Abstract
In the face of the worldwide threat of severe acute respiratory syndrome (SARS) to human life, some of the most urgent challenges are to develop fast and accurate analytical methods for early diagnosis of this disease as well as to create a safe anti-viral vaccine for prevention. To these ends, we investigated the antigenicity of the spike protein (S protein), a major structural protein in the SARS-coronavirus (SARS-CoV). Based upon the theoretical analysis for hydrophobicity of the S protein, 18 peptides were synthesized. Using Enzyme-Linked Immunosorbent Assay (ELISA), these peptides were screened in the sera from SARS patients. According to these results, two fragments of the S gene were amplified by PCR and cloned into pET-32a. Both S fragments were expressed in the BL-21 strain and further purified with an affinity chromatography. These recombinant S fragments were confirmed to have positive cross-reactions with SARS sera, either by Western blot or by ELISA. Our results demonstrated that the potential epitope regions were located at Codons 469–882 in the S protein, and one epitope site was located at Codons 599–620. Identification of antigenic regions in the SARS-CoV S protein may be important for the functional studies of this virus or the development of clinical diagnosis.
Key words: SARS, the spike protein, expression, antigenicity
Introduction
Severe acute respiratory syndrome (SARS) has been considered a worldwide threat to health since it was initially reported in Guangdong province at the end of 2002 (1). To control the spread of SARS, the World Health Organization (WHO) coordinated extraordinary collaborations among laboratories around the world to identify the causative agent of SARS. In April 2003, WHO announced that a new pathogen, the SARS-coronavirus (SARS-CoV), a member of the coronavirus family never seen before in humans, was the cause of SARS 2., 3.. Subsequently, a number of laboratories unraveled the genetic information of the SARS-CoV 4., 5., 6.. The immense challenge remains to elucidate the mechanisms by which the virus functions and causes illness based on the viral genomic information. Fortunately, outbreaks of SARS around the world have begun to dwindle. However, we must realize that our knowledge of this disease is still very limited (7). It is still unknown how such a new deadly coronavirus emerged in humans. There is no explanation for what viral or host factors make the disease so infectious, and no approved anti-SARS-CoV drugs have been developed. In short, SARS research is still in its early phase, but is facing urgent clinical demands.
The SARS-CoV has four structural proteins, spike (S), envelope (E), membrane (M) and nucleocapsid (N), which function during host cell entry, virion morphogenesis and release (8). The S protein, with its size of 1,255 amino acids and 24 glycosylation sites, is the largest structural protein in the SARS-CoV (5). Since the S protein is a major viral component of the SARS-CoV to directly attach to host cells, details about its antigenicity can enhance our understanding of pathogenesis as well as the development of diagnostic approaches (9).
The amino acid sequence of the S protein is significantly different from other coronaviruses, with only 24% pairwise amino acid identity (4). The topology of the S protein in SARS-CoV is similar to that of other coronavirus, which usually have a huge amount of ectodomains and short transmembrane spans (10). The majority of amino acids of the SARS-CoV S protein at the N-terminus (residues 14 to 1,195) were found on the outside of the viral particle, while only 13 amino acids at the C-terminus were embedded in the viral membrane (5). The configuration of the SARS-CoV S protein exposed on the viral surface may be different from those of other coronaviruses. In numerous coronaviruses, a furin-like protease cleaves the full-length S protein into two similar-sized fragments, a peripheral S1 and a membrane anchored S2, during viral maturation at the trans-Golgi network. The two S fragments play different roles during viral infection. The peripheral S1 portion can independently bind cellular receptors while the integral membrane S2 portion is required to mediate fusion of vial and cellular membranes (11). Noncovalent interactions coordinate S1 and S2 to attack a target cell. It has been widely accepted that the formation of the S1-S2 complex is a key event where coronavirial virion is recognized by the membrane of the host cell (12). For instance, the S protein of murine hepatitis virus (MHV) was not captured by its receptors until the S1-S2 oligomers were formed (13). Interestingly, there is no protease cleavage site in the SARS-CoV S protein; thus, this protein is probably not cleaved into S1 and S2 subunits (4). Does this mean that the SARS-CoV may conduct different infection mechanisms to host cells without intermolecular cleavage? Is it possible to form an uncleaved S oligomer in the SARS-CoV S protein during infection? To address these questions, immunoassay is a powerful tool that uses specific antibodies to define different regions in the S protein. Thus, we embarked on the studies to assess the S antigenic regions.
A study was done to investigate the S antigenic regions with a strategy developed in our lab. According to theoretical estimation, we first synthesized 18 peptides derived from the S protein. These peptides were screened in the sera from SARS patients using ELISA measurement. Based on the ELISA data, two truncated S recombinants were designed and expressed in the BL-21 strain. With confirmation of immunoactivities for the two recombinant S proteins, we have demonstrated that an epitope site is located around Codons 599–620 in the S protein.
