Abstract
Expressed Sequence Tag (EST) analysis has pioneered genome-wide gene discovery and expression profiling. In order to establish a gene expression index in the rice cultivar indica, we sequenced and analyzed 86,136 ESTs from nine rice cDNA libraries from the super hybrid cultivar LYP9 and its parental cultivars. We assembled these ESTs into 13,232 contigs and leave 8,976 singletons. Overall, 7,497 sequences were found similar to the existing sequences in GenBank and 14,711 are novel. These sequences are classified by molecular function, biological process and pathways according to the Gene Ontology. We compared our sequenced ESTs with the publicly available 95,000 ESTs from japonica, and found little sequence variation, despite the large difference between genome sequences. We then assembled the combined 173,000 rice ESTs for further analysis. Using the pooled ESTs, we compared gene expression in metabolism pathway between rice and Arabidopsis according to KEGG. We further profiled gene expression patterns in different tissues, developmental stages, and in a conditional sterile mutant, after checking the libraries are comparable by means of sequence coverage. We also identified some possible library specific genes and a number of enzymes and transcription factors that contribute to rice development.
Key words: EST, expression profile
Introduction
Rice (Oryza sativa) is one of the most important crops in the world. Identifying rice genes and gene expression patterns is important for the understanding of rice biology as well as for the study of traits such as high yield, disease resistance and stress resistance. The most effective approach to identify large number of genes is expressed sequence tag (EST) sequencing, which complements genomic DNA sequencing by explicitly identifying transcribed regions 1., 2.. EST sequencing has also been employed to identify genes expressed in particular tissues and to identify genes that are differentially expressed under various conditions 1., 2., 3., 4.. Care must be taken to use EST frequencies only as a rough estimate, not an exact measure, of gene expression levels.
Up to December 2001, researchers have reported about 95,000 rice EST sequences, the majority of which are from Nipponbare, a japonica variety (5). In this paper, we report the sequencing of a total of 86,136 ESTs from a new set of rice varieties and environmental and developmental circumstances that have not been previously studied. Furthermore, we analyzed both this set of EST sequences and the total set of over 173,000 EST sequences including public EST sequences. We calibrated the sequence clustering methodology with three different algorithms, and confirmed that our EST assembly was of very high quality. We report the new rice genes identified, especially those involved in key pathways, a new look at the gene “landscape” of rice, and genes identified to have highly different levels of gene expression between different rice varieties, environments and development stages. We hope to uncover genes that contribute to traits including high yield.
Results
We summarized our analysis result of the 86,136 good quality rice ESTs. We firstly checked that our EST libraries and sequences are of good quality. Secondly, we made sequencing progress monitor to get an overview of rice gene discovery through EST projects. Thirdly, we assembled those high quality ESTs into contigs, and did necessary re-assembling, splitting and merging. Then we managed to find complete ORFs, using GC content gradient as an additional criterion. After that, we assigned annotations to our contigs/ESTs using BLASTN and BLASTX, and thus classified these contigs into different catalogues assorted by Gene Ontology (GO). Finally we did expression profile to find the genes most differentially expressed between different libraries.
Library information and quality check
We evaluated the quality of our rice EST libraries (See Table 1). We found very low rRNA content (around 1%), no mitochondrial mRNA, few chimeric clones as detected by BLAST searches, and relatively constant expression of constitutive housekeeping genes such as G3PD. Those high quality contigs/ESTs was organized for further analysis.
Table 1.
Quality Assessment of the cDNA Libraries
| Library | rRNA | Mitochondria mRNA | G3PD | Actin | Tubulin | MADS |
|---|---|---|---|---|---|---|
| Lib 1 | 0.25% | 4.90% | 0.56% | 0.29% | 0.09% | 0.06% |
| Lib 2 | 0.66% | 0.78% | 0.71% | 0.20% | 0.20% | 0.00% |
| Lib 3 | 1.99% | 0.18% | 0.50% | 0.36% | 0.19% | 0.06% |
| Lib 4 | 0.09% | 0.31% | 0.78% | 0.76% | 0.83% | 0.34% |
| Lib 5 | 0.64% | 0.65% | 0.76% | 0.50% | 1.10% | 0.00% |
| Lib 6 | 0.40% | 0.22% | 0.44% | 0.66% | 1.04% | 0.13% |
| Lib 7 | 0.20% | 0.30% | 0.55% | 0.59% | 1.31% | 0.10% |
| Lib 8 | 0.18% | 0.31% | 0.92% | 0.62% | 2.25% | 0.40% |
| Lib 9 | 0.35% | 0.31% | 0.78% | 0.17% | 0.20% | 0.10% |
| Mean | 0.53% | 0.88% | 0.67% | 0.46% | 0.80% | 0.13% |
| STDEV | 0.58% | 1.52% | 0.16% | 0.21% | 0.72% | 0.14% |
| STDEV/Mean | 0.24 | 0.46 | 0.89 | 1.08 | ||
Table 1 The evaluations of the library qualities. We found very low rRNA and mitochondrial mRNA content (most less than 1%), and relatively constant expression of constitutive housekeeping genes such as G3PD compared to Tublin and MADS. We calculated the mean and standard deviation to compare the expression variety of these genes. The good quality of the libraries allows for our further analyses.
Sequence quality check
We collected a total of 86,136 EST sequences after quality assessment (trimmed at Q20, Phred scores) and follow-up filters. Fig. 1, Fig. 2 show the length and quality distribution, and clone duplication check of the sequences. We found no sequences with name duplication, and 6 sequences that have less than 100 bp nucleotides left after masking of vector sequences. These sequences had all been filtered out subsequently. To do library clone duplication check, we ran pairwise sequence comparison within each library using BLASTN, and grouped sequences that have more than 90% overall similarity. Five non-normalized libraries, constructed by Krizman protocol 1 (Lib281), LTI non-normalized (Lib6346), Soares non-normalized (Lib185) and Krizman protocol 2 (Lib675 and Lib774), were used as controls. We believe our libraries are quite good compared to the controls.
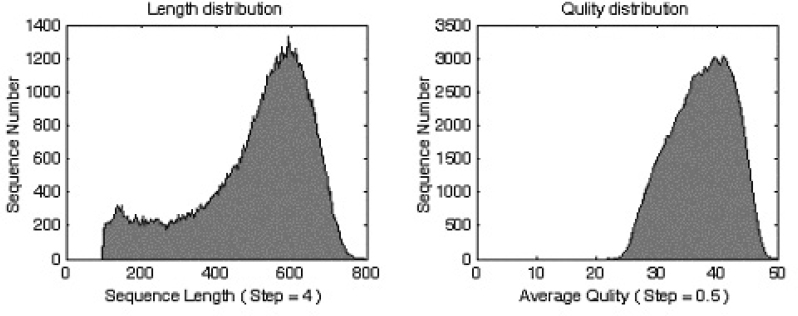
Fig. 1.
