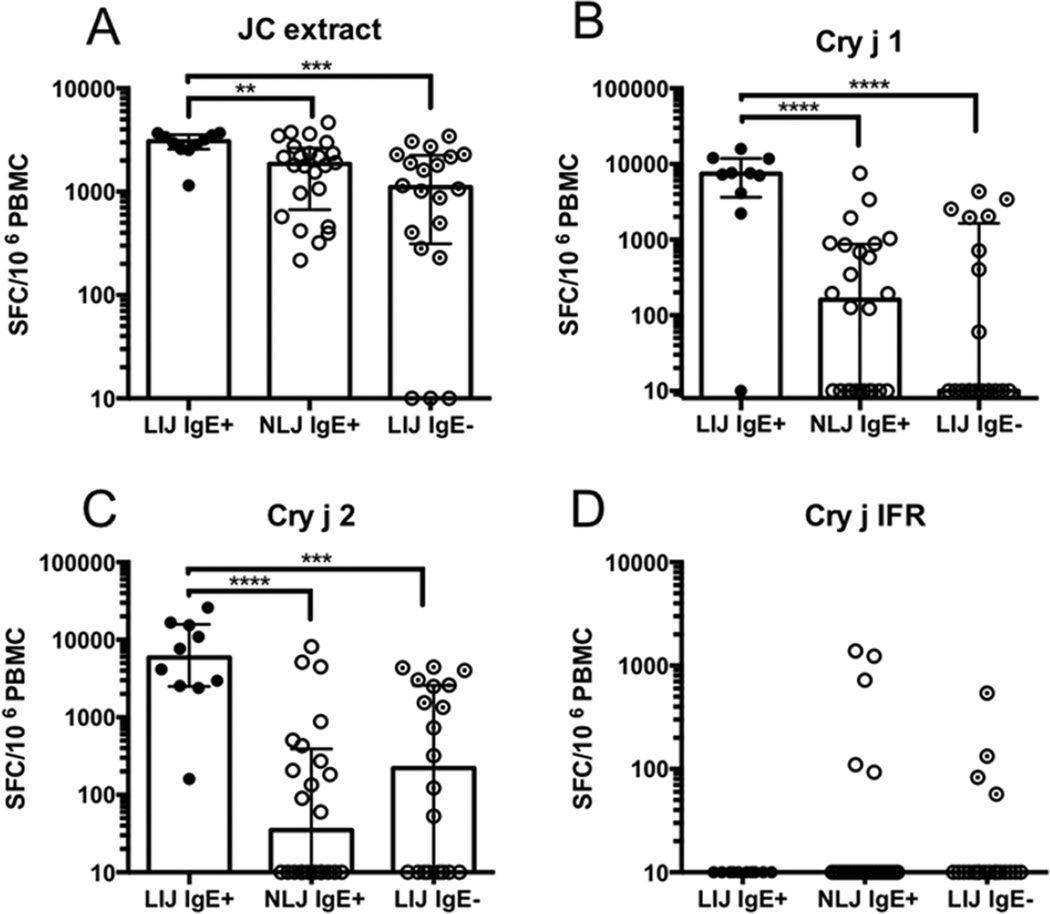
We then determined T cell reactivity (expressed as the sum of IL-5 and IFNg producing cells) to JC extract, and the Cry j 1, 2 and Cry j IFR allergens in all 3 cohorts (Figure 2). Panels of 16-mer peptides, overlapping by 8 residues and spanning the Cry j allergen sequences, were generated and screened in pools of ~10 for IFNγ and IL-5 produced by in vitro JC pollen extract expanded PBMC in ELISPOT assays. Positive pools were deconvoluted to identify individual epitopes. For each allergen, overall T cell reactivity is expressed as the sum of individual peptide responses observed in each donor (Figure 2). Responses to medium and PHA stimulation are shown in Supplemental Figure 1.
Figure 2.
Bar graphs depicting median values of total allergen-specific T cell responses (sum of IL-5 and IFNg responses to extract or individual peptides) to A) JC extract, B) Cry j 1, C) Cry j 2 and D) Cry j IFR in all 3 cohorts tested. Each symbol represents a single donor. Error bars indicate interquartile range. Statistical analysis was performed by Mann-Whitney test (non-parametric, two-tailed), **- p<0.01, ***- p <0.001, ****- p <0.0001. LIJ IgE+ cohort: N=10; NLJ IgE+ cohort: N=24; LIJ IgE− cohort: N=20
In the case of the JC pollen extract, responses were highest in the LJI IgE+ cohort, with all patients responding with a median magnitude of 3080 SFC (Figure 2A). In the NLJ IgE+ cohort extract also elicited detectable T cell reactivity in all patients tested with a median response magnitude of 1857 SFC (Figure 2A). As expected, in the non-allergic LIJ cohort (LJI IgE−), T cell response magnitudes and frequencies to JC pollen extract were lower than in the NLJ IgE+ and LJI IgE+ cohorts, eliciting positive responses in 17/20 (85%) donors with a median response of 1103 SFC (Figure 2A).
Similar to extract T cell responses, reactivity to allergen-derived peptides was also high. For Cry j 1 (Figure 2B), 90% of the LJI IgE+ cohort donors responded, with a median response across the entire cohort of 7430 SFC. In the NLJ IgE+ cohort, T cell responses were significantly lower. Responses were detected in 14/24 donors (58%) with a median magnitude of 160 SFC (Figure 2B). In the LIJ IgE− cohort, 8/20 (40%) patients responded to Cry j 1 with magnitudes ranging from 60–3411 SFC (Figure 2B).
For Cry j 2, 100% of LJI IgE+ donors responded with a median response of 5899 SFC (Figure 2C). In the NLJ IgE+ cohort, 12/24 (50%) patients responded with a median magnitude of 35 SFC. Surprisingly, 12/20 (60%) of LJI IgE− donors responded to Cry j 2 with a median magnitude of 222 SFC (Figure 2C). Finally, no T cell responses against Cry j IFR were detected in the LJI IgE+ cohort (Figure 2D). T cell responses were observed in only 5/24 (21%) of NLJ IgE+ donors with magnitudes ranging from 93 to 1383 SFC, whereas reactivity was detected in 4/20 (20%) of the LIJ IgE− donors (magnitudes ranged from 57–543 SFC) (Figure 2D).