Results and Discussion
The S protein is one of the major antigenic proteins in SARS structural proteins
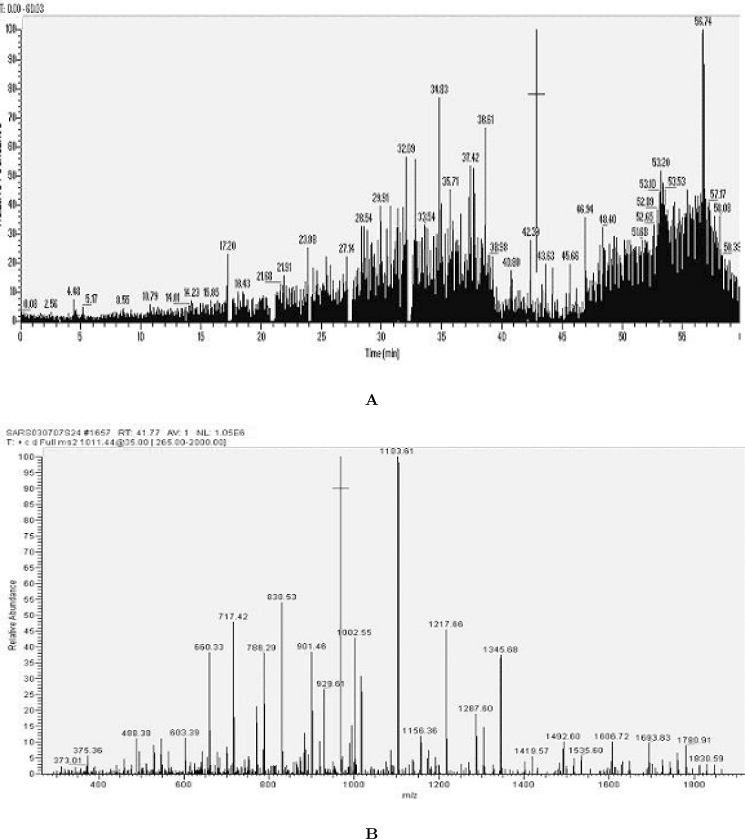
The extracted proteins from Vero-E6 cells infected by the SARS-CoV were separated by SDS-PAGE (12% polyacrylamide), transblotted onto a PVDF (Polyvinylidene Fluoride) membrane and immuno recognized by antibodies in the sera from SARS patients. As shown in Figure 1, the cell extracts were analyzed by Western blot using three SARS sera. Although individual patient sera display different immunoreactivities against the same infected cell lysate, the major immunostaining bands are located around two regions (Figure 1), 50 kDa and 120–240 kDa. Because the molecular weight (MW) of the N protein is 46 kDa, the protein band near to 50 kDa do reasoned to be the N protein. The N protein bands examined in this study have shown a constant expression; nevertheless, the immunostained bands around 120–240 kDa do not display a constant pattern. These bands are likely to be the S protein because the masses of these protein bands are over 120 kDa. Spike proteins have been reported to have multiple glycosylation sites (14). Therefore, it is not surprising that the SARS-CoV S proteins may show apparently different molecular weights due to variable extents in glycosylated modifications during infection. To confirm the bands around 120–240 kDa, these bands were excised and completely digested by trypsin. The tryptic fragments were analyzed by mass spectrometry to identify the composition of peptides (Figure 2). The results from searching the peptide demonstrated that the S proteins are the major components in these gel bands. The sequence coverage based on the data of matched peptides is 72.2% for the S protein.
Fig. 1.
The Western blot patterns of the immunoreactive proteins in Vero-E6 cells infected by the SARS-CoV. Each lane was loaded with 50 mg of the protein from the lysate of Vero-E6 cells. Lane 1: control, the serum from a healthy donor as the primary antibody; Lane 2–4: SARS-CoV, the sera from SARS patients as primary antibodies; Lane 5: standard protein markers.
Fig. 2.
Analysis of mass spectrometry for the protein bands located between 124 to 209 kDa and recognized by SARS-CoV sera in Western blot. A. Base peak ion chromatogram of trypsinized peptides from a piece of the excised gel after in-gel digestion. B. The tandem mass spectra (MS/MS) for parent ion 1011.44 (sequence: QLSSNFGAISSVLNDILSR).
Serological studies on coronaviruses have shown the existence of at least three antigenic groups within the family. For instance, the human coronavirus (HCV)−229E and the canine coronavirus (CCV) be long to Group I, MHV to Group II and infectious bronchitis virus (IBV) to Group III (15). Viruses within each subgroup have partial antigenic crossreactivity based on the S proteins (16). Koch et al. analyzed the antigenic structure of the S protein from IBV, in which six antigenic sites were located at S1 and two were at S2 (17). Since the S1 protein is responsible for the attachment to cellular host membranes, it is not surprising that this protein is stronger in antigenicity than the S2 protein. Therefore, in many coronaviruses, the S1 protein has been confirmed to correlate with serotype (18). Identification of antigenic sites on the S1 protein has shown its significance for virus detection and classification (18). In our Western blot experiment, the SARS-CoV S protein was also found to be a major antigenic protein. Does the SARS-CoV S protein have any homology to other coronavirus S proteins in the antigenic structure? Does the antigenic site on the SARS-CoV S protein appear biological significance during SARS infection? To address these questions, we extended our studies to search for the antigenic site(s) of this protein.