The figure on the left is the length distribution of all the EST sequences passing our quality check. The X-axis is the sequence length. The Y-axis is the number of sequences within the range of sequence length indicated by X-axis with an increase step of 4 bp. Note that in our filter we discarded all ESTs shorter than 100 bp after head/tail trimming and vector masking. The figure on the right is the average quality distribution of all the EST sequences passed our quality check. The X-axis is the average sequence quality score. The Y-axis is the number of sequences within the range of sequence quality score indicated by X-axis with an increase step of 0.5.
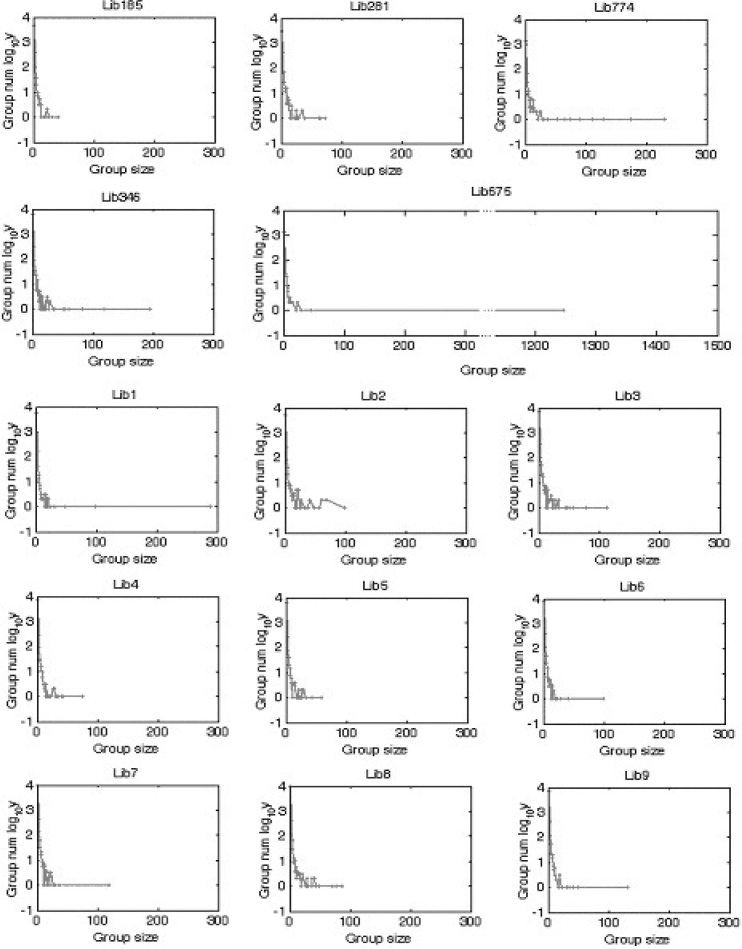
Fig. 2.
We ran pairwise sequence comparison within each library using BLASTN, and grouped sequences that have more than 90% overall similarity. The X-axis stands for the group size or the number of sequences in one group. The Y-axis is the log of the group numbers for every group size. We’ve done the check to all the 9 libraries (Lib1—Lib9). And we used 5 libraries from CGAP (http://cgap.nci.nih.gov/) as controls. These libraries are non-normalized constructed by Krizman protocol 1 (Lib281), LTI non-normalized (Lib6346), Soares non-normalized (Lib185) and Krizman protocol 2 (Lib675 and Lib774). We believe our libraries are quite good compared to the controls.
To find out contribution of our EST data to the discovery of novel rice genes, we compared our ESTs to all available 106,724 public rice ESTs and mRNAs retrieved from NCBI Entrez. 41,076 ESTs have more than 80% overall identity to public rice sequences (BLASTN, E-value 1E-15), and thus about 45,000 ESTs may be considered novel. With the addition of our EST sequences nearly doubling the total number of available rice sequences, it is interesting to take a new look at the gene “landscape” of rice. We pooled together a total of 180,602 sequences from our ESTs and public rice ESTs, and assembled them into 31,543 contigs, 19,279 of which contain two or more ESTs and 12,246 remain singletons. In our gene sequencing process analysis, the rates of rice gene discovery have been plateaued when we look at the alignment length of our EST contigs aligned to rice indica genome working draft, suggesting that the effectiveness of gene finding by further EST sequencing is reduced. This was done by progressively aligning rice EST contigs with rice indica genomic scaffolds, and with Arabidopsis genes from TAIR database (The Arabidopsis Information Resource, (6)). To avoid the error derived from genome duplication, which is common in Arabidopsis and very likely in rice, each contig/EST could only be aligned with genomic sequence once. Although the curve of the rice contig number slightly went down, it still needs tens of thousands sequences to get plateaued, probably because ESTs are random samples of gene sequences, especially when we pooled all available ESTs, there are 5’ and 3’ sequencing that would double the contig numbers. But when we align the consensi to genome, or another control data set such as Arabidopsis genes, we would see the curve become flat a little bit earlier than the EST contig numbers (Fig. 3).
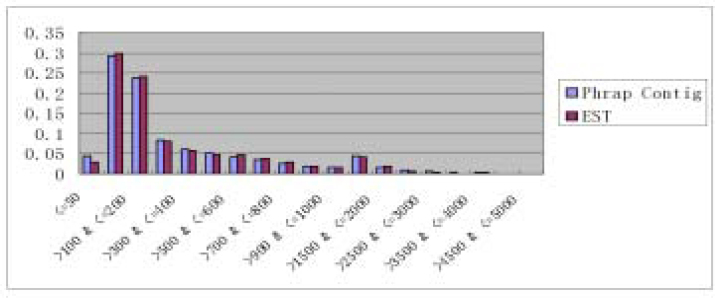
Fig. 3.

Contribution of our EST data to rice gene set. The ESTs in this study were combined with public rice ESTs and then aligned with rice indica genomic scaffolds by BLASTN. The BLAST threshold was set at E-value less than 1E-5. Y-axis of the circles represents the total matched genomic sequence length. Y-axis of the stars represents the contig number of the progressive assembly. The rectangles are the number of the progressively generated contigs that Arabidopsis gene hits. They share the same Y-axis with the stars, but note that we increased the Y value of hit number by 10 folds to make the points easy to read. To avoid the error derived from genome duplication, which is common in Arabidopsis and very likely in rice, each contig/EST could only be aligned with genomic sequence once.