T cell epitope recognition and immunodominance in the 3 donor cohorts
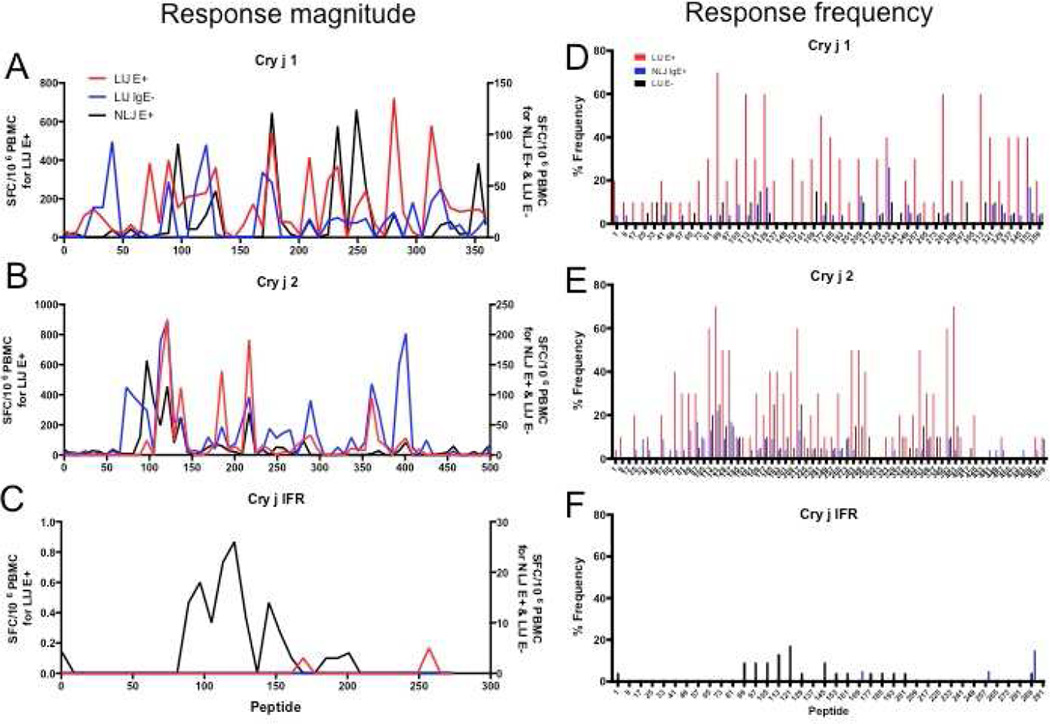
Next, we further analyzed individual T cell epitopes from the three allergens for each donor cohort. Figure 3 A–C shows the magnitude of recognition of the three Cry j allergens and Figure 3D–F shows the corresponding frequencies. Due to the differences in response magnitudes between cohorts, data in Figure 3 A–C is plotted using two different scales (the LIJ IgE+ cohort is plotted according to the scale shown on the left axis, while data from the NLJ IgE+ and LIJ IgE− cohorts are plotted according to the scale shown on the right axis) to facilitate the visualization of T cell dominance.
Figure 3.
Average epitope-specific T cell responses (A–C) (sum of IL-5 and IFNg) and response frequency (D–F) for each individual peptide spanning A and D) Cry j 1, B and E) Cry j 2 and C and F) Cry j IFR in LIJ IgE+, NLJ IgE+ and LIJ IgE− donors. Due to the differences in response magnitudes between cohorts, data in panels A–C is plotted using two different scales. The LIJ IgE+ cohort is plotted according to the scale shown on the left axis, data from the NLJ IgE+ and LIJ IgE− cohorts are plotted according to the scale shown on the right axis). LIJ IgE+ cohort: N=10; NLJ IgE+ cohort: N=24; LIJ IgE− cohort: N=20
Overall, we identified 117 T cell reactive peptides (43 from Cry j 1, 57 from Cry j 2 and 17 from Cry j IFR). To the best of our knowledge, 27 of these epitopes have never been reported before (3 from Cry j 1, 12 from Cry j 2 and 12 from Cry j IFR; no matching entry in the Immune Epitope Database34). Different epitopes were dominant within each cohort. The strongest responses were observed in the LIJ IgE+ cohort, with the most dominant 11 peptides (inducing an average response >400 SFC) accounting for 40% of the total response in that cohort. Responses in the NLJ IgE+ and LIJ IgE− cohorts were similarly dominated by a small number of peptides. In both cohorts the 6 strongest peptides, eliciting an average of 90 SFC or higher, accounted for 40% or more of the total response. Sequences, average response magnitudes and frequencies for all peptides for each allergen are summarized in Table 2.
Table 2.
Summary of average response magnitudes and frequencies for each peptide spanning Cryj 1, Cry j 2 and Cryj IFR for each donor cohort.
| Average response magnitude (SFC) | Average response frequency (%) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptide start |
Antigen | Sequence | LIJ IgE+ |
SD | NLJ IgE+ |
St. Dev |
LIJ IgE− |
SD | LIJ IgE+ |
NLJ IgE+ |
LIJ IgE− |
| 1 | Cry j 1 | MDSPCLVALLVFSFVI | 30 | 68 | 3 | 17 | 0 | 0 | 20 | 4 | 0 |
| 9 | Cry j 1 | LLVFSFVIGSCFSDNP | 9 | 28 | 4 | 19 | 0 | 0 | 10 | 4 | 0 |
| 17 | Cry j 1 | GSCFSDNPIDSCWRGD | 110 | 349 | 0 | 0 | 0 | 0 | 10 | 0 | 0 |
| 25 | Cry j 1 | IDSCWRGDSNWAQNRM | 147 | 466 | 0 | 0 | 29 | 129 | 10 | 0 | 5 |
| 33 | Cry j 1 | SNWAQNRMKLADCAVG | 97 | 308 | 0 | 0 | 29 | 90 | 10 | 0 | 10 |
| 41 | Cry j 1 | KLADCAVGFGSSTMGG | 39 | 122 | 7 | 35 | 93 | 404 | 20 | 4 | 10 |
| 49 | Cry j 1 | FGSSTMGGKGGDLYTV | 18 | 58 | 0 | 0 | 0 | 0 | 10 | 0 | 0 |
| 57 | Cry j 1 | KGGDLYTVTNSDDDPV | 64 | 203 | 8 | 41 | 0 | 0 | 10 | 4 | 0 |
| 65 | Cry j 1 | TNSDDDPVNPAPGTLR | 16 | 50 | 0 | 0 | 6 | 28 | 10 | 0 | 5 |
| 73 | Cry j 1 | NPAPGTLRYGATRDRP | 384 | 928 | 0 | 0 | 0 | 0 | 20 | 0 | 0 |
| 81 | Cry j 1 | YGATRDRPLWIIFSGN | 72 | 141 | 3 | 15 | 0 | 0 | 30 | 4 | 0 |
| 89 | Cry j 1 | LWIIFSGNMNIKLKMP | 401 | 490 | 4 | 18 | 54 | 200 | 70 | 4 | 10 |
| 97 | Cry j 1 | MNIKLKMPMYIAGYKT | 153 | 322 | 91 | 444 | 0 | 0 | 20 | 4 | 0 |
| 105 | Cry j 1 | MYIAGYKTFDGRGAQV | 210 | 448 | 8 | 30 | 0 | 0 | 30 | 9 | 0 |
| 113 | Cry j 1 | FDGRGAQVYIGNGGPC | 218 | 280 | 8 | 37 | 54 | 199 | 60 | 4 | 10 |
| 121 | Cry j 1 | YIGNGGPCVFIKRVSN | 231 | 438 | 22 | 75 | 90 | 251 | 30 | 9 | 15 |
| 129 | Cry j 1 | VFIKRVSNVIIHGLYL | 363 | 495 | 45 | 146 | 3 | 13 | 60 | 17 | 5 |
| 137 | Cry j 1 | VIIHGLYLYGCSTSVL | 29 | 65 | 0 | 0 | 0 | 0 | 20 | 0 | 0 |
| 145 | Cry j 1 | YGCSTSVLGNVLINES | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 153 | Cry j 1 | GNVLINESFGVEPVHP | 67 | 130 | 0 | 0 | 0 | 0 | 30 | 0 | 0 |
| 161 | Cry j 1 | FGVEPVHPQDGDALTL | 172 | 368 | 0 | 0 | 0 | 0 | 20 | 0 | 0 |
| 169 | Cry j 1 | QDGDALTLRTATNIWI | 54 | 134 | 0 | 0 | 63 | 176 | 30 | 0 | 15 |
| 177 | Cry j 1 | RTATNIWIDHNSFSNS | 543 | 832 | 121 | 593 | 53 | 175 | 50 | 4 | 10 |
| 185 | Cry j 1 | DHNSFSNSSDGLVDVT | 82 | 150 | 15 | 74 | 0 | 0 | 40 | 4 | 0 |
| 193 | Cry j 1 | SDGLVDVTLTSTGVTI | 77 | 167 | 3 | 15 | 0 | 0 | 30 | 4 | 0 |
| 201 | Cry j 1 | LTSTGVTISNNLFFNH | 13 | 40 | 0 | 0 | 0 | 0 | 10 | 0 | 0 |
| 209 | Cry j 1 | SNNLFFNHHKVMLLGH | 414 | 852 | 16 | 45 | 18 | 60 | 30 | 13 | 10 |
| 217 | Cry j 1 | HKVMLLGHDDAYSDDK | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 225 | Cry j 1 | DDAYSDDKSMKVTVAF | 298 | 796 | 3 | 14 | 16 | 71 | 30 | 4 | 5 |