Screening the S antigenic sites with synthesized peptides
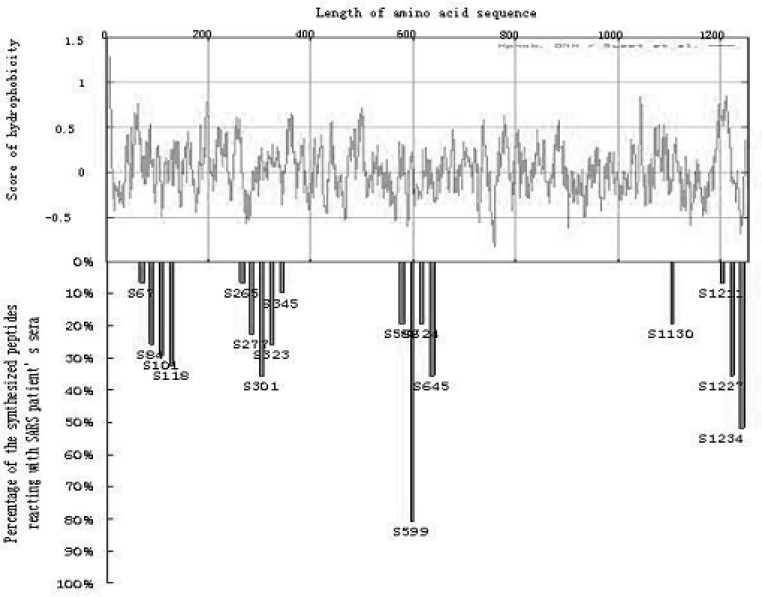
Naturally, a key question for searching for antigenic sites is how to select potential fragments among over 1,000 amino acids in the S protein. To select the candidates for the antigenic peptide, we used several theoretical calculations to estimate the biophysical parameters of the peptides along the S protein sequences. Specifically, we looked for high local hydrophilicity, the charged residues on the exposed protein surface, and accessible surface area. From Figure 3 (upper panel), the hydropathy plot indicates that most regions of the S protein are very hydrophobic. Hence, we only selected 18 peptides derived from the S protein. These peptides, sized from 16–25 amino acid residues, possess relatively low hydrophobicity scores compared to the other regions of the S protein. The synthesized peptides were coated on microtiter plates as antigens and screened in the SARS sera (31 cases) as antibodies in ELISA experiments.
Fig. 3.
Evaluation of the detection rates of 18 synthesized peptides spanning the SARS-CoV S protein. The upper graph shows hydropathy plots and the lower graph indicates the detection rates of synthesized peptides against 31 sera from SARS patients by using ELISA.
The correlations between hydrophobicity and immunoreactive detection rates are shown in Figure 3. Although 18 peptides were tested, only the peptide S599 located at Codons 599–620 (QDVNCTDVSTAIHADQLTPAWR) shows a significant immunoreactivity to SARS sera. This peptide reacted to over 80% of patient sera and responded negative to all of the normal sera. The fact that the major synthesized peptides had low immunoresponses against SARS sera could be resulted from two possibilities, 1) the S protein may contain few epitope sites; 2) some epitope sites may have been missed by our experimental design.
According to the cleavage model of S proteins, the S2 protein is quite conserved, whereas the S1 protein contains an extensive heterogeneity in its amino acid sequence (19). In some coronaviruses, the cellular receptor binding regions with the hypervariable sequences are located at the N-terminus of the S1 protein. The tropism determinant of the MHV S1 protein is located in the region of amino acid residues from 456 to 592 nt (20). The neutralization site of the feline infectious peritonitis virus (FIPV) is located in the region of amino acid residues from 480 to 649 nt (21). Interestingly, these sites were also found to act as antigenic sites in these viruses. MHV 4 is a highly neurotropic virus that produces acute fatal encephalitis in mice. Once amino acid residues in this region are singly or multiply deleted, the neurovirulence would be significantly reduced (20). Therefore, developing a specific antibody (monoclonal antibodies) against these regions could help to further understand the pathogenesis of coronaviruses. Though the SARS-CoV S protein could not be cleaved into two subunits during infection, its antigenic region(s) may be somehow similar to other coronavirus S proteins depending on its folding state. More importantly, the S peptide located at Codons 599–620 is in a region that exists in both MHV and FIPV with antigenic and neutralized functions. Whether this site could bind to a host cell and cause the pathogenesis of the SARS-CoV is a tempting topic for further studies.