Clustering
To minimize the EST assembly error, we first compared the effectiveness of three assembly algorithms: Phrap, CAP3 (2) and CAT 7., 8.. Both the consensi of CAP3 and Phrap have higher alignment percentage, which indicates that the clustering step had overcome some sequence errors in raw EST data. When we used a specific clustering tool, higher alignment percentage was found when we compare ESTs/contigs and genome sequences from the same subspecies. CAP3 gave contig consensi that were aligned to the genomic scaffolds the best when sequence number went up to more than eighty thousands (Table 2). We chose Phrap after considering the trade-offs among consensus quality, clustering time, and memory requirement. We chose Phrap after considering the trade-offs among consensus quality, clustering time, and memory requirement.
Table 2.
EST Assembly Evaluation
|
Oryza sativa L. ssp. japonica Genome: hsp/query |
Oryza sativa L. ssp. indica Genome: hsp/query |
|
|---|---|---|
| EST | ||
| BGI | 85.93% | 90.33% |
| japonica (Nipponbare) | 91.56% | 89.56% |
| indica (93-11) | 86.07% | 90.76% |
| Consensus CAT | ||
| BGI | 77.73% | 79.62% |
| japonica | 80.50% | 80.55% |
| indica | 79.88% | 81.47% |
| Consensus CAP3 | ||
| BGI | 87.02% | 92.28% |
| japonica | 86.42% | 88.74% |
| indica | 88.76% | 90.10% |
| Consensus Phrap | ||
| BGI | 87.89% | 90.89% |
| japonica | 91.82% | 89.67% |
| indica | 88.61% | 91.17% |
We aligned all ESTs with contig consensi by BLASTN to automatically detect chimeric contigs, and reran Phrap with those EST sequences that were in chimeric contigs. In our 32,489 contigs of 86,136 ESTs, we re-ran Phrap on 5,618 ESTs or 157 contigs and increased the contig number by 167. To evaluate the assembly error rate, we aligned ESTs and EST contig consensi to rice indica genomic scaffold (9) using BLASTN and Sim4 10., 11.. Assuming the gaps between BLASTN HSPs are mostly introns, we found that there was no significant difference between HSP gap size distribution of contig consensi and of EST sequences (Fig. 4), which indicated the EST assembly was quite good. We further identified individual chimeric contigs using their BLAST subject sequence annotation. An EST contig was suspected chimeric if a part of it was aligned with several known sequences (in NCBI none-redundant or Swissprot databases), and another part of it was aligned with some other known sequences. A following manual check indicated that there are almost no chimeric contigs.
Fig. 4.
To check out chimerics, we aligned both raw data (ESTs) and contig consensi to rice genome. This figure shows the putative intron length distribution by aligning ESTs and contig consensi with rice indica genomic scaffolds using BLASTN (E-value 1E-15). HSPs with identity length greater than 70% of the contigs/ESTs were chosen. The gaps between two HSPs were putative introns. We found that 524 contigs have introns longer than 2 kb but shorter than 5 kb, and 237 contigs have introns longer than 5 kb.
During alignment of the contig consensi to rice indica genome by BLASTN, a forced joint was made if two contigs have overlap region on the genome. A total of 3,926 contigs were merged, resulting in reduction of our contig numbers by 32,489 to 30,222. This is validated by 963 rice cDNAs (complete CDS) from GenBank.
Complete ORF finding
We did ORF finding in assembled contig/ESTs, and extracted the longest complete ORFs in each contig. Totally 28,088 potential ORFs (length>99 bp) were found, and the maximum length was 2,790 bp.
Function assignment and classification
To assign annotation to contig sequences, we first used BLASTN to search the NCBI non-redundant (nr) database (E-values 1E-15). The same algorithm developed in Uniblast (Bioinformatics accepted, 2002) was used to figure out a gene symbol in the description lines of the hits. And we used BLASTX to search the Swissprot database. If BLASTX returned one or more sequences with E-value less than 1E-10, then the annotation of the highest scoring sequence was assigned to the rice contig. 4,407 contigs/ESTs were assigned annotations by BLASTN and 5,881 contigs/ESTs were assigned annotations by BLASTX, 24,807 contigs/ESTs could not be annotated by either BLASTN or BLASTX. After all, we annotated 7,682, or 23.6% of all the 32,489 rice EST contigs (sequenced in Beijing Genomics Institute (BGI)).
We classified 32,489 contigs of 86,136 ESTs sequenced in our center to GO (12) catalogues using the GO indices for Swissprot proteins and the GO indices for Arabidopsis proteins. We compared the results to the classification of 53,398 predicted genes of rice indica genome (9) using the same method. First, we found that though the percentage of genes or EST contigs classified into each category changed very slightly (data not shown), the actual classified contig numbers changed a lot. Generally speaking, less EST contigs were classified into GO categories when we used indices for Arabidopsis proteins. To be more specific, 792, 2,486 and 1,221 EST contigs were classified in cell component, molecular function and biological process through the GO indices for Arabidopsis proteins. And 4,354, 4,457 and 4,451 were classified in the same catalogues using Swissprot indices. Second, we found that most of the GO categories contained more genome predicted genes than EST contigs. This is not a surprise because EST projects only detect active (expressed) genes (Fig. 5).
Fig. 5.
The comparison between different GO (12) catalogues of predicted genes on rice indica genome (total 53,398 genes) classified by GO indices for Swissprot proteins, EST contigs (total 32,489 contigs, 86,136 ESTs) classified by GO indices for Swissprot proteins, and EST contigs classified by GO indices for Arabidopsis proteins. The Y-axis stands for different GO categories in molecular function and biological process. The X-axis was the gene/contig numbers linked to the specific category. To make the figures readable, log numbers were used here.
Table 2 ESTs and contig consensi generated by three algorithms were aligned to Syngenta’s published rice japonica genome sequence and indica genomic contigs (9) by BLASTN. The percentage of the identical alignment was calculated as the aligned length divided by the total EST/contig length. BGI stands for our 9 libraries with more than eighty thousand ESTs. 93 – 11 stands for the indica library we sequenced, which has 8,190 ESTs (Table 1). Japonica ESTs are those Nipponbare libraries containing 66,728 sequences Oryza sativa L. ssp. japonica genome comes from (www.tmri.org, (13)), which includes 42,109 sequences with 389,809,244 total nucleotides. Oryza sativa L. ssp. indica genome includes 127,550 sequences and 359,419,680 nucleotides (9). Both the consensi of CAP3 and Phrap have higher alignment percentage, which indicates that the clustering step had overcome some sequence errors in raw EST data. When we use a specific clustering tool, higher alignment percentage was found when we compare ESTs/contigs and genome sequences from the same subspecies. The alignment percentage was slightly lower than the number in Oryza sativa L. ssp. indica genome paper (90.3% vs. 92.0%) (9), because we’ve taken out the overlap regions of HSPs returned by BLAST this time.