| 233 | Cry j 1 | SMKVTVAFNQFGPNCG | 369 | 634 | 108 | 315 | 19 | 57 | 40 | 26 | 10 |
| 241 | Cry j 1 | NQFGPNCGQRMPRARY | 0 | 0 | 0 | 0 | 14 | 60 | 0 | 0 | 5 |
| 249 | Cry j 1 | QRMPRARYGLVHVANN | 157 | 441 | 124 | 440 | 14 | 62 | 20 | 9 | 5 |
| 257 | Cry j 1 | GLVHVANNNYDPWTIY | 238 | 524 | 49 | 239 | 18 | 81 | 30 | 4 | 5 |
| 265 | Cry j 1 | NYDPWTIYAIGGSSNP | 64 | 202 | 0 | 0 | 0 | 0 | 10 | 0 | 0 |
| 273 | Cry j 1 | AIGGSSNPTILSEGNS | 8 | 25 | 0 | 0 | 15 | 67 | 10 | 0 | 5 |
| 281 | Cry j 1 | TILSEGNSFTAPNESY | 722 | 963 | 22 | 106 | 24 | 107 | 60 | 4 | 5 |
| 289 | Cry j 1 | FTAPNESYKKQVTIRI | 127 | 332 | 0 | 0 | 0 | 0 | 20 | 0 | 0 |
| 297 | Cry j 1 | KKQVTIRIGCKTSSSC | 27 | 58 | 0 | 0 | 34 | 134 | 20 | 0 | 10 |
| 305 | Cry j 1 | GCKTSSSCSNWVWQST | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 313 | Cry j 1 | SNWVWQSTQDVFYNGA | 579 | 746 | 0 | 0 | 35 | 115 | 60 | 0 | 10 |
| 321 | Cry j 1 | QDVFYNGAYFVSSGKY | 254 | 348 | 12 | 46 | 47 | 151 | 40 | 9 | 10 |
| 329 | Cry j 1 | YFVSSGKYEGGNIYTK | 152 | 394 | 15 | 61 | 8 | 34 | 20 | 9 | 5 |
| 337 | Cry j 1 | EGGNIYTKKEAFNVEN | 132 | 185 | 3 | 14 | 12 | 52 | 40 | 4 | 5 |
| 345 | Cry j 1 | KEAFNVENGNATPQLT | 140 | 253 | 4 | 20 | 0 | 0 | 40 | 4 | 0 |
| 353 | Cry j 1 | GNATPQLTKNAGVLTC | 144 | 222 | 72 | 290 | 9 | 38 | 40 | 17 | 5 |
| 359 | Cry j 1 | LTKNAGVLTCSLSKRC | 118 | 228 | 13 | 65 | 18 | 78 | 20 | 4 | 5 |
| 1 | Cry j 2 | MAMKLIAPMAFLAMQL | 8 | 24 | 8 | 37 | 0 | 0 | 0 | 4 | 0 |
| 9 | Cry j 2 | MAFLAMQLIIMAAAED | 19 | 60 | 0 | 0 | 0 | 0 | 10 | 0 | 0 |
| 17 | Cry j 2 | IIMAAAEDQSAQIMLD | 9 | 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 25 | Cry j 2 | QSAQIMLDSWEKYLR | 26 | 62 | 3 | 16 | 0 | 0 | 20 | 4 | 0 |
| 33 | Cry j 2 | SVVEKYLRSNRSLRKV | 0 | 0 | 7 | 25 | 0 | 0 | 0 | 9 | 0 |
| 41 | Cry j 2 | SNRSLRKVEHSRHDAI | 11 | 36 | 6 | 27 | 0 | 0 | 10 | 4 | 0 |
| 49 | Cry j 2 | EHSRHDAINIFNVEKY | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 57 | Cry j 2 | NIFNVEKYGAVGDGKH | 28 | 61 | 9 | 29 | 0 | 0 | 20 | 9 | 0 |
| 65 | Cry j 2 | GAVGDGKHDCTEAFST | 0 | 0 | 3 | 12 | 0 | 0 | 0 | 4 | 0 |
| 73 | Cry j 2 | DCTEAFSTAWQAACKN | 450 | 787 | 2 | 12 | 0 | 0 | 40 | 4 | 0 |
| 81 | Cry j 2 | AWQAACKNPSAMLLVP | 391 | 783 | 2 | 12 | 0 | 0 | 30 | 4 | 0 |
| 89 | Cry j 2 | PSAMLLVPGSKKFVVN | 340 | 946 | 30 | 102 | 0 | 0 | 30 | 13 | 0 |
| 97 | Cry j 2 | GSKKFWNNLFFNGPC | 295 | 809 | 157 | 656 | 24 | 107 | 30 | 17 | 5 |
| 105 | Cry j 2 | NLFFNGPCQPHFTFKV | 20 | 62 | 89 | 413 | 0 | 0 | 10 | 9 | 0 |
| 113 | Cry j 2 | QPHFTFKVDGIIAAYQ | 760 | 785 | 49 | 201 | 136 | 351 | 60 | 13 | 20 |
| 121 | Cry j 2 | DGIIAAYQNPASWKNN | 888 | 840 | 114 | 358 | 226 | 549 | 70 | 22 | 25 |
| 129 | Cry j 2 | NPASWKNNRIWLQFAK | 216 | 400 | 20 | 79 | 24 | 66 | 50 | 9 | 15 |
| 137 | Cry j 2 | RIWLQFAKLTGFTLMG | 246 | 597 | 62 | 196 | 112 | 345 | 50 | 17 | 15 |
| 145 | Cry j 2 | LTGFTLMGKGVIDGQG | 14 | 44 | 6 | 20 | 14 | 46 | 10 | 9 | 10 |
| 153 | Cry j 2 | KGVIDGQGKQWWAGQC | 29 | 91 | 0 | 0 | 0 | 0 | 10 | 0 | 0 |
| 161 | Cry j 2 | KQWWAGQCKWVNGREI | 18 | 58 | 5 | 26 | 0 | 0 | 10 | 4 | 0 |
| 169 | Cry j 2 | KWVNGREICNDRDRPT | 118 | 333 | 13 | 61 | 3 | 11 | 30 | 4 | 5 |
| 177 | Cry j 2 | CNDRDRPTAIKFDFST | 41 | 88 | 18 | 71 | 20 | 62 | 20 | 9 | 10 |
| 185 | Cry j 2 | AIKFDFSTGLIIQGLK | 187 | 442 | 15 | 63 | 140 | 416 | 40 | 9 | 25 |
| 193 | Cry j 2 | GLIIQGLKLMNSPEFH | 47 | 69 | 7 | 35 | 9 | 42 | 40 | 4 | 5 |
| 201 | Cry j 2 | LMNSPEFHLVFGNCEG | 77 | 160 | 5 | 26 | 14 | 47 | 30 | 4 | 10 |
| 209 | Cry j 2 | LVFGNCEGVKIIGISI | 225 | 628 | 0 | 0 | 4 | 16 | 40 | 0 | 5 |
| 217 | Cry j 2 | VKIIGISITAPRDSPN | 384 | 686 | 70 | 273 | 192 | 459 | 60 | 13 | 25 |
| 225 | Cry j 2 | TAPRDSPNTDGIDIFA | 9 | 30 | 0 | 0 | 2 | 10 | 10 | 0 | 5 |
| 233 | Cry j 2 | TDGIDIFASKNFHLQK | 33 | 78 | 9 | 46 | 16 | 70 | 20 | 4 | 5 |
| 241 | Cry j 2 | SKNFHLQKNTIGTGDD | 176 | 329 | 0 | 0 | 9 | 40 | 30 | 0 | 5 |
| 249 | Cry j 2 | NTIGTGDDCVAIGTGS | 113 | 357 | 13 | 61 | 0 | 0 | 10 | 4 | 0 |
| 257 | Cry j 2 | CVAIGTGSSNIVIEDL | 146 | 461 | 12 | 59 | 5 | 22 | 10 | 4 | 5 |
| 265 | Cry j 2 | SNIVIEDLICGPGHGI | 166 | 468 | 3 | 13 | 4 | 17 | 30 | 4 | 5 |
| 273 | Cry j 2 | ICGPGHGISIGSLGRE | 0 | 0 | 5 | 18 | 16 | 53 | 0 | 9 | 10 |
| 281 | Cry j 2 | SIGSLGRENSRAEVSY | 104 | 147 | 23 | 112 | 25 | 64 | 50 | 4 | 15 |
| 289 | Cry j 2 | NSRAEVSYVHVNGAKF | 363 | 548 | 0 | 0 | 33 | 108 | 50 | 0 | 15 |
| 297 | Cry j 2 | VHVNGAKFIDTQNGLR | 130 | 234 | 0 | 0 | 10 | 32 | 40 | 0 | 10 |
| 305 | Cry j 2 | IDTQNGLRIKTWQGGS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 313 | Cry j 2 | IKTWQGGSGMASHIIY | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 321 | Cry j 2 | GMASHIIYENVEMINS | 50 | 158 | 0 | 0 | 0 | 0 | 10 | 0 | 0 |
| 329 | Cry j 2 | ENVEMINSENPILINQ | 6 | 19 | 2 | 10 | 0 | 0 | 10 | 4 | 0 |
| 337 | Cry j 2 | ENPILINQFYCTSASA | 115 | 329 | 0 | 0 | 11 | 35 | 20 | 0 | 10 |
| 345 | Cry j 2 | FYCTSASACQNQRSAV | 26 | 81 | 0 | 0 | 9 | 38 | 10 | 0 | 5 |
| 353 | Cry j 2 | CQNQRSAVQIQDVTYK | 86 | 237 | 0 | 0 | 8 | 34 | 20 | 0 | 5 |