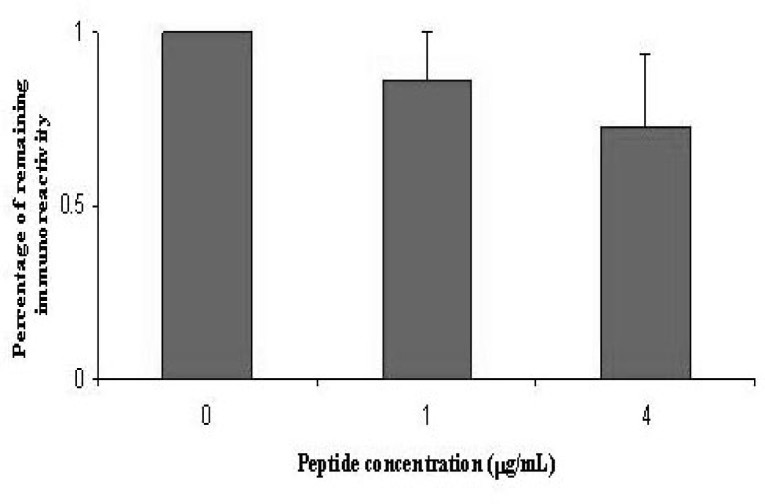
To further confirm the immunoreactive function of the peptide S599, we examined whether this peptide could compete with the SARS-CoV to bind the antibodies in SARS sera. If a peptide really mimics an epitope site in this virus, it should elicit an immunoresponse and neutralize the immunoreaction in ELISA. In previous ELISA tests, the peptide S599 with different concentrations was mixed with the patient sera for 30 min. The extracted proteins from the SARS-CoV were coated onto microtiter plates followed by incubation of the mixed sera for ELISA measurement. As shown in Figure 4, the ELISA reactivities were attenuated with the increase of the peptide concentrations in the SARS sera. Thus, this competition experiment has strengthened our argument that the peptide S599 represents one epitope site in the S protein.
Fig. 4.
Peptide competition experiment in ELISA. Prior to ELISA, the sera from SARS patients were incubated with the peptide S599 in different concentrations. Then the mixtures were incubated with the extraction of the SARS-CoV coated on microtiter plates for further ELISA measurement. The competition experiment at each concentration was tested in parallel for four times, and four patient sera were used for the test.
Cloning and expression of the S fragments around S599
As described above, the peptide S599 is 22 amino acids long and reacts 80% of the SARS sera. Obviously, it is not an ideal antigen because about 20% positive cases were missed during ELISA detection. How can more sensitive antigen(s) be developed with the knowledge we have gained about S599? Coronavirus has a number of antigenic sites that often form a cluster around certain regions. For instance, two vicinal antigenic domains in bovine coronavirus (BCV) span from amino acids 351 to 768 nt (22) and the FIPV antigenic regions cross from 480 to 649 nt (21). Therefore, we have designed to express two large S fragments with MW around 30 kDa. In these fragments, the peptide S599 is a core antigenic element. Centered on this sequence, the recombinant S469-749 extends 130 amino acids toward the N-terminus of the S protein, and S602-882 expresses 262 amino acids to the C-terminus of the S protein.
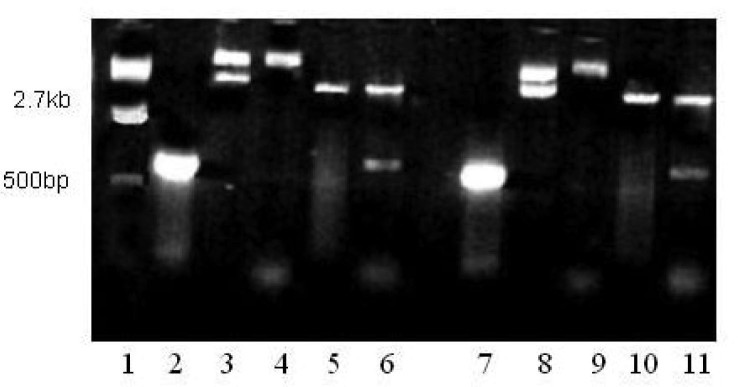
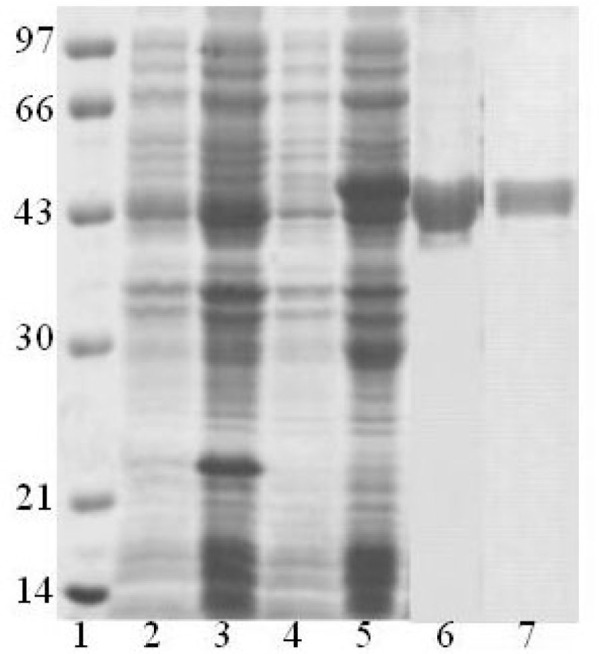
To amplify the S gene fragments, the cDNA reversely transcripted from the SARS-CoV RNA was used as the template, and two pairs of primers, S469-749 and S602—882, were synthesized (see Materials and Methods). Figure 5 shows that the sizes of PCR products coincide well with the theoretical estimation based upon the genomic sequence, 844 and 846 bp, for S469-749 and S602-882, respectively. Both PCR products were amplified with two restriction sites, BamH I at 5′ and Xho I at 3′. These fragments were further digested by enzymes and inserted into a bacterial expression vector pET32a to generate the expression vectors pET32a-S469-749 and pET32a-S602-882. The ligation products were examined by restriction digestion (Figure 5) and further confirmed by DNA sequencing.
