Abstract
The sudden outbreak of severe acute respiratory syndrome (SARS) in 2002 prompted the establishment of a global scientific network subsuming most of the traditional rivalries in the competitive field of virology. Within months of the SARS outbreak, collaborative work revealed the identity of the disastrous pathogen as SARS-associated coronavirus (SARS-CoV). However, although the rapid identification of the agent represented an important breakthrough, our understanding of the deadly virus remains limited. Detailed biological knowledge is crucial for the development of effective countermeasures, diagnostic tests, vaccines and antiviral drugs against the SARS-CoV. This article reviews the present state of molecular knowledge about SARS-CoV, from the aspects of comparative genomics, molecular biology of viral genes, evolution, and epidemiology, and describes the diagnostic tests and the anti-viral drugs derived so far based on the available molecular information.
Key words: severe acute respiratory syndrome, SARS-CoV, genome, phylogenetics, human leukocyte antigen (HLA) system, molecular epidemiology
Introduction
The first SARS case was reported in late 2002 in China’s Guangdong Province (1). The disease was contagious and spreaded rapidly, resulting in a SARS outbreak in Hong Kong in mid-February 2003, and other outbreaks elsewhere in the world. At the end of March 2003, a virus of the Coronaviridae family was identified as the causative agent of the disease 2., 3., 4.. This identification has been confirmed by the World Health Organization, and the virus concerned has been designated as the SARS-associated coronavirus (SARS-CoV). During the SARS outbreaks in 2002 and 2003, SARS cases were identified in 19 countries, and in total 8,605 individuals became infected, of whom 774 died (http://www.who.int/csr/sars/country/table2003_09_23/en/).
In addition to its cost in human lives, the SARS outbreak also had a great impact on the health care system and economy of Hong Kong and other infected regions. In Hong Kong, the estimated economic loss was about HK$46 billion (US$5.9 billion; ref. 5). The possibility that SARS-CoV transmission can occur between human beings without reinforcement from the animal reservoir (5) and the capability of the virus to infect multiple cell types (6) and animals (7) further increased the epidemiological burden of the SARS pandemic. Although the spread of the virus had seemed to be confined by July 2003 through rigorous quarantine measures (http://www.who.int/csr/sars/country/table2003_09_23/en/), it may still be circulating in the animal reservoir and it is impossible to say that it will not return 8., 9., 10.. Because of this possibility, better monitoring of SARS outbreaks through accurate diagnostic tests and the development of effective anti-viral therapies are urgently required. These in turn depend on better molecular knowledge about the SARS-CoV. Such research is therefore of vital importance if the community is to be properly prepared for a possible recurrence of the SARS pandemic.
Molecular Biology of SARS-CoV
Molecular characterization of the SARS-CoV genome
The etiological entity of a viral infection relies on both molecular and traditional virological methods including serological techniques, virus isolation by cell culture, and electron microscopy 2., 10.. Both molecular approaches and conventional approaches were employed for the initial characterization of the SARS pathogen (2). Peiris et al (2) firstly isolated the virus from in vitro tissue culture and subsequently yielded a 646-bp genomic fragment by RT-PCR using degenerate primers, which showed more than 50% homology to the RNA polymerase gene of bovine coronavirus (BCV) and murine hepatitis virus (MHV). The use of gene chip further confirmed the coronavirus as a possible cause of SARS (11).
Soon after the identification of the SARS-CoV, laboratories started to investigate the phylogenetic relationship between the virus and the other members of the same family through extensive comparison of their genome sequences. In mid-April 2003, the British Columbia Cancer Agency (BCCA) Genome Science Center in Canada (12), the Center of Disease Control in the United States (13) and the University of Hong Kong (14) announced at nearly the same time that the complete genome sequence of the SARS-CoV had been isolated in the corresponding areas (15). The results of independent sequencing of the SARS-CoV genome all indicated that it was a polyadenylated genomic RNA of 29.7 Kb in length. Comparative analysis of the genome with other coronaviruses suggested that the virus genome was very similar to previously characterized coronaviruses, with the order (starting from the N-terminal): replicase (R), spike (S), envelope (E), membrane (M) and nucleocapsid (N) gene, where there are few accessory genes or motifs spanning between the structural genes and at the 3’ UTR (untranslated region), which may not be necessary for viral replication (12). The replicase gene, with two open reading frames (ORF) 1a and 1b, covering more than two thirds of the genome, is predicted to encode only two proteinases 12., 13., 14. that regulate both the replication of the positive-stranded genomic RNA and the subsequent transcription of a nested set of eight subgenomic (sg) mRNAs (Table 1; ref. 16), which is a common transcription strategy adopted by coronavirus members 17., 18., 19., 20., 21..