| 361 | Cry j 2 | QIQDVTYKNIRGTSAT | 474 | 693 | 2 | 10 | 95 | 306 | 50 | 4 | 15 |
| 369 | Cry j 2 | NIRGTSATAAAIQLKC | 298 | 650 | 7 | 24 | 24 | 78 | 30 | 9 | 10 |
| 377 | Cry j 2 | AAAIQLKCSDSMPCKD | 70 | 148 | 0 | 0 | 15 | 48 | 30 | 0 | 10 |
| 385 | Cry j 2 | SDSMPCKDIKLSDISL | 19 | 61 | 0 | 0 | 0 | 0 | 10 | 0 | 0 |
| 393 | Cry j 2 | IKLSDISLKLTSGKIA | 602 | 848 | 7 | 26 | 17 | 56 | 60 | 9 | 10 |
| 401 | Cry j 2 | KLTSGKIASCLNDNAN | 809 | 832 | 21 | 103 | 28 | 78 | 70 | 4 | 15 |
| 409 | Cry j 2 | SCLNDNANGYFSGHVI | 49 | 156 | 0 | 0 | 0 | 0 | 10 | 0 | 0 |
| 417 | Cry j 2 | GYFSGHVIPACKNLSP | 0 | 0 | 0 | 0 | 9 | 39 | 0 | 0 | 5 |
| 425 | Cry j 2 | PACKNLSPSAKRKESK | 99 | 262 | 0 | 0 | 0 | 0 | 20 | 0 | 0 |
| 433 | Cry j 2 | SAKRKESKSHKHPKTV | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 441 | Cry j 2 | SHKHPKTVMVENMRAY | 0 | 0 | 2 | 11 | 0 | 0 | 0 | 4 | 0 |
| 449 | Cry j 2 | MVENMRAYDKGNRTRI | 0 | 0 | 4 | 18 | 0 | 0 | 0 | 4 | 0 |
| 457 | Cry j 2 | DKGNRTRILLGSRPPN | 24 | 76 | 14 | 71 | 0 | 0 | 10 | 4 | 0 |
| 465 | Cry j 2 | LLGSRPPNCTNKCHGC | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 473 | Cry j 2 | CTNKCHGCSPCKAKLV | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 481 | Cry j 2 | SPCKAKLVIVHRIMPQ | 0 | 0 | 5 | 27 | 0 | 0 | 0 | 4 | 0 |
| 489 | Cry j 2 | IVHRIMPQEYYPQRWI | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 497 | Cry j 2 | EYYPQRWICSCHGKIY | 7 | 21 | 13 | 65 | 0 | 0 | 10 | 4 | 0 |
| 499 | Cry j 2 | YPQRWICSCHGKIYHP | 63 | 199 | 14 | 47 | 0 | 0 | 10 | 9 | 0 |
| 1 | Cry j IFR | MGGSRVLIIGGTGYIG | 0 | 0 | 4 | 19 | 0 | 0 | 0 | 4 | 0 |
| 9 | Cry j IFR | IGGTGYIGRHVTNASL | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | Cry j IFR | RHVTNASLAQGHPTFL | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 25 | Cry j IFR | AQGHPTFLLVREITPS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 33 | Cry j IFR | LVREITPSNPEKAQLL | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 41 | Cry j IFR | NPEKAQLLESFTSKGA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 49 | Cry j IFR | ESFTSKGATLVQGSID | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 57 | Cry j IFR | TLVQGSIDDHASLVAA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 65 | Cry j IFR | DHASLVAALKKVDVVI | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 73 | Cry j IFR | LKKVDWISTLGAPQI | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 81 | Cry j IFR | STLGAPQIADQFNLIK | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 89 | Cry j IFR | ADQFNLIKAIKEVGTI | 0 | 0 | 14 | 49 | 0 | 0 | 0 | 9 | 0 |
| 97 | Cry j IFR | AIKEVGTIKRFFPSEF | 0 | 0 | 18 | 61 | 0 | 0 | 0 | 9 | 0 |
| 105 | Cry j IFR | KRFFPSEFGNDVDKHH | 0 | 0 | 10 | 36 | 0 | 0 | 0 | 9 | 0 |
| 113 | Cry j IFR | GNDVDKHHAVEPMKSM | 0 | 0 | 22 | 67 | 0 | 0 | 0 | 13 | 0 |
| 121 | Cry j IFR | AVEPMKSMFDLKIKLR | 0 | 0 | 26 | 66 | 0 | 0 | 0 | 17 | 0 |
| 129 | Cry j IFR | FDLKIKLRRTIEAEGI | 0 | 0 | 11 | 53 | 0 | 0 | 0 | 4 | 0 |
| 137 | Cry j IFR | RTIEAEGIPHTYWPH | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 145 | Cry j IFR | PHTYVVPHCFAGYFLT | 0 | 0 | 14 | 52 | 0 | 0 | 0 | 9 | 0 |
| 153 | Cry j IFR | CFAGYFLTNLAQLGLA | 0 | 0 | 8 | 38 | 0 | 0 | 0 | 4 | 0 |
| 161 | Cry j IFR | NLAQLGLAAPPRDKIV | 0 | 0 | 3 | 15 | 0 | 0 | 0 | 4 | 0 |
| 169 | Cry j IFR | APPRDKIVIYGDGTTK | 0 | 0 | 0 | 0 | 3 | 13 | 0 | 0 | 5 |
| 177 | Cry j IFR | IYGDGTTKAVYMKEED | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 |
| 185 | Cry j IFR | AVYMKEEDIGTFTIKA | 0 | 0 | 3 | 12 | 0 | 0 | 0 | 4 | 0 |
| 193 | Cry j IFR | IGTFTIKAVDDPRTLN | 0 | 0 | 3 | 14 | 0 | 0 | 0 | 4 | 0 |
| 201 | Cry j IFR | VDDPRTLNKTLYLKPP | 0 | 0 | 4 | 19 | 0 | 0 | 0 | 4 | 0 |
| 209 | Cry j IFR | KTLYLKPPANTISTND | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 217 | Cry j IFR | ANTISTNDLVALWEAK | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 225 | Cry j IFR | LVALWEAKIGKTLEKV | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 233 | Cry j IFR | IGKTLEKVYLSEEQVL | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 241 | Cry j IFR | YLSEEQVLKLLQDTPF | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 249 | Cry j IFR | KLLQDTPFPGTFMVSI | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 257 | Cry j IFR | PGTFMVSIFHTIYVKG | 0 | 0 | 0 | 0 | 5 | 20 | 0 | 0 | 5 |
| 265 | Cry j IFR | FHTIYVKGDQTNFQIG | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 273 | Cry j IFR | DQTNFQIGPDGVEASA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 281 | Cry j IFR | PDGVEASALYPDVKYT | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 289 | Cry j IFR | LYPDVKYTTVEEYISA | 0 | 0 | 8 | 39 | 26 | 74 | 0 | 4 | 15 |
| 291 | Cry j IFR | PDVKYTTVEEYISAFV | 0 | 0 | 0 | 0 | 8 | 34 | 0 | 0 | 5 |
SD- standard deviation