Fig. 5.
Construction of the S truncated recombinants into an expression vector, pET30a. Lane 1: DNA ladder; Lanes 2 and 7: the S fragments amplified by PCR, S469-749 and S602-882, respectively; Lanes 3 and 8: pET30a vector only; Lanea 4 and 9: pET30a-S469-749 and pET30a-S602-882 vectors, respectively; Lanes 5 and 10: pET30a digested by Xho I and BamH I; Lanes 6 and 11: pET30a-S469-749 and pET30a-S602-882 vectors digested by Xho I and BamH I, respectively.
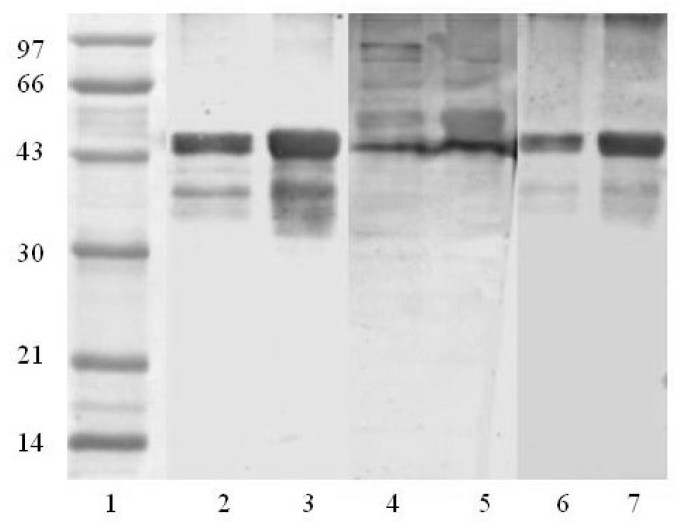
The two expression vectors were transformed into the BL-21 strain and the proteins were expressed by inducement of IPTG (isopropyl-β-D-thiogalactopyranoside). Since the S protein is quite hydrophobic, its recombinant may not be easily soluble in bacterial cytosol. A tag of thioredoxin was added at the N-terminus of S fragments to induce the formation of soluble S recombinant proteins. Both S recombinants were successfully expressed in BL-21 when induced by IPTG (Figure 6). However, the expression products were in inclusionbodies even with the soluble tag of thioredoxin. Figure 7 displays that the molecular weights of the expressed S proteins are approximately 50 kDa, which are much higher than the values of theoretical estimation, 30 kDa, for both S469-749 and S602-882. Since there is no protease digestion after expression and purification, the apparent MW representing the fusion protein consisting of S fragment and thioredoxin is not surprising. To obtain a high protein yield, the bacterial pellets were treated with 8 M urea followed by a strong probe sonication to completely dissolve the inclusions. Since the denatured proteins retained an affinity to Ni2+, a Ni-NTA column was effectively applied to the purification of these recombinants. As shown in Figure 7, the recombinant proteins purified by Ni-NTA were of high purity (>95%).
Fig. 6.
SDS-PAGE analysis of expression and purification of the recombinant S proteins. Lane 1: protein ladder; Lanes 2 and 4: the lysate of BL-21 containing pET30a-S469-749 and pET30a-S602-882 vectors without IPTG inducement, respectively; Lanes 3 and 5: the lysate of BL-21 containing pET30a-S469-749 and pET30a-S602-882 vectors under IPTG inducement, respectively; Lanes 6 and 7: purified recombinant S proteins, S469-749 and S602-882, respectively.