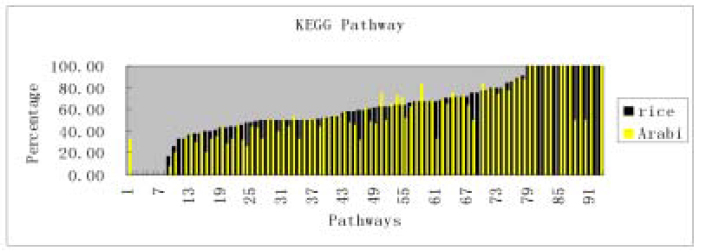
We further compared the frequencies of both rice and Arabidopsis ESTs that were assigned to 93 metabolism pathways defined by KEGG (http://www.genome.ad.jp/kegg/, 13., 14.). A total of 180,602 rice ESTs had been used here, which include both public and our new ESTs, different cultivas (LYP9, PA64s, 93 – 11), tissues (leaf, panicle) and different development stages (trefoil, tillering, booting). A total of 99,426 Arabidopsis ESTs had been used here, which include different tissues (Dry seeds, green siliques, inflorescence) and different development stages (cycling cells, greenhouse plants, two to six-week old). We chose non-normalized libraries to make sure the results are comparable. 2,4-Dichlorobenzoate degradation, Biphenyl degradation, Blood group glycolipid biosynthesis—lact series, Blood group glycolipid biosynthesis—neolact series, Fluorene degradation, Retinol metabolism and Xylene degradation did not have any matches either in rice or in Arabidopsis. Besides, 1,4-Dichlorobenzene degradation did not have matches in rice. D-Alanine metabolism, Chondroitin / Heparin sulfate biosynthesis, Atrazine degradation, Glycosylphosphatidylinositol (GPI)-anchor biosynthesis and Tetrachloroethylene degradation did not have matches in Arabidopsis. To find matches we ran BLASTX of rice and Arabidopsis ESTs against full-length cDNAs defined in KEGG with threshold E-value 1E-10 and overall identity 30% (Fig. 6).
Fig. 6.
The coverage difference between Arabidopsis thaliana and rice ESTs. A total of 180,602 rice ESTs had been used here, which include different cultivars (LYP9, PA64s, 93 – 11), tissues (leaf, panicle) and different development stages (trefoil, tillering, booting). A total of 99,426 Arabidopsis ESTs had been used here, which include different tissues (Dry seeds, green siliques, inflorescence) and different development stages (cycling cells, greenhouse plants, two- to six-week old). We chose non-normalized libraries to make sure the results are comparable. Each column stands for a metabolism pathway defined in KEGG. The height of the bar means the percentage of the enzymes that found matches in Arabidopsis thaliana (light) and rice (black) ESTs of that pathway. To find matches we ran BLASTX of rice and Arabidopsis ESTs against full length CDS defined in KEGG with threshold E-value 1E-10 and overall identity 30%.
Table 3 The genes with the greatest differences in relative EST abundance between the 93 – 11 parental variety and the high-yield variety LYP9 in tillering stage. The genes that show the largest differences include those involved in photosynthesis and protein synthesis. Table 4 shows LYP9 genes with the greatest differences in relative EST abundance between the tillering and trefoil development stages. Table 4 shows the genes with the greatest differences in relative EST abundance in the conditional sterile mutant PA64s with short exposure to sunlight (fertile) versus when grown with extended exposure to sunlight (sterile). The table columns indicate the contig/EST names, the annotation returned by BLASTN (E-value 1E-15, overall identity > 30%, and BLASTX (E-value 1E-10, overall identity > 25%), the change folds and the P-value of Chi-square test. Only the contigs/ESTs having Chi-square P-value less than 1E-6 were listed. You will find multiple entries in the same cell of ‘contig name’, because we’ve merged the contigs/ESTs if they share the position on rice indica genome. Note that change folds less than 1 indicate down-regulated genes in the second libraries in the comparisons.
Table 3.
Genes Most Differentially Expressed between 93-11 (Lib 5) and LYP9(Lib 3) Varieties
| Contig Name | BLASTN Annotation | BLASTX Annotation | Change Folds | Chi-square Test |
|---|---|---|---|---|
| Contig13918 Contig5428 Contig13594 Contig13445 Contig3126 siceg_11012.y1.abd contig13769 |
Avena sativa fructose 1,6-bisphosphate aldolase precursor, mRNA, complete cds; nuclear gene for chloroplast product | (Q40677) Fructose-bisphosphate aldolase, chloroplast precur | 0.24 | 1.74E-10 |
| Contig6621 Contig13700 | Oryza sativa mRNA for ribonuclease, complete cds | Unkown | 39.30 | 5.6E-10 |
| Contig13245 Contig13907 Contig58 rsiceg_5696.y1.abd |
Unkown | (P51327) Cell division protein ftsH homolog (EC 3.4.24.-) | 0.11 | 4.96E-09 |
| Contig13893, Contig698 | Oryza sativa mRNA for the small subunit of ribulose-1,5-bisphosphate carboxylase, complete cds, clone pOSSS2106 | (P18566) Ribulose bisphosphate carboxylase small chain A | 0.03 | 7.15E-09 |
| Contig13906 | Oryza sativa Zn-induced protein (RezA) mRNA, complete cds | Unkown | 33.45 | 1.13E-08 |
| Contig13704 | Oryza sativa Zn-induced protein (RezA) mRNA, complete cds | Unkown | 12.82 | 1.33E-08 |
| Contig13764 rsicek_0875.y1.abd Contig11940 Contig10877 rsiceg_7521.y1.abd |
Oryza sativa hsp70 gene for heat shock protein 70 | (P27322) Heat shock cognate 70 kDa protein 2 | 0.12 | 5.36E-08 |
| Contig13913 | Oryza sativa 25S ribosomal RNA gene | Unkown | 11.43 | 1.12E-07 |
| Contig13767 | Zea mays chloroplast rRNA-operon | Unkown | 2.72 | 2.73E-07 |
| Contig13736 | Polygonum tinctorium mRNA for transketolase, complete cds | (Q43848) Transketolase, chloroplast precursor (EC 2.2.1.1) | 0.11 | 3.02E-07 |
| Contig13914 | Oryza sativa light-induced mRNA | (Q03200) Light regulated protein precursor | 8.57 | 3.83E-07 |
| Contig13680 | Unkown | Unkown | 0.04 | 1.87E-06 |
| Contig13695 | Oryza sativa hsp70 gene for heat shock protein 70 | (P22953) Heat shock cognate 70 kDa protein 1 (Hsc70.1) | 0.08 | 2.58E-06 |
| Contig13920 | Oryza sativa OsrcaA2 mRNA for RuBisCO activase small isoform precursor, complete cds | (P93431) Ribulose bisphosphate carboxylase/oxygenase activa | 0.17 | 3.23E-06 |
| Contig13613 | Unkown | Unkown | 20.90 | 7.49E-06 |
| Contig727 Contig9659 Contig12248 Contig13927 Contig10986 |
Oryza sativa chlorophyll a/b binding protein (kcdl895) mRNA, complete cds | (P06671) Chlorophyll A-B binding protein, chloroplast precu | 0.34 | 8.61E-06 |
| Contig13708 rsiceg_11507.y1.abd |
Triticum aestivum RNA for phosphoribulokinase | (P26302) Phosphoribulokinase, chloroplast precursor (EC 2.7) | 0.20 | 9.26E-06 |
Table 4.