In the T cell response of LIJ IgE+ patients to Cry j 1, the most dominant peptide with respect to response magnitude was Cry j 1281 inducing average response of 722 SFC, recognized in 60% of LIJ IgE+ patients. The most frequently recognized peptide was Cry j 189, recognized in 70% of donors. In the case of NLJ IgE+ patients, Cry j 1249 (average of 124 SFC) induced the most dominant response but Cry j 1233 was most frequently recognized (positive in 26% of patients). In the LIJ IgE− cohort, Cry j 141 was mostly strongly recognized (93 SFC on average) and Cry j 1121 and Cry j 1169 were highest in recognition frequency (15%).
In the case of Cry j 2, the most dominant peptide in the LIJ IgE+ patients, Cry j 2121, was shared in the LIJ IgE− cohort, inducing an average T cell response of 888 and 226 SFC, respectively. It was among the most frequently recognized peptides in both cohorts (recognized in 25% of the LIJ IgE− cohort and 70% in the LIJ IgE+ cohort). The second most dominant peptide in the LIJ IgE+ patients, Cry j 2401, did not elicit strong responses in either of the other 2 cohorts. In the NLJ IgE+ patients, Cry j 297 elicited most prominent responses (average of 157 SFC, 17% recognition frequency) but the most dominantly recognized peptide in NLJ IgE+ patients was also Cry j 2121, which was seen in 22% of patients.
For Cry j IFR, responses were much weaker overall and no significant overlap of T cell epitope repertoire was observed (Table 2). Detectable T cell responses against Cry j IRF peptides were only observed in NLJ IgE+ patients, with the most dominant peptide, Cry j IFR121, triggering an average response of 26 SFC, being reactive in 17% of patients.
T cell responses from JC pollen allergic and non-allergic patients have a similar hierarchy of epitope recognition
Despite the fact that the most reactive epitopes differed in the three cohorts, several epitopes were strongly recognized in multiple cohorts (e.g. Cry j 1177, Cry j 1233, Cry j 2121, Cry j 2217 and Cry j 2361; Table 2). To address this further, we compared the T cell epitope repertoire in each cohort for each of the 3 Cry j allergens by performing a Spearman’s rank correlation analysis. Reactivity to each peptide within each cohort can be found in the Immune Epitope Database (IEDB submission ID 1000703; www.iedb.org). Analysis of T cell reactivity against Cry j 1-derived peptides revealed that the hierarchy of reactivity against the various peptides in all three cohorts correlates significantly (Figure 3A). Patterns of epitope recognition were most similar between LIJ IgE+ and NLJ IgE+ patients (R2= 0.34 and p<0.001, all values shown in Table 3). The second strongest overlap in T cell reactive regions was observed between LIJ IgE+ and IgE− patients (R2= 0.19 and p=0.003). Epitopes recognized by NLJ IgE+ and LIJ IgE− were least similar but the correlation still reached statistical significance (R2= 0.09 and p=0.043, Table 3). Similarly to Cry j 1, correlations of peptide recognition for Cry j 2 between cohorts were significant overall (Table 3), however the hierarchies of overlap were distinct from Cry j 1 (Figure 3B). The greatest overlap in epitope repertoire was observed between the LIJ IgE+ and IgE− cohorts (R2= 0.4 and p<0.001), followed by the LIJ IgE+ and NLJ IgE+ cohorts (R2= 0.16 and p=0.001) and lastly the NLJ IgE+ and LIJ IgE− cohorts (R2= 0.14 and p=0.002).
Table 3.
Correlation of T cell responses between cohorts for Cry j 1, Cry j 2 and Cry j IFR was performed by Spearman’s rank correlation analysis. P<0.05 is considered significant.
|
Cry j 1 R2 values |
Cohort | Cry j 1 p values | Cohort | ||||||
| LIJ E+ |
NLJ E+ |
LIJ IgE− |
LIJ E+ |
NLJ E+ |
LIJ IgE− |
||||
| Cohort | LIJ E+ | 0.34 | 0.19 | Cohort | LIJ E+ | <0.001 | 0.003 | ||
| NLJ E+ | 0.34 | 0.09 | NLJ E+ | <0.001 | 0.043 | ||||
| LIJ IgE− | 0.19 | 0.09 | LIJ IgE− | 0.003 | 0.043 | ||||
|
Cry j 2 R2 values |
Cohort | Cry j 2 p values | Cohort | ||||||
| LIJ E+ |
NLJ E+ |
LIJ IgE− |
LIJ E+ |
NLJ E+ |
LIJ IgE− |
||||
| Cohort | LIJ E+ | 0.16 | 0.4 | Cohort | LIJ E+ | 0.001 | <0.001 | ||
| NLJ E+ | 0.16 | 0.14 | NLJ E+ | 0.001 | 0.002 | ||||
| LIJ IgE− | 0.4 | 0.14 | LIJ IgE− | <0.001 | 0.002 | ||||
|
Cry j IFR R2 values |
Cohort | Cry j IFR p values | Cohort | ||||||
| LIJ E+ |
NLJ E+ |
LIJ IgE− |
LIJ E+ |
NLJ E+ |
LIJ IgE− |
||||
| Cohort | LIJ E+ | n.a. | n.a. | Cohort | LIJ E+ | n.a. | n.a. | ||
| NLJ E+ | n.a. | 0.03 | NLJ E+ | n.a. | 0.293 | ||||
| LIJ IgE− | n.a. | 0.03 | LIJ IgE− | n.a. | 0.293 | ||||
Inferred HLA restrictions of dominant epitopes
We and others previously reported that allergen epitopes are rather heterogeneous in terms of HLA restrictions31, 35. That is, while DR-restricted responses were the most prevalent, DQ and DP restrictions were also identified, with some epitopes restricted by multiple loci. Here, to identify potential HLA restrictions, and to facilitate the design and use of HLA tetramers, we used the RATE program33 to calculate the relative frequency and significance of association between all the epitopes/regions and HLA alleles (or combinations thereof) expressed in responding donors.