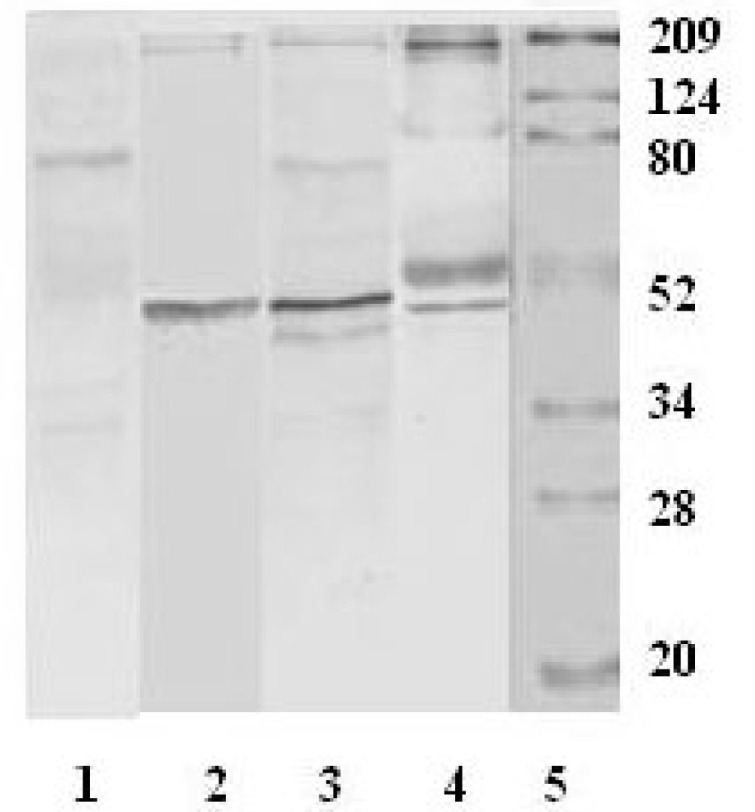
Fig. 7.
Western blot analysis for the recombinant S proteins using SARS sera as primary antibodies. Lane 1: Protein ladder. Lanes 2, 4 and 6 are loaded with the S469-749 protein. Lanes 3, 5 and 7 are loaded with S602-882 protein. Three patient sera were used for this experiment. Of these lanes, 2 and 3, 4 and 5, and 6 and 7 share the same patient serum, respectively.
Immunoassay for the S recombinant proteins by Western blot and ELISA
The S recombinants were further examined on their immunoresponses against the antibodies raised from SARS sera. In Western blot, both recombinant proteins were recognized by all of the three patient sera. Notably, the two S proteins showed slightly different immunoresponses, in which the S602-882 protein had significantly higher immunostaining intensity than that of the S469-749 protein. Similar phenomenon was observed in all the three tested patient sera, indicating that the S602-882 protein may contain a strong epitope side to elicit the immunoresponse. The two S recombinants share almost 50% amino acid sequence, that is, 147 amino acids at the C-terminus of S469-749 are identical with the N-terminus of S602-882. The sequence of the peptide S599 is mainly located within S602-882. Hence, this result provides more evidence that the site around Codons 599–620 is an antigenicity region. Furthermore, S469-749 does display positive immunoreaction to SARS sera even though it only contains three amino acids from the peptide 599. Normally, a short peptide with three amino acids is impossible to form an antigenic site. A logical deduction is that the region neighboring the three amino acids is antigenic or there is another epitope site(s) in the S469-749 protein.
The two S recombinant proteins were screened by SARS sera with ELISA. Six sera, four from SARS patients and two from controls, were examined. The ELISA data revealed that both S469-749 and S602-882 proteins had strong positive reactions to patient sera but negative to normal sera.
Materials and Methods
The E. coli strains, DH5α and BL-21 (λCE6), were purchased from Shanghai Dingguo Co. All primers were synthesized by Shanghai Ding’an Co. The bacterial protein expression vector, pET-32a, was purchased from Novagen (Darmstadt, Germany). Restriction enzymes and nitro-blue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3′-indolyphosphate ptoluidine salt (BCIP) were obtained from Promega (Madison, USA). Taq DNA polymerase and the related PCR reagents were from Invitrogen (Carlsbad, USA). Ni-NTA resin was purchased from Qiagen (Hilden, Germany). The antibody anti-human IgG conjugated with alkaline phosphatase was purchased from Beijing Zhongshan Company.
Sera of SARS patients were from hospitals in Beijing and the control sera were obtained from healthy volunteers. The clinical diagnostic criteria for SARS followed the Clinical Description of SARS were released by WHO.
The SARS-CoV BJ01 strain cultured by the Microbe Epidemic Institute in Chinese Academy of Military Medical Sciences was used in this research. Its genome was sequenced by Beijing Genomics Institute (BGI). All of the S protein fragments were designed based upon this genomic sequence (http://www.ncbi.nlm.nih.gov, GI: 30275666).
The SARS virus was propagated on Vero-E6 cells as described before in the Microbe Epidemic Institute (23). After viral propagation, the cells were harvested and placed at 70°C for 2 h to inactivate the virus. The inactivated infected Vero-E6 cells in culture medium were concentrated by polyethylene glycol (PEG) 20,000 followed by cell lysis and protein denatureation with 8 M urea. The cell lysate was further sonicated with a probe sonicator and centrifuged at 13,000 g remove the insoluble debris. The supernatant was used for Western blot and ELISA determinations.