LYP9 Genes Mostly Differentially Expressed between Tillering (Lib 3) and Trefoil (Lib 2) Stages
| MasterContig Name | BLASTN Annotation | BLASTX Annotation | Change Folds | Chi-sqare Test |
|---|---|---|---|---|
| Contig13767 | Zea mays chloroplast rRNA-operon | Unkown | 0.10 | 6.34E-15 |
| Contig13718 Contig13638 Contig12674 |
Oryza sativa mRNA for ferredoxin, complete cds | (P00228) Ferredoxin, chloroplast precursor | 9.24 | 3.57E-13 |
| Contig13727 Contig12420 rsiced_10341.y1.abd rsiced_4570.y1.abd |
Hordeum vulgare chloroplast photosystem I PSK-I subunit mRNA, complete cds | (P36886) Photosystem I reaction center subunit X, chloropla | 8.56 | 1.00E-10 |
| Contig27 Contig13694 rsiced_11896.y1.abd Contig5628 rsiced_3479.y1.abd |
Unkown | Unkown | 15.73 | 2.84E-10 |
| Contig6621 Contig13700 | Oryza sativa mRNA for ribonuclease, complete cds | Unkown | 0.05 | 7.59E-09 |
| Contig13904 | Unkown | (Q40070) Photosystem II 10 kDa polypeptide, chloroplast pre | 10.84 | 8.03E-09 |
| Contig13906 | Oryza sativa Zn-induced protein (RezA) mRNA, complete cds | Unkown | 0.03 | 3.20E-08 |
| Contig13913 | Oryza sativa 25S ribosomal RNA gene | Unkown | 0.06 | 8.52E-08 |
| Contig13751 | Oryza sativa chloroplast carbonic anhydrase mRNA, complete cds | (P40880) Carbonic anhydrase, chloroplast precursor (EC 4.2.) | 4.98 | 8.85E-08 |
| Contig13911 | Oryza sativa chlorophyll a/b binding protein (kcdl895) mRNA, complete cds | (P06671) Chlorophyll A-B binding protein, chloroplast pre | 11.90 | 9.67E-08 |
| Contig13920 | Oryza sativa OsrcaA2 mRNA for RuBisCO activase small isoform precursor, complete cds | (P93431) Ribulose bisphosphate carboxylase/oxygenase activa | 7.23 | 1.00E-07 |
| Contig13704 | Oryza sativa Zn-induced protein (RezA) mRNA, complete cds | Unkown | 0.11 | 1.42E-07 |
| Contig13901 | Oryza sativa mRNA for the small subunit of ribulose-1,5-bisphosphate carboxylase, complete cds, clone pOSSS1139 | (P18567) Ribulose bisphosphate carboxylase small chain C | 22.95 | 3.23E-06 |
| Contig13546 Contig7253 rsiced_4290.y1.abd |
Oryza sativa mRNA for precursor of 22 kDa protein of photosystem II (PSII-S), complete cds | (P54773) Photosystem II 22 kDa protein, chloroplast pre | 12.75 | 4.24E-06 |
| Contig12144 Contig13905 rsiceg_15548.y1.abd |
Oryza sativa chlorophyll a-b binding protein mRNA, complete cds | (P27523) Chlorophyll A-B binding protein of LHCII type III | 7.65 | 4.82E-06 |
| Contig13926 | Oryza sativa chlorophyll a/b binding protein (RCABP89) mRNA, nuclear gene encoding chloroplast protein, complete cds | (P27519) Chlorophyll A-B binding protein, chloroplast pre | 2.59 | 5.92E-06 |
| Contig13723 Contig150 rsicee_817.y1.abd |
Unkown | Unkown | 0.09 | 7.37E-06 |
| Contig13715 Contig1541 | Unkown | (P27522) Chlorophyll A-B binding protein 8, chloroplast pre | 5.74 | 7.54E-06 |
| Contig13576 | Oryza sativa mRNA for RicMT, complete cds | Unkown | 12.11 | 8.20E-06 |
| Contig13914 | Oryza sativa light-induced mRNA | (Q03200) Light regulated protein precursor | 0.19 | 9.04E-06 |
| Contig4926 Contig13475 siced_4355.z1.abd |
Oryza sativa RNase S-like protein mRNA, complete cds | (P42815) Ribonuclease 3 precursor (EC 3.1.27.1) | 7.33 | 9.11E-06 |
| Contig13604 | Oryza sativa mRNA for RicMT, complete cds | Unkown | 7.33 | 9.11E-06 |
| Contig727 Contig9659 Contig12248 Contig13927 Contig10986 |
Oryza sativa chlorophyll a/b binding protein (kcdl895) mRNA, complete cds | (P06671) Chlorophyll A-B binding protein, chloroplast pre | 2.98 | 9.64E-06 |
Expression profile analysis
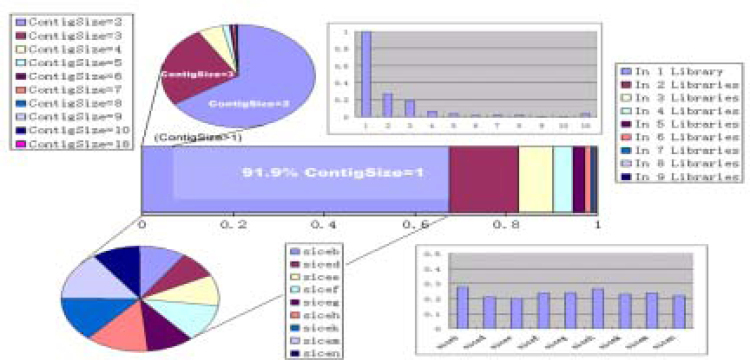
We profiled gene expression in different cultivars, developmental stages, and growth conditions, using EST abundance as an approximation. ESTs in each library were assembled and analyzed separately (Table 6). Noted that our sequence number may not be enough to cover all the genes expressed in a particular EST library, we firstly drew a whole picture of the gene expression pattern (Fig. 7). We found that nearly 65% of the genes existed uniquely in our nine libraries, so we offset the EST copy numbers by one for every gene discovered to make these genes comparable. And we found these 9 libraries contributed almost equally to those uniquely existing genes, which implies these libraries are comparable by means of sequence coverage. This result encouraged us to go further to the library-library expression profile comparison.
Table 6.