A detailed account of the results of the RATE analysis is shown in Table 4, which gives the number of donors that responded (R+) or did not respond (R−) to a given peptide, and expressed (A+) or did not express the given HLA(s) (A−). For example, the Cry j 1233 epitope has 100% of the responders express the HLA molecules DPB1*05:01, DRB5*01:01 or DRB1*04:01, while only 7/16 (44%) of the non-responders express the any of these same HLAs (p=0.001). This analysis allowed inference of potential restrictions for a majority of the main epitopes (Table 4). Of the 20 cases where restrictions could be inferred, all were promiscuous, i.e. the epitope is inferred to be potentially restricted by multiple HLAs, thus confirming and extending the previous results31.
Table 4.
Inferred HLA allele restriction analysis performed using the RATE analysis tool. A- Allele, R- Responder, RF- relative frequency, OR-odds ratio. P<0.05 is considered significant.
| Number of donors | Percentage of of donors | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antigen | Start | Peptide | Allele(s) of inferred restriction | A+R+ | A− R+ |
A+R− | A− R− |
A+R+ | A− R+ |
A+R− | A− R− |
No. of donors |
RF | OR | p- value |
| Cry j 1 | 233 | SMKVTVAFNQFGPNCG | DPB1*05:01,DRB5*01:01,DRB1*04:01 | 9 | 0 | 7 | 16 | 28% | 0% | 22% | 50% | 32 | 2.0 | inf | 0.001 |
| Cry j 2 | 121 | DGIIAAYQNPASWKNN | DRB1*15:02,DRB5*01:01,DRB5*01:02,DRB5*02:02 | 8 | 2 | 3 | 19 | 25% | 6% | 9% | 59% | 32 | 2.3 | 25.3 | 0.001 |
| Cry j 2 | 113 | QPHFTFKVDGIIAAYQ | DPB1*02:01,DPB1*09:01,DRB1*15:02,DPB1*01:01 | 7 | 0 | 8 | 17 | 22% | 0% | 25% | 53% | 32 | 2.1 | inf | 0.002 |
| Cry j 2 | 393 | IKLSDISLKLTSGKIA | DPB1*02:01,DRB1*15:02,DRB5*01:02 | 6 | 0 | 6 | 20 | 19% | 0% | 19% | 63% | 32 | 2.7 | inf | 0.001 |
| Cry j 2 | 217 | VKIIGISITAPRDSPN | DRB1*15:02,DRB1*04:03,DRB5*01:01,DRB5*01:02,DRB1*14:01 | 6 | 1 | 5 | 20 | 19% | 3% | 16% | 63% | 32 | 2.5 | 24.0 | 0.003 |
| Cry j 2 | 401 | KLTSGKIASCLNDNAN | DRB1*04:04,DQB1*03:03,DRB1*15:02,DRB1*04:01 | 5 | 1 | 4 | 22 | 16% | 3% | 13% | 69% | 32 | 3.0 | 27.5 | 0.003 |
| Cry j 2 | 129 | NPASWKNNRIWLQFAK | DRB1*15:01,DPB1*18:01,DPB1*11:01,DRB1*15:02 | 5 | 1 | 4 | 22 | 16% | 3% | 13% | 69% | 32 | 3.0 | 27.5 | 0.003 |
| Cry j 2 | 89 | PSAMLLVPGSKKFVVN | DRB1*13:03,DRB1*15:02,DRB1*03:02,DRB5*01:02 | 5 | 1 | 1 | 25 | 16% | 3% | 3% | 78% | 32 | 4.4 | 125.0 | 0.000 |
| Cry j 1 | 321 | QDVFYNGAYFVSSGKY | DPB1*18:01,DPB1*11:01,DPB1*05:01,DRB3*01:01 | 5 | 0 | 6 | 21 | 16% | 0% | 19% | 66% | 32 | 2.9 | inf | 0.002 |
| Cry j 2 | 137 | RIWLQFAKLTGFTLMG | DPB1*18:01,DPB1*11:01,DRB1*15:02,DRB5*01:02,DPB1*09:01,DPB1*03:01 | 5 | 2 | 2 | 23 | 16% | 6% | 6% | 72% | 32 | 3.3 | 28.8 | 0.002 |
| Cry j 1 | 281 | TILSEGNSFTAPNESY | DRB1*04:05,DRB1*15:02,DRB1*04:01 | 5 | 1 | 4 | 22 | 16% | 3% | 13% | 69% | 32 | 3.0 | 27.5 | 0.003 |
| Cry j 1 | 129 | VFIKRVSNVIIHGLYL | DRB1*14:04,DRB1*04:04,DRB1*15:02,DRB5*01:02 | 5 | 3 | 1 | 23 | 16% | 9% | 3% | 72% | 32 | 3.3 | 38.3 | 0.002 |
| Cry j IFR |
121 | AVEPMKSMFDLKIKLR | DRB1*16:02,DRB5*02:02,DRB1*13:01,DRB1*03:01 | 4 | 0 | 2 | 26 | 13% | 0% | 6% | 81% | 32 | 5.3 | inf | 0.000 |
| Cry j 2 | 81 | AWQAACKNPSAMLLVP | DRB1*15:02,DRB1*13:03,DRB1*03:01 | 4 | 0 | 5 | 23 | 13% | 0% | 16% | 72% | 32 | 3.6 | inf | 0.004 |
| Cry j 1 | 257 | GLVHVANNNYDPWTIY | DRB1*04:05,DRB1*15:02,DQB1*05:03 | 4 | 0 | 5 | 23 | 13% | 0% | 16% | 72% | 32 | 3.6 | inf | 0.004 |
| Cry j 1 | 353 | GNATPQLTKNAGVLTC | DRB3*03:01,DRB1*15:02,DRB1*13:03,DRB1*13:02 | 4 | 4 | 2 | 22 | 13% | 13% | 6% | 69% | 32 | 2.7 | 11.0 | 0.023 |
| Cry j 1 | 89 | LWIIFSGNMNIKLKMP | DPB1*11:01,DRB1*15:02,DPB1*09:01,DRB1*04:03,DRB5*01:02,DRB1*14:01 | 4 | 2 | 1 | 25 | 13% | 6% | 3% | 78% | 32 | 4.3 | 50.0 | 0.002 |
| Cry j 2 | 361 | QIQDVTYKNIRGTSAT | DRB1*15:01,DRB1*15:02,DRB5*01:02,DRB1*04:01 | 4 | 0 | 6 | 22 | 13% | 0% | 19% | 69% | 32 | 3.2 | inf | 0.006 |
| Cry j 1 | 209 | SNNLFFNHHKVMLLGH | DRB1*08:01,DRB1*04:01,DRB1*03:01 | 4 | 1 | 3 | 24 | 13% | 3% | 9% | 75% | 32 | 3.7 | 32.0 | 0.004 |
| Cry j 1 | 313 | SNWVWQSTQDVFYNGA | DRB1*09:01,DRB1*15:02,DRB1*04:01,DPB1*09:01 | 4 | 0 | 5 | 23 | 13% | 0% | 16% | 72% | 32 | 3.6 | inf | 0.004 |
Discussion
In the present study, we compared patterns of immunodominance in T cell recognition to several Cry j allergens in a cohort of sensitized (IgE+) individuals that have resided in Japan for a minimum of 12 months versus sensitized individuals from Southern California who, to the best of our knowledge never resided in Japan. We find that the two cohorts have commonalities in terms of T cell immunodominance at the antigen level, and even at the level of epitope recognition. This finding was contrary to our expectations, because the two cohorts differ in many important aspects, such as ethnicity, JC-specific IgE titers, pattern of polysensitization to other allergens, and most importantly the presumed level exposure to JC pollen.