The amino acid sequence of the S protein was downloaded into the ProtScale program at the Swiss Institute of Bioinformatics (SIB) to analyze the physical characteristics of the proteins, such as hydrophilicity, hydrophobicity, accessible residues, buried residues, MW, and pI values. Forty-one peptides ranging in size from 16 to 25 amino acid residues were selected for synthesis. All of the peptides were synthesized commercially by Chinese Peptide Co., Hangzhou. The synthesized peptides were characterized by HPLC and mass spectrometry.
The viral genomic RNA was prepared using TRIzol reagent (Invitrogen). First-strand cDNA synthesis was carried out as described in the company manual (Superscript™ kit). Two pairs of primers were designed to amplify S469-749 and S602-882 fragments based upon the open reading frame (ORF) of the S gene from BJ01 SARS genome. Specifically, the sequences of these primers are, 5′ CGGATCCCCACCTGCTCTTAATTGTTA (S469-749 forward with a BamH I restriction site at the 5′ end), 5′CCGCTCGAGGAGTGCACGATTTAGTTGTGT (S469-749 reverse with a Xho I restriction site at the 3′ end), 5′CGGGATCCAACTGCACTGAGTTTCTACA (S602-882 forward with a BamH I restriction site at the 5′ end), and CCGCTCGAGGCATAGCAAAAGGTATTTGAAG (S602-882 reverse with a Xho I restriction site at the 3′ end). PCR products and pET-32a vector were excised with BamH I and Xho I overnight at 37°C, purified from agarose gel and ligated with each other at room temperature for 3 h. The ligation products were transformed into DH5α strains. The expression vectors with S gene insertions were checked first by restriction digestion and further confirmed by DNA sequencing. The correct vectors were transformed into BL-21 (λCE6) for protein expression (24).
Two transformed BL-21 stains containing the expression vectors, pET32a-S469-749 and PET32a-S469-749, were inoculated into 500 mL of LB broth containing kanamycin of 100 µg/mL. Being shaken, the cultures grew to an optical density of 0.6–0.8 at 600 nm at 37C, and IPTG was added to a final concentration of 1 mM. The bacteria were incubated at 37°C for additional 4–6 h and harvested, followed by centrifugation at 4,000 g for 10 min get the bacterial pellets. The pellets were resuspended in a 10-mL binding buffer containing 20 mM Tris-HCl, pH 7.9, 200 mM NaCl, and 5 mM imidazole. The buffer was then lysed by sonication for 3 min. The bacterial pellets containing inclusion bodies were dissolved in the binding buffer containing 8 M urea. The recombinant proteins were purified with an affinity chromatography, Ni-NTA (Qiagen), with 100 mM imidazole.
The recombinant proteins were separated by SDS-PAGE (10%) and transferred onto PVDF membranes. The membranes were blocked by 3% BSA in Tris 100 mM, NaCl 120 mM, 0.1% Tween-20, pH 7.9 (TTBS), and then were incubated with the sera from SARS patients as the primary antibody. An anti-human IgG conjugated with alkaline phosphatase was used as the second antibody. The immunoprecipitated bands were developed using a substrate mixture of NBT and BCIP.
ELISA experiments were carried out according to conventional protocol. In brief, the purified fusion protein was mixed with sample dilution buffer, embedded in 96-well ELISA plates, and incubated at 37°C for 30 min. Then the wells were washed for five times with washing buffer and incubated with the sera from either SARS patients or normal controls. Each well was washed and incubated with peroxidase-conjugated goat anti-human IgG at 37°C for 20 min. Finally the wells were washed again with PBS containing 0.5% Tween-20. The peroxidase reaction was visualized using the o-pheylenediamine solution as substrate. After 10 min of incubation at 37°C, the reaction was stopped by the addition of 50 µL of 4 M sulphuric acid, and optical density at 450 nm with a reference wavelength of 630 nm was measured by using an automatic ELISA plate reader (Multiskan Microplate Photometer, Finland).
On SDS-PAGE, the protein bands stained by Commassie Blue corresponding to the immunostaining signals in Western blot were excised, successively destained and dehydrated with 50% acetonitrile. The proteins were reduced with 10 mM DTT at 56°C for 1 h and alkylated by 55 mM iodoacetamide in dark at room temperature for 45 min. Finally, the gel slices were thoroughly washed with 25 mM ammonium bicarbonate in water/acetonitrile (50/50) solution and were completely dried in a Speedvac. Proteins were digested in 25 µL of modified trypsin solution (10 ng/µL in 25 mM ammonium bicarbonate) by incubation overnight at 37°C.