Table 1.
Features of SARS-CoV Genome Sequence and Subgenomic Transcripts
| g/sg mRNA | ORF |
Start-End | No. of a.a. | No. of Bases | Frame | |||
|---|---|---|---|---|---|---|---|---|
| Thiel et al. | Zeng et al. | Marra et al. | Rota et al. | |||||
| mRNA 1 | ORF 1a | ORF 1a | ORF 1a | ORF 1a | 265-13,398 | 4,382 | 13,149 | +1 |
| mRNA 1 | ORF 1b | ORF 1b | ORF 1b | ORF 1b | 13,398-21,485 | 2,628 | 7,887 | +3 |
| mRNA 2 | S protein | S protein | S protein | S protein | 21,492-25,259 | 1,255 | 3,768 | +3 |
| mRNA 3 | ORF 3a | X1 | ORF 3 | X1 | 25,268-26,092 | 274 | 825 | +2 |
| mRNA 3 | ORF 3b | N/R | ORF 4 | X2 | 25,689-26,153 | 154 | 465 | +3 |
| mRNA 4 | E protein | N/R | E protein | E protein | 26,117-26,347 | 76 | 231 | +2 |
| mRNA 5 | M protein | M protein | M protein | M protein | 26,398-27,063 | 221 | 666 | +1 |
| mRNA 6 | ORF 6 | N/R | ORF 7 | X3 | 27,074-27,265 | 63 | 192 | +2 |
| mRNA 7 | ORF 7a | X2 | ORF 8 | X4 | 27,273-27,641 | 122 | 369 | +3 |
| mRNA 7 | ORF 7b | N/R | ORF 9 | N/R | 27,638-27,772 | 44 | 135 | +2 |
| mRNA 8 | ORF 8a | X3 | ORF 10 | N/R | 27,779-27,898 | 39 | 120 | +2 |
| mRNA 8 | ORF 8b | N/R | ORF 11 | X5 | 27,864-28,118 | 84 | 255 | +3 |
| mRNA 9 | N protein | N protein | N protein | N protein | 28,120-29,388 | 422 | 1,269 | +1 |
| mRNA 9 | ORF 9b | N/R | ORF 13 | N/R | 28,130-28,426 | 98 | 297 | +2 |
SARS-CoV protein products
5′ and 3′ UTR
The 5′ UTR of the SARS-CoV genome was characterized by 5′ Rapid Amplification of cDNA Ends (5′ RACE; ref. 14) and Northern blot assay 13., 16., 22.. These procedures elucidated the leader sequence and the transcription regulatory sequence (TRS). The leader sequence found in the viral sg mRNA transcripts is at least 72 nucleotides long. Through the alignment of the leader sequence at the 5’ end of the eight sg mRNAs, there is a minimal consensus TRS, namely, 5’-ACGAAC-3’, which participates in the discontinuous synthesis of sg mRNAs as a signaling sequence. The degree of sequence variance flanking the TRS showed no clear relationship with the abundance of the sg mRNAs (22). A highly conserved s2m motif with 32 nucleotides was also identified in the 3’ region of the genome, which had also been described in avian infectious bronchitis virus (AIBV; ref. 12., 14.).
Replicase Gene
The replicase gene of the SARS-CoV encodes for at least two proteins as a consequence of the proteolytic processing of the large polyprotein (ORF 1a and 1b; ref. 16). The translation of segment 1b of such polyprotein is interrupted by the −1 ribosomal frame shifting by a putative “slippery” sequence and a putative pseudoknot structure (16). Two functional domains—papain-like cysteine proteinase (PL2PRO) and 3C-like cysteine proteinase (3CLPRO), were identified experimentally and were responsible for the proteolytic processing of the polyprotein into 16 subunits 16., 22., 23.. A 375-a.a. SARS-CoV unique domain was identified upstream of the PL2PRO domain, which is unparalleled in any other known coronaviruses (16). In addition, seven more putative regions encoding RNA processing enzymes were identified, namely, RNA-dependent RNA polymerase (RDRP), RNA helicase (HEL) poly (U)-specific endoribonuclease (XendoU), 30-to-50 exonuclease (ExoN), S-adenosylmethionine-dependent ribose 20-O-methyltransferase (20-O-MT), adenosine diphosphate-ribose 100-phosphatase (ADRP), and a cyclic phosphodiesterase (CPD; ref. 16).
The translation of two polyproteins from ORF 1a and 1b starts the genome expression. The two proteinases, PCL2PRO and 3CLPRO, are then coupled with the proteolytic processing of the two polyproteins into 16 units. PCL2PRO is responsible for the N-proximal cleavage and 3CLPRO is responsible for the C-proximal cleavage. The helicase is then released. ATPase activity and DNA duplex-unwinding activity were demonstrated by purified helicase, indicating that the protein has RNA polymerase activity 16., 24..