Description of the Surveyed Rice cDNA Libraries and the Number of EST Sequenced in Each Library
| Library | Tissue | Cultival | Stage | Condition | Phenotype | Sequences | Contigs (size>l) | Chimeric | Singletons | Annotated | Novel |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lib 1 | leaf | PA64s | trefoil | 7,074 | 801 | 3 | 3,568 | 848 | 3,521 | ||
| Lib 2 | whole plant | LYP9 | trefoil | 7,682 | 940 | 1 | 3,462 | 947 | 3,455 | ||
| Lib 3 | whole plant | LYP9 | tillering | 9,795 | 1,406 | 1 | 4,355 | 1,233 | 4,520 | ||
| Lib 4 | panicle | PA64s | heading/flowering | high temperature, long sunlight | sterile | 9,483 | 1,213 | 0 | 5,032 | 1,041 | 5,204 |
| Lib 5 | whole plant | 93-11 | tilering | 8,190 | 1,015 | 5 | 4,403 | 958 | 4,460 | ||
| Lib 6 | panicle | PA64s | heading/lowering | high temperature, short sunlight | fertile | 10,003 | 1,355 | 2 | 5,569 | 893 | 6,031 |
| Lib 7 | panicle | PA64s | heading/flowering | high temperature, short sunlight | fertile | 12,053 | 1,827 | 0 | 5,443 | 1,106 | 6,164 |
| Lib 8 | panicle | PA64s | heading/flowering | high temperature, long sunlight | sterile | 12,708 | 1,948 | 0 | 5,796 | 1,210 | 6,534 |
| Lib 9 | whole plant | LYP9 | booting | 9,148 | 1,386 | 0 | 4,393 | 946 | 4,833 | ||
| Total | 86,136 | 11,891 | 12 | 42,021 | 9,182 | 44,722 | |||||
Fig. 7.
An overview of the expression patterns of every gene in the nine libraries we’ve sequenced. The bar in the middle shows the percentage of gene expressions in one library only and two libraries and so on to 9 libraries. Not surprisingly, about 91.9% of the uniquely expressed genes are singletons. Unique genes that have more than one EST are showed in the upper pie. Relative abundance of unique expressed genes in genes having the same contig size is showed in the upper bar chart. The X-axis is the contig size, or the ESTs in the contigs, the Y-axis is the number of uniquely expressed genes divided by the total number of the genes having the same contig size. Not surprisingly, the singletons or the contigs with size one are a hundred percent unique genes. The lower pie chart shows the contributions (contig numbers) of libraries to uniquely expressed genes. The lower bar chart is the relative contribution of each library. The X-axis stands for libraries, the Y-axis is the unique gene numbers divided by the total EST numbers in that library.
Discussion
Oryza sativa L. ssp. indica and japonica are two subspecies close to reproduction separation. They have 16% of genomic sequence difference (9). However, ESTs from indica and japonica align to indica genomic scaffolds and japonica genome data with very little difference in percentage of similarity (Table 2), indicating that the sequence variation of gene transcripts between these two subspecies is insignificant. This suggests that intergenic regulatory regions play important roles that remain to be uncovered.
Overall, the gene “landscape” in Fig. 5 is similar to that reported for rice and Arabidopsis by the Institute for Genomic Research (TIGR, http://www.tigr.org/tdb/ogi/GO/GO.html). The differences among the relative proportions in each class may be attributable to several factors, including the difference among japonica, indica, and Arabidopsis, the greater number of rice ESTs in our study, additional annotation, and the use of different tissues and development stages. One needs to take caution in interpreting the number of genes in each GO category. The assignment of genes to the GO function hierarchies is based on the annotation of known genes with similar sequences. This annotation may not reflect the true function of the rice gene in some cases. In addition, about one third of the genes were not sufficiently similar to any known gene and were not assigned any annotation; once the functions of these genes are determined, they will also likely change the relative numbers in each category.
Fig. 7 provides a whole view of gene expression in 9 libraries, in which nearly 65% of the represented genes existed in one library only. Among those genes, 91.9% are actually singletons, which most likely to be the result of random sampling rather than library specific. The upper pie chart in Fig. 7 grouped only-library contigs by their contig size. The contigs have more ESTs are considered to be more likely to be library specific genes. The lower pie chart shows the 9 libraries contributed equally to the ‘unique’ genes, which implies these libraries are comparable by means of sequence coverage. This result encouraged us to go further to the library-library expression profile comparison.
Because EST abundance is an imperfect approximation of gene expression level, we only look for genes for which the relative EST abundance is highly varied between the libraries, in which case the true gene expression levels are more likely to be different. Genes that are expressed at low levels or have smaller changes may also contribute to the phenotypic differences, though they are not detected by this experimental method.
In the comparison of paternal 93 – 11 with the high-yield F1 LYP9 (Table 3), the elevation of Fructose-bisphosphate aldolase in the LYP9 library may indicate the increased photosynthesis activities. FtsH is a cell division protein that seems to act as an ATP-dependent zinc metallopeptidase. Its increased expression in the LYP9 library may also indicate an accelerated cell division. Phosphoribulokinase is a Calvin cycle related protein that is light-regulated via thioredoxin by reversible oxidation/reduction of sulfhydryl/disulfide groups. Its elevation in the LYP9 cultivars libraries may explain the increased protein synthesis.
In the comparison of LYP9 in trefoil stage versus tillering stage (Table 4), the genes that show the largest differences overlap the genes in Table 4. This may indicate that these genes are mostly involved in plant growth, resulting in either high yield or maturation. We expected, from previous studies, that the transcription factor ERF would appear late in development, and that the MADS box containing transcription factors would appear during flower development 15., 16.. MADS box genes play important roles in flower formation and floral organ identity determination. Most MADS genes are expressed in the treproductive phases; very few are expressed in the vegetative phases. These expectations were confirmed in the comparison of the developmental stages. MADS gene contents in the libraries Lib 6, 7, 8 and 8 (all heading/flowering stages) are five or more fold greater than in Lib 1, 2 and 3 (trefoil and tillering stages).
Materials and Methods
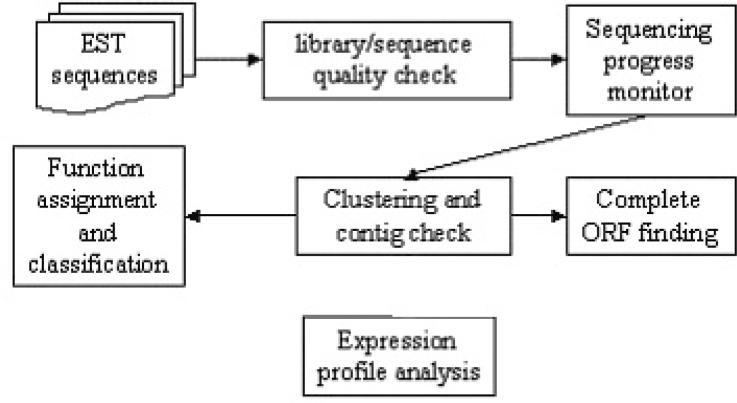
We describe in detail the materials used and methods developed in the sequencing and analysis of rice ESTs. Fig. 8 shows an overall workflow of the primary components of our analyses.