The digested peptides were directly loaded and analyzed by LC-MS/MS using a LCQ DecaXP ion trap mass spectrometer (ThermoFinnigan, Finland). A linear gradient elution program was conducted to separate the peptides, delivering buffer A (0.1% formic acid in water) from 98% to 20% and buffer B (0.1% formic acid in acetonitrile) from 2% to 60% within 66 min. After HPLC, the eluant was split and introduced to the mass spectrometer with a flow rate of about 2 µL/min. The spray voltage was 3.2 kV and the heated desolvation capillary was set to 150°C. The m/z range from 400-2,000 was scanned in 1.2 sec, and the ions were detected with a high energy Conversion Dynode detector. The LC-MS/MS data were converted into DTA-format files and submitted to the searching software, TurboSEQUEST. Protein searches were performed by comparing experimental data with the protein database of SARS-coronavirus generated by Beijing Genomics Institute.
References
- 1.World Health Organization Severe acute respiratory syndrome (SARS) Wkly. Epidemiol. Rec. 2003;78:81–83. [PubMed] [Google Scholar]
- 2.Peiris J.S. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ksiazek T.G. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 4.Rota P.A. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 5.Marra M.A. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 6.Qin E.D. A complete sequence and comparative analysis of a SARS-associated virus (Isolate BJ01) Chin. Sci. Bull. 2003;48:941–948. doi: 10.1007/BF03184203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drosten C. Severe acute respiratory syndrome: identification of the etiological agent. Trends. Mol. Med. 2003;9:325–327. doi: 10.1016/S1471-4914(03)00133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan H.L. Coronavirus in severe acute respiratory syndrome (SARS) Trends. Mol. Med. 2003;9:323–325. doi: 10.1016/S1471-4914(03)00135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruan Y.J. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haring J., Perlman S. Mouse hepatitis virus. Curr. Opin. Microbiol. 2001;4:462–466. doi: 10.1016/S1369-5274(00)00236-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sturman L.S. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: activation of cell-fusing activity of virions by trypsin and separation of two different 90K cleavage fragments. J. Virol. 1985;56:904–911. doi: 10.1128/jvi.56.3.904-911.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taguchi F., Shimazaki Y.K. Functional analysis of an epitope in the S2 subunit of the murine coronavirus spike protein: involvement in fusion activity. J. Gen. Virol. 2000;81:2867–2871. doi: 10.1099/0022-1317-81-12-2867. [DOI] [PubMed] [Google Scholar]
- 13.Krueger D.K. Variations in disparate regions of the murine coronavirus spike protein impact the initiation of membrane fusion. J. Virol. 2001;75:2792–2802. doi: 10.1128/JVI.75.6.2792-2802.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallagher T.M. Murine coronavirus spike glycoprotein. Receptor binding and membrane fusion activities. Adv. Exp. Med. Biol. 2001;494:183–192. [PubMed] [Google Scholar]
- 15.Dea S. Antigenic and genomic relationships among turkey and bovine enteric coronaviruses. J. Virol. 1990;64:3112–3118. doi: 10.1128/jvi.64.6.3112-3118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavanagh D., Davis P.J. Evolution of avian coronavirus IBV: sequence of the matrix glycoprotein gene and intergenic region of several serotypes. J. Gen. Virol. 1988;69:621–629. doi: 10.1099/0022-1317-69-3-621. [DOI] [PubMed] [Google Scholar]
- 17.Koch G. Antigenic domains on the peplomer protein of avian infectious bronchitis virus: correlation with biological functions. J. Gen. Virol. 1990;71:1929–1935. doi: 10.1099/0022-1317-71-9-1929. [DOI] [PubMed] [Google Scholar]
- 18.Ignjatovic J., Galli L. Structural proteins of avian infectious bronchitis virus: role in immunity and protection. Adv. Exp. Med. Biol. 1993;342:449–453. doi: 10.1007/978-1-4615-2996-5_71. [DOI] [PubMed] [Google Scholar]
- 19.Schikora B.M. Genetic diversity of avian infectious bronchitis virus California variants isolated between 1988 and 2001 based on the S1 subunit of the spike glycoprotein. Arch. Virol. 2003;148:115–136. doi: 10.1007/s00705-002-0904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker S.E. Sequence analysis reveals extensive polymorphism and evidence of deletions within E2 glycoprotein gene of several strains of murine hepatitis virus. Virology. 1989;173:664–673. doi: 10.1016/0042-6822(89)90579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kida K. Selection of antigenic variants of the S glycoprotein of feline infectious peritonitis virus and analysis of antigenic sites involved in neutralization. J. Vet. Med. Sci. 1999;61:935–938. doi: 10.1292/jvms.61.935. [DOI] [PubMed] [Google Scholar]
- 22.Yoo D., Deregt D. A single amino acid change within antigenic domain II of the spike protein of bovine coronavirus confers resistance to virus neutralization. Clin. Diagn. Lab. Immunol. 2001;8:297–302. doi: 10.1128/CDLI.8.2.297-302.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Q.Y. Isolation and identification of a novel coronavirus from patients with SARS. J. Chin. Biotech. 2003;23:106–112. [Google Scholar]
- 24.Sambrook J. Third Edition. Cold Spring Harbor Laboratory Press; New York, USA: 2000. Molecular Cloning: A Laboratory Manual. [Google Scholar]