S Gene
Together with the M protein, the spike protein is believed to be incorporated into the viral envelope before the mature virion is released (17). Initial analysis of the 1255-a.a. peplomer protein of the virus reveals the possible existence of a signal peptide that would likely be cleaved between residues 13 and 14 (12). The whole structure is predicted to contain a receptor-binding unit (S1) in the N-terminus 14., 25., 26., 27. and a transmembrane unit (S2) in the C-terminus 13., 14., 25., 27.. Molecular modeling of the S1 and S2 subunits of the spike glycoprotein 26., 28. suggested that the former unit is consisted of mainly anti-parallel β-sheets with dispersed α and β regions, in addition to the three domains identified in the S2 unit. The confidence level of the predicted molecular models was strengthened by the good correlation between predicted accessibility and hydropathy profiles and by the correct locations of the N/O-glycosylation sites and most of the disulfide bridges. Whether the experimentally determined N-glycosylated sites from purified spike protein treated by tryptic digest together with PNGase followed by time-of-flight (TOF) mass spectrometry (29) are correctly located in the proposed model remains to be clarified. In the aspect of biological activities, receptors for the binding of the SARS-CoV remain mysterious, as comparative genomics did not point out any significant similarity with the S1 domain of other human coronaviruses, implying that these viruses are using different receptors for cell entry (12). Subsequently, angiotensinconverting enzyme 2 (ACE2) was demonstrated to be a functional receptor for the SARS-CoV in vitro. Synctia was observed in cell culture expressing ACE2 and the SARS-CoV S1 domain, which could be inhibited by anti-ACE2 antibody (30). Fine mapping on the N-terminal unit of the spike protein indicates that the receptor-binding domain is probably located between the residues 303 and 537 (31).
ORF 3a
The sequence of the gene product from ORF 3a shows no homology to any known proteins 12., 14.. Signal peptide or a cleaved site is likely to be present in the protein except three predicted transmembrane domains (12). The exact function of the protein is yet to be determined, though the C-terminal of the protein may be involved in ATP-binding properties (12).
E Gene
The envelope protein of the SARS-CoV is thought to be the component of the virus envelope. Topology prediction suggested that the E protein is a type II membrane protein with the C-terminus hydrophilic domain exposed on the virion surface. Comparative protein sequence analysis suggested the SARS-CoV E protein resembles the protein connected with MHV 12., 32., 33..
M Gene
The matrix glycoprotein is not likely to be cleaved (12) and contains three putative transmembrane domains 12., 13., 14.. Its hydrophilic domain is believed to interact with the nucleocapsid protein and is located inside the virus particle (12). Linear epitope mapping of the M protein using synthetic peptides revealed that amino acid residues 2,137-2,158 interacted with SARS patient sera by ELISA assay, implying the potential capability of the M protein to induce immune response 34., 35..
ORF 7a and 8a
Like ORF 3a, sequence homology search yielded no significant result for any existing proteins, but the existence of a cleavage site (between residues 15 and 16) and a transmembrane helix were predicted. For ORF 7a, it is a putative type I membrane protein (12).
N gene
The N gene sequence showed high homology with the nucleocapsid protein of other coronaviruses. A putative short lysine-rich nuclear localization signal (KTFPPTEPKKDKKKKTDEAQ) was identified (12). A potential and well-conserved RNA interaction domain was also identified at the middle region of the gene, in which its basic nature may assist its role 12., 14.. The N protein was reported to activate the AP-1 signal transduction pathway, indicating that the protein may play a role in the regulation of the host cell cycle (36). Apart from the possible role in pathogenicity, N gene was also believed to be the most abundant antigen in the host during the course of infection, making it an excellent candidate for diagnostic purposes. The linear epitopes of the protein have been mapped 35., 37., 38., and the possibility of using these antigenic peptides or recombinant proteins in the diagnosis was discussed.
Phylogenetic analysis of the SARS-CoV
Protein sequence based on individual ORFs
The phylogenetic relationship by the comparison of the deduced amino acid sequences of the replicase gene and four structural genes (S, E, M, N) with other coronaviruses was described 12., 13., 14.. The conclusions drawn by the different research groups were similar, with the observation that SARS-CoV itself forms a distinct cluster—the fourth group of Coronaviridae, a notion supported by the high bootstrap values (above 90%). As a result, it has been concluded that the SARS-CoV is phylogenetically equidistant from all other known coronaviruses. Moreover, no detectable recombination event was concluded in the similarity plot on the whole genome alignment with other coronaviruses (14). The above findings suggest that the SARS-CoV is neither a mutant nor a recombinant of existing coronaviruses, and that the possibility of such a virus emerging as a product of genetic engineering can be excluded, as it is unlikely to generate an infectious coronavirus with 50% of its genome different from the existing coronaviruses (9).