Fig. 8.
An overview of the relationship of our EST sequence analysis methods. After library and sequence quality check, high quality EST sequences of good libraries went through sequencing progress monitor to make sure enough sequences had been collected. Then high quality none-redundant dataset were generated by clustering and contig check. Complete ORF search, function assignment and classification and expression profile analysis were performed on those carefully checked EST contigs.
Library information
We sequenced nine cDNA libraries from three cultivars of Oryza sativa. Table 6 describes these nine directional cDNA libraries in detail. The three cultivars include a super high-yield hybrid Liang-You-Pei-Jiu (LYP9), its paternal variety 93-11 (indica), its maternal variety Pei-Ai 64s (PA64s, an indica-japonica hybrid). Oryza sativa L. ssp. indica is a common rice subspecies grown as field crops in China and many other Asian-pacific regions. We prepared whole plant, panicle, and leaf libraries at the trefoil, tillering, and heading/flowering developmental stages. In addition, we made libraries from the maternal variety PA64s grown at high temperature (27-28 °C). At high temperature, this conditional mutant is fertile when grown with short exposure to sunlight (12 h/d) and sterile when grown with extended exposure to sunlight (14.5 h/d). The libraries were not normalized in order to provide a rough estimate of the gene expression levels. The EST sequences are available through http://rice.genomics.org.cn/.
Sequence quality check
Clones from the libraries were randomly selected for single-pass, mostly 5’ sequencing to yield ESTs. The libraries were not normalized in order to preserve the random nature of the original expression patterns for quantitative analysis. We used the Phred program for base calling (17), Cross_match for vector sequences masking, and Phrap for sequence assembly. To do library clone duplication check we ran self sequence comparison within each library using BLASTN, and grouped sequences that have more than 90% overall similarity. Five publicly available human non-normalized EST libraries, constructed by Krizman protocol 1 (Lib281), LTI non-normalized (Lib6346), Soares non-normalized (Lib185) and Krizman protocol 2 (Lib675 and Lib774), were used as controls.
106,724 public rice ESTs and mRNAs were retrieved from NCBI Entrez. We used them to check the contribution of our EST data by BLASTN (E-value 1E-15, overall identity 80%). In public rice sequences, 94,466 were ESTs. We pooled these ESTs with our 86,136 ESTs, which resulted in a total of 180,602 sequences. Sequencing process analysis was done by progressively and randomly sampling these rice EST sequences and clustering them by Phrap (Phill Green, unpublished). We used a loose parameter to allow sequence variation between subspecies. Further more, we aligned the contig consensi, clustered in each library, with rice indica genomic scaffolds (9) and with Arabidopsis genes from TAIR database (The Arabidopsis Information Resource, (6)) to avoid the potential problem of double counting genes because there were both 5’ and 3’ sequencing for public rice ESTs. To avoid the potential error derived from genome duplication, which is common in Arabidopsis and very likely in rice, each contig/EST could only be aligned with genomic sequence once.
Table 6 Nine cDNA libraries from three cultivars of Oryza sativa. The three cultivars include a super high-yield hybrid Liang-You-Pei-Jiu (LYP9), its paternal variety 93 – 11 (indica) and its maternal variety PA64s. We prepared whole plant, panicle, and leaf libraries at the trefoil, tillering, and heading/flowering developmental stages. In addition, we made libraries from the maternal variety PA64s grown at high temperature (27-28 °C). The libraries were not normalized in order to provide a rough estimate of the gene expression levels. We collected a total number of 86,136 EST sequences after quality assessment and trimming at Q20 (Phred scores). Sequences in each library were assembled by PHRAP. The contig (containing more than one EST) number, the singleton number, the annotated contig/singleton number and the novel contig/singleton number are listed. Annotation was done by BLASTN to NCBI none-redundant database with threshold of E-value 1E-15 and overall identity 30% and BLASTX to Swissprot protein database with threshold of E-value 1E-15 and over all identity 25%.
Clustering
To minimize the EST assembly error, we compared the effectiveness of three assembly algorithms: Phrap, CAP3 (2) and CAT 7., 8.. We finally chose Phrap after considering trade-offs among consensus quality, clustering time, and memory requirement. After Phrap assembly, we aligned all ESTs with contig consensi by BLASTN to automatically detect chimeric contigs, and reran Phrap with those EST sequences that were in chimeric contigs. We further identified individual chimeric contigs using their BLAST subject sequence annotation. An EST contig was suspected chimeric if a part of it was aligned with several known sequences (in NCBI none-redundant or Swissprot databases), and the other part of it aligned with some other known sequences. To evaluate the assembly error rate, we aligned ESTs and EST contig consensi to rice indica genomic scaffold (9) using BLASTN and Sim4 10., 11.. A forced joint was made if two contigs have overlap region on the genome.
Complete ORF finding
GetORF (http://www.hgmp.mrc.ac.uk/Software/EMBOSS/) was used to search potential Open Reading Frame (ORF) in assembled contigs/ESTs. GC content gradient feature is used to check the completeness of the ORF. If the start codon of a potential ORF has GC gradient feature, it was considered more likely to be a complete CDS.
Function assignment and classification
To assign annotation to contig sequences, we first used BLASTN to search the NCBI non-redundant (nr) database (E-values 1E-15). The same algorithm developed in UniBlast (Bioinformatics accepted, 2002) was used to figure out a gene symbol in the description lines of the hits. And we used BLASTX to search the Swissprot database. If BLASTX returned one or more sequences with E-value less than 1E-10, then the annotation of the highest scoring sequence was assigned to the rice contig. If neither BLASTN nor BLASTX returned a sequence that passed the criteria, then the rice contig sequences or ESTs was not assigned any annotation, but was subject to Pfam to search for functional domains. If contig/EST had annotation, it’ll be classified into Gene Ontology categories (http://www.geneontology.org/, (12)). We further compared the frequencies of both rice and Arabidopsis ESTs that were assigned to 93 metabolism pathways defined by KEGG (http://www.genome.ad.jp/kegg/, 13., 14.). To find matches we ran BLASTX of rice and Arabidopsis ESTs against full-length cDNAs defined in KEGG with threshold E-value 1E-10 and overall identity 30%.
Our annotation and classification was based on data collected and extracted from the following public databases, data and files:
GenBank release 129.0
SWISSPROT release 40.0.