To the best of our knowledge, ours is the first side-by-side comparison of human T cell reactivity to various Cry j allergens in JC-allergic patients who have lived in Japan versus allergic patients who have not. We found that T cell reactivity correlates with the reported dominance of IgE responses against the same allergens, in that IFR IgE reactivity is observed less frequently (76%)15 compared to Cry j 1 and Cry j 2 (> 90%)11. Indeed despite the reports of Kawamoto et al.15, who show IgE reactivity in a majority of individuals tested, T cell reactivity to this allergen was negligible in all 3 cohorts.
With respect to the magnitude of immunological reactivity to JC pollen, we found that the NLJ IgE+ donors exhibited much lower immune-reactivity both at the serological and T cell level. This lower reactivity may reflect low/infrequent exposure (travel to Japan, exposure to Japanese cedar plants cultivated in the USA, and/or cross-reactive species). The substantial difference in reactivity between the two cohorts is underlined by the drastic difference in the pattern of polysensitization, which indicates that NLJ IgE+ patients have much higher IgE titers to grasses and weeds, whereas LIJ IgE+ patients are mostly IgE reactive to JC-related tree pollens.
Comprehensive mapping of T cell epitopes for each allergen revealed that overall patters of reactivity overlap significantly between both IgE+ cohorts irrespectively of their geographical location, and even IgE− control donors. These commonalities were somewhat surprising, given the many differences between the various cohorts. This finding can be reconciled in light of several reports that highlight how a significant overlap exists between different HLA class II allelic variants, and that epitopes capable of binding multiple HLAs (promiscuous epitopes) account for a significant fraction of overall T cell reactivity31.
Despite the significant overlap, for each allergen and each cohort, unique dominant peptides were also identified. These differences are potentially explained by differences in the frequency of different HLA class II allelic variants in the different cohorts, and also magnified by the relative small number of donors tested in each cohort.
Several studies have been conducted to identify T cell epitopes from Cry j 1 and 2, however these studies have largely been focused on JC pollen allergic individuals from Japan. As has been reported before22, Cry j-specific T cell reactivity is also detected in non-sensitized individuals, albeit at a much lower magnitude and frequency. Analyzing the T cell response to these allergens in non-sensitized individuals and patients who have JC pollen-specific IgE but have never lived in Japan allowed to define sets of epitopes that are of broad potential utility, as they would be active in different cohorts of individuals, associated with large differences in exposure and ethnicity. Our data emphasizes the value of performing studies evaluating antigens and epitopes in different geographical settings, as has been done previously in different disease settings36, 37.
Here we defined 117 Cry j-derived epitopes, 27 of which have to the best of our knowledge never been reported in the literature before. The identification of dominant T cell epitopes provides a tool that can be used to study the immunological characteristics and modulatory changes of the allergic T cell response before and after therapy, using it as immunological assessment for treatment efficacy as has been done in the Timothy grass system38, 39. To further facilitate antigen-specific T cell studies, we used the RATE program, which can infer restriction elements for the more dominant epitopes. This data will allow the production of tetrameric staining reagents to be used in future studies characterizing Cry j-specific T cell responses in context of allergic disease and immunotherapy.
This comprehensive characterization of T cell reactivity in Japanese and non-Japanese allergic and non-allergic individuals is of high relevance for the development of immunotherapeutic approaches. A clear understanding of similarities and differences in the T cell response in patients with true sensitization versus co-recognition due to sensitization to a cross-reactive allergen40 is essential to project efficacy of different therapeutic reagents in these distinct cohorts.
Supplementary Material
Acknowledgments
Funding: Funding was provided in part by ALK-Abello A/S (Horsholm, Denmark) and with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under grant number U19 AI100275.
Conflict of interest
Alessandro Sette and Bjoern Peters are consultants for ALK-Abelló A/S Bøge Allé 6 DK-2970 Hørsholm, Denmark.
Abbreviations
- IFR
isoflavone reductase
- PBMC
Perioheral blood mononuclear cells
- JC
Japanese cedar
- LIJ
lived in Japan
- NLJ
not lived in Japan
- IFNg
interferon gamma
- IL
interleukin
- ASHI
American society for histocompatibility and immunogenetics
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaneko Y, Motohashi Y, Nakamura H, Endo T, Eboshida A. Increasing prevalence of Japanese cedar pollinosis: a meta-regression analysis. Int Arch Allergy Immunol. 2005;136:365–371. doi: 10.1159/000084256. [DOI] [PubMed] [Google Scholar]
- 2.Okuda M. Epidemiology of Japanese cedar pollinosis throughout Japan. Ann Allergy Asthma Immunol. 2003;91:288–296. doi: 10.1016/S1081-1206(10)63532-6. [DOI] [PubMed] [Google Scholar]
- 3.Sakashita M, Hirota T, Harada M, et al. Prevalence of allergic rhinitis and sensitization to common aeroallergens in a Japanese population. Int Arch Allergy Immunol. 2010;151:255–261. doi: 10.1159/000242363. [DOI] [PubMed] [Google Scholar]
- 4.Araki H. Studies on pollinosis. II. Sensitization with pollens. Arerugi. 1961;10:354–370. [PubMed] [Google Scholar]
- 5.Horiguchi S, Saito Y. Discovery of Japanese Cedar Pollinosis in Nikko, Ibaraki Prefecture. Arerugi. 1964;13:16–18. [PubMed] [Google Scholar]
- 6.Saito Y. Japanese cedar pollinosis: discovery, nomenclature, and epidemiological trends. Proc Jpn Acad Ser B Phys Biol Sci. 2014;90:203–210. doi: 10.2183/pjab.90.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamada T, Saito H, Fujieda S. Present state of Japanese cedar pollinosis: the national affliction. J Allergy Clin Immunol. 2014;133:632–639. e5. doi: 10.1016/j.jaci.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Ito Y, Hattori R, Mase H, Watanabe M, Shiotani I. Forecasting models for sugi (Cryptomeria japonica D. Don) pollen count showing an alternate dispersal rhythm. Allergol Int. 2008;57:321–329. doi: 10.2332/allergolint.O-07-520. [DOI] [PubMed] [Google Scholar]
- 9.Yasueda H, Yui Y, Shimizu T, Shida T. Isolation and partial characterization of the major allergen from Japanese cedar (Cryptomeria japonica) pollen. J Allergy Clin Immunol. 1983;71:77–86. doi: 10.1016/0091-6749(83)90550-x. [DOI] [PubMed] [Google Scholar]
- 10.Sakaguchi M, Inouye S, Taniai M, et al. Identification of the second major allergen of Japanese cedar pollen. Allergy. 1990;45:309–312. doi: 10.1111/j.1398-9995.1990.tb00501.x. [DOI] [PubMed] [Google Scholar]
- 11.Hashimoto M, Nigi H, Sakaguchi M, et al. Sensitivity to two major allergens (Cry j I and Cry j II) in patients with Japanese cedar (Cryptomeria japonica) pollinosis. Clin Exp Allergy. 1995;25:848–852. doi: 10.1111/j.1365-2222.1995.tb00027.x. [DOI] [PubMed] [Google Scholar]
- 12.Fujimura T, Futamura N, Midoro-Horiuti T, et al. Isolation and characterization of native Cry j 3 from Japanese cedar (Cryptomeria japonica) pollen. Allergy. 2007;62:547–553. doi: 10.1111/j.1398-9995.2007.01331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Futamura N, Tani N, Tsumura Y, et al. Characterization of genes for novel thaumatin-like proteins in Cryptomeria japonica. Tree Physiol. 2006;26:51–62. doi: 10.1093/treephys/26.1.51. [DOI] [PubMed] [Google Scholar]
- 14.Futamura N, Ujino-Ihara T, Nishiguchi M, et al. Analysis of expressed sequence tags from Cryptomeria japonica pollen reveals novel pollen-specific transcripts. Tree Physiol. 2006;26:1517–1528. doi: 10.1093/treephys/26.12.1517. [DOI] [PubMed] [Google Scholar]
- 15.Kawamoto S, Fujimura T, Nishida M, et al. Molecular cloning and characterization of a new Japanese cedar pollen allergen homologous to plant isoflavone reductase family. Clin Exp Allergy. 2002;32:1064–1070. doi: 10.1046/j.1365-2222.2002.01405.x. [DOI] [PubMed] [Google Scholar]
- 16.Futamura N, Mukai Y, Sakaguchi M, et al. Isolation and characterization of cDNAs that encode homologs of a pathogenesis-related protein allergen from Cryptomeria japonica. Biosci Biotechnol Biochem. 2002;66:2495–2500. doi: 10.1271/bbb.66.2495. [DOI] [PubMed] [Google Scholar]
- 17.Fujimura T, Shigeta S, Suwa T, et al. Molecular cloning of a class IV chitinase allergen from Japanese cedar (Cryptomeria japonica) pollen and competitive inhibition of its immunoglobulin E-binding capacity by latex C-serum. Clin Exp Allergy. 2005;35:234–243. doi: 10.1111/j.1365-2222.2005.02167.x. [DOI] [PubMed] [Google Scholar]
- 18.Ibrahim AR, Kawamoto S, Aki T, et al. Molecular cloning and immunochemical characterization of a novel major Japanese cedar pollen allergen belonging to the aspartic protease family. Int Arch Allergy Immunol. 2010;152:207–218. doi: 10.1159/000283026. [DOI] [PubMed] [Google Scholar]
- 19.Ibrahim AR, Kawamoto S, Nishimura M, et al. A new lipid transfer protein homolog identified as an IgE-binding antigen from Japanese cedar pollen. Biosci Biotechnol Biochem. 2010;74:504–509. doi: 10.1271/bbb.90685. [DOI] [PubMed] [Google Scholar]
- 20.Ibrahim AR, Kawamoto S, Mizuno K, et al. Molecular cloning and immunochemical characterization of a new Japanese cedar pollen allergen homologous to plant subtilisin-like serine protease. World Allergy Organ J. 2010;3:262–265. doi: 10.1097/WOX.0b013e318201d81d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sone T, Morikubo K, Miyahara M, et al. T cell epitopes in Japanese cedar (Cryptomeria japonica) pollen allergens: choice of major T cell epitopes in Cry j 1 and Cry j 2 toward design of the peptide-based immunotherapeutics for the management of Japanese cedar pollinosis. J Immunol. 1998;161:448–457. [PubMed] [Google Scholar]
- 22.Hashiguchi S, Hino K, Taniguchi Y, et al. Immunodominance of seven regions of a major allergen, Cry j 2, of Japanese cedar pollen for T-cell immunity. Allergy. 1996;51:621–632. doi: 10.1111/j.1398-9995.1996.tb04682.x. [DOI] [PubMed] [Google Scholar]
- 23.Ikagawa S, Matsushita S, Chen YZ, Ishikawa T, Nishimura Y. Single amino acid substitutions on a Japanese cedar pollen allergen (Cry j 1)-derived peptide induced alterations in human T cell responses and T cell receptor antagonism. J Allergy Clin Immunol. 1996;97:53–64. doi: 10.1016/s0091-6749(96)70283-x. [DOI] [PubMed] [Google Scholar]
- 24.Panzani R, Yasueda H, Shimizu T, Shida T. Cross-reactivity between the pollens of Cupressus sempervirens (common cypress) and of Cryptomeria japonica (Japanese cedar) Ann Allergy. 1986;57:26–30. [PubMed] [Google Scholar]
- 25.Hinz D, Seumois G, Gholami AM, et al. Lack of allergy to timothy grass pollen is not a passive phenomenon but associated with allergen-specific modulation of immune reactivity. Clin Exp Allergy. 2015 doi: 10.1111/cea.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebner C, Schenk S, Najafian N, et al. Nonallergic individuals recognize the same T cell epitopes of Bet v 1, the major birch pollen allergen, as atopic patients. J Immunol. 1995;154:1932–1940. [PubMed] [Google Scholar]
- 27.Smith KA, Gray NJ, Saleh F, et al. Characterisation of CD154+ T cells following ex vivo allergen stimulation illustrates distinct T cell responses to seasonal and perennial allergens in allergic and non-allergic individuals. BMC Immunol. 2013;14:49. doi: 10.1186/1471-2172-14-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teranishi YK H, Katoh T, Kasuya M, Oura E, Taira H. Possible role of climate change in the pollen scatter of Japnese cedar Cryptomeria japonica in Japan. Climate Research. 2000;14:65–70. [Google Scholar]
- 29.Taira H, T H, Kenda Y. Preseasonal scattering of Cryptomeria japonica pollen in Japan, with reference to the dormancy of the male flowers. Allergology International. 2000;49:263–268. [Google Scholar]
- 30.Radauer C, Nandy A, Ferreira F, et al. Update of the WHO/IUIS Allergen Nomenclature Database based on analysis of allergen sequences. Allergy. 2014;69:413–419. doi: 10.1111/all.12348. [DOI] [PubMed] [Google Scholar]
- 31.Oseroff C, Sidney J, Kotturi MF, et al. Molecular determinants of T cell epitope recognition to the common Timothy grass allergen. J Immunol. 2010;185:943–955. doi: 10.4049/jimmunol.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pham J, Oseroff C, Hinz D, et al. Sequence conservation predicts T cell reactivity against ragweed allergens. Clin Exp Allergy. 2016 doi: 10.1111/cea.12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paul S, Dillon MB, Lindestam Arlehamn CS, et al. A population response analysis approach to assign class II HLA-epitope restrictions. J Immunol. 2015;194:6164–6176. doi: 10.4049/jimmunol.1403074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vita R, Overton JA, Greenbaum JA, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43:D405–D412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jahn-Schmid B, Wopfner N, Hubinger G, et al. The T-cell response to Amb a 1 is characterized by 3 dominant epitopes and multiple MHC restriction elements. J Allergy Clin Immunol. 2010;126:1068–1071. 71, e1–e2. doi: 10.1016/j.jaci.2010.05.038. [DOI] [PubMed] [Google Scholar]
- 36.Thomas WR. Geography of house dust mite allergens. Asian Pac J Allergy Immunol. 2010;28:211–224. [PubMed] [Google Scholar]
- 37.Carpenter C, Sidney J, Kolla R, et al. A side-by-side comparison of T cell reactivity to fifty-nine Mycobacterium tuberculosis antigens in diverse populations from five continents. Tuberculosis (Edinb) 2015;95:713–721. doi: 10.1016/j.tube.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schulten V, Tripple V, Sidney J, et al. Association between specific timothy grass antigens and changes in TH1- and TH2-cell responses following specific immunotherapy. J Allergy Clin Immunol. 2014;134:1076–1083. doi: 10.1016/j.jaci.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schulten V, Tripple V, Aasbjerg K, et al. Distinct modulation of allergic T cell responses by subcutaneous versus sublingual allergen-specific immunotherapy. Clin Exp Allergy. 2015 doi: 10.1111/cea.12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asero R, Wopfner N, Gruber P, Gadermaier G, Ferreira F. Artemisia and Ambrosia hypersensitivity: co-sensitization or co-recognition? Clin Exp Allergy. 2006;36:658–665. doi: 10.1111/j.1365-2222.2006.02477.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.