And the following files are from Gene Ontology Consortium:
gene_association.tair version 1.3, 10/10/2001
gene_association.goa version 1.3, 10/10/2001
function.ontology version 1.311, 28/03/2002
component.ontology version 1.311, 28/03/2002
process.ontology version 1.311, 28/03/2002
Expression profile analysis
Genes expressed in two libraries were compared using a master gene set created from all the 86,136 ESTs clustered and annotated in this study. After carefully clustering the ESTs into contigs, we check EST sequence names in each contig to find out their library origin. Because all libraries are not normalized, and we may miss genes with low expression level in most EST projects, we offset the EST copy number by one for all genes. Then we subtracted the expression of the same master gene in each library to produce the differential value of that gene. Finally these differential values were ranked to produce the up-regulated and down-regulated gene list.
Table 5.
Genes Mostly Differentially Expressed in PA64s between Short Sunlight (Fertile, Lib 6 and 7) and Long Sunlight (Sterile, Lib 4 and 8)
| MasterContig Name | BLASTN Annotation | BLASTX Annotation | Change Folds | Chi-square Test |
|---|---|---|---|---|
| Contig8033 Contig13583 Contig13282 Contig12566 Contig10517 Contig11574 Contig13743 Contig12532 Contig1611 |
Oryza sativa mRNA for novel protein, osr40c1 | Unknown | 10.52 | 5.93E-23 |
| Contig13748 | Unknown | Unknown | 5.76 | 2.27E-09 |
| Contig13702 Contig972 Contig716 Contig12348 Contig12780 |
Oryza sativa APXb mRNA for L-ascorbate peroxidase, complete cds | (Q05431) L-ascorbate peroxidase, cytosolic (EC 1.11.1.11) | 6.46 | 4.67E-09 |
| Contig13724 Contig964 Contig13691 rsicek_1248.y1.abd |
Oryza sativa mRNA for sucrose synthase | (P30298) Sucrose synthase 1 (EC 2.4.1.13) | 0.24 | 5.11E-09 |
| Contig13413 Contig13912 | Oryza sativa GF14-C protein mRNA, complete cds | (Q9SP07) 14-3-3-like protein | 4.47 | 1.13E-08 |
| Contig13637 Contig13705 Contig13584 Contig12438 |
Zea mays plasma membrane integral protein ZmPIP2-l mRNA, complete cds | (P42767) Aquaporin | 3.01 | 2.04E-08 |
| Contig5735 Contig13762 Contig13678 rsicef_9381.y1.abd Contig13184 Contig944 Contig10283 rsicee_1225.y1.abd rsiced_2665.y1.abd rsiceh_22549.y1.abd rsicef_12473.y1.abd rsiceh_20108.y1.abd |
Oryza sativa mRNA for aquaporin, complete cds | (Q08733) Plasma membrane intrinsic protein 1C | 2.61 | 5.32E-08 |
| Contig13617 | Unknown | (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor | 0.10 | 4.93E-07 |
| Contig11991 Contig13740 Contig917 Contig9907 Contig13488 Contig12338 Contig12990 Contig8794 |
Zea mays methionine synthase mRNA, partial cds | (Q42699) 5-methyltetrahydropteroyl-triglutamate—homocystein | 0.32 | 1.04E-06 |
| Contig13681 rsiceg_11955.y1.abd Contig3125 |
Oryza sativa mRNA for ribosomal protein S4 | (O22424) 40S ribosomal protein S4 | 5.63 | 3.33E-06 |
| Contig8371 Contig12088 Contig13877 rsicen_21644.y1.abd Contig10603 Contig13875 Contig3069 rsiceh_8473.y1.abd rsicek_11431.y1.abd siceh_0191.z1.abd Contig12354 Contig13930 Contig13919 Contig47 rsicef_6881.y1.abd sicef_0294.z1.abd rsicef_6584.y1.abd |
Oryza sativa mRNA for EF-1 alpha, complete cds | (O64937) Elongation factor 1-alpha (EF-1-alpha) | 0.63 | 5.76E-06 |
| Contig13741 Contig12901 Contig13527 Contig430 rsicef_2367.y1.abd |
Oryza sativa mRNA for gamma-Tip, complete cds | (P50156) Tonoplast intrinsic protein, gamma (Gamma TIP) | 3.16 | 6.89E-06 |
| Contig13766 Contig12761 Contig12457 Contig13394 Contig12389 Contig6925 |
Oryza sativa gene for heat shock protein 82 HSP82 | (P33126) Heat shock protein 82 | 0.47 | 7.7E-06 |
| Contig13917 | Oryza sativa high mobility group protein (HMG) mRNA, complete cds | Unknown | 4.83 | 9.8E-06 |
References
- 1.Adams M.D. 3,400 new expressed sequence tags identify diversity of transcripts in human brain. Nat. Genet. 1993;4:256–267. doi: 10.1038/ng0793-256. [DOI] [PubMed] [Google Scholar]
- 2.Huang G.M. Prostate cancer expression profiling by cDNA sequencing analysis. Genomics. 1999;59:178–186. doi: 10.1006/geno.1999.5822. [DOI] [PubMed] [Google Scholar]
- 3.McCombie W.R. Caenorhabditis elegans expressed sequence tags identify gene families and potential disease gene homologues. Nat. Genet. 1992;1:124–131. doi: 10.1038/ng0592-124. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y.H. EST analysis of gene expression in early cleavage-stage sea urchin embryos. Development. 1999;126:3857–3867. doi: 10.1242/dev.126.17.3857. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto K., Sasaki T. Large-scale EST sequencing in rice. Plant Mol. Biol. 1997;35:135–144. [PubMed] [Google Scholar]
- 6.Huala E. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001;29:102–105. doi: 10.1093/nar/29.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou A., Burke J. CRAWview: for viewing splicing variation, gene families, and polymorphism in clusters of ESTs and full-length sequences. Bioinformatics. 1999;15:376–381. doi: 10.1093/bioinformatics/15.5.376. [DOI] [PubMed] [Google Scholar]
- 8.Burke J. d2_cluster: a validated method for clustering EST and full-length cDNA sequences. Genome Res. 1999;9:1135–1142. doi: 10.1101/gr.9.11.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 10.Altschul S.F. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Florea L. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner M. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakao M. Genome-scale Gene Expression Analysis and Pathway Reconstruction in KEGG. Genome Inform. Ser. Workshop Genome Inform. 1999;10:94–103. [PubMed] [Google Scholar]
- 14.Ogata H. Computation with the KEGG pathway database. Biosystems. 1998;47:119–128. doi: 10.1016/s0303-2647(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 15.Chung Y.Y. Early flowering and reduced apical dominance result from ectopic expression of a rice MADS box gene. Plant. Mol. Biol. 1994;26:657–665. doi: 10.1007/BF00013751. [DOI] [PubMed] [Google Scholar]
- 16.Jack T. Plant development going MADS. Plant Mol. Biol. 2001;46:515–520. doi: 10.1023/a:1010689126632. [DOI] [PubMed] [Google Scholar]
- 17.Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]