Protein sequence based on functional domain of the replicase gene
Snijder et al (22) conducted an extensive phylogenetic analysis concerning the replicase gene of the SARS-CoV by using torovirus as an outgroup. These authors criticized the phylogram construction based on different SARS-CoV proteins as unconvincing, and suggested the possibility that the SARS-CoV can be clustered into an existing group. As the structural and other accessory genes can either be gained or lost throughout the evolutionary process and in view of their low level of conservation, the author decided to target the replicase gene to perform the phylogenetic analysis. For this reason, the phylogenetic relationship was reconstructed through a rooted tree. The construction of the phylogram was done with the fused replicase gene with manual adjustment and exclusion of poorly conserved region. The resulting tree reveals that the gene was mostly related to group 2 coronaviruses and was assigned as a subgroup 2b. The author further pointed out that the SARS-CoV contains homologues of domains that are unique for group 2 coronaviruses, in the region of nsp1 and nsp3 (PL2PRO), in addition to the differences in the sequence and arrangements of the 3’-located ORFs, and the lack of antigenic cross-reactivity do not contradict their conclusion, as such a phenomenon was also observed in group 1 coronaviruses.
Using Bayesian phylogenic inference approach, a recombination break point within the SARS-CoV RDRP was identified at protein sequence level (39). Phylogenetic analysis on the 5’ end of the domain indicated that it might originate from the common ancestor of all existing coronaviruses, while the same analysis on the 3’ end gave another tree topology that suggests a sister relationship with group 3 avian coronaviruses. These results suggested that a recombination event occurred between the common ancestor of the SARS-CoV and that of other coronaviruses, or alternatively that the 5’ fragment of the SARS-CoV diverged before the one between or within other known coronaviruses and the 3’ fragment diverged more recently (39).
Genome organization
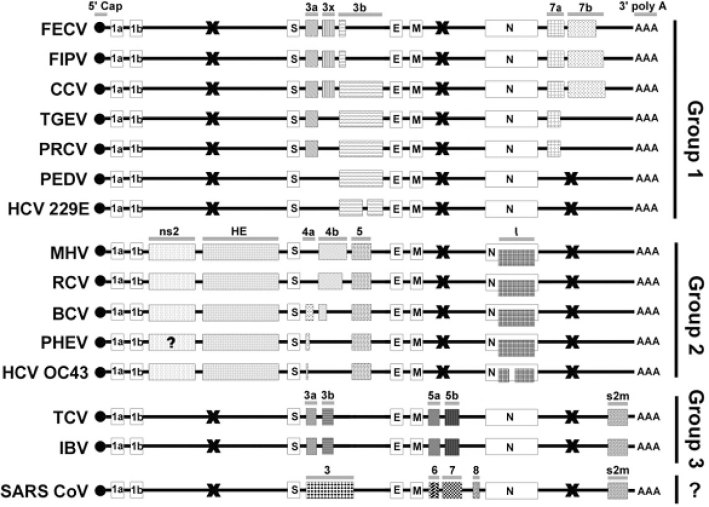
Based on the antigenic cross reactivity and genome characteristics, existing coronaviruses are generally classified into three subgroups (40). All coronaviruses share a very similar organization in their functional and structural genes, but the arrangement of the so-called non-essential genes is remarkably different among the subgroups. Group 1 coronaviruses are mainly characterized by the presence of ORFs following the N gene. Group 2 coronaviruses have two additional ORFs, non-structural protein 2 (ns2) and HE gene, located between ORF 1b and the S gene. Only group 3 species have ORFs located between the M and N gene, and a conserved stem-loop motif s2m at their 3’ UTR (Figure 1). Accessory ORFs are found between the S and E genes in all of the subgroups. However, these accessory ORFs within the S-E intergenic region do not seem to be homologous between the subgroups, though they are conserved within subgroups. The rate of evolution of these accessory genes is obviously higher than that of the essential genes, which provides an alternative to access the phylogeny of the coronavirus family.
Fig. 1.
Comparison of accessory genes among all known coronaviruses. The open boxes represent essential ORFs (not drawn to scale) while the shaded boxes represent accessory ORFs/motifs. Homologous ORFs are shaded with the same pattern. The names of the group-specific accessory ORFs were unified and denoted on the top of the corresponding subgroup ORFs. The X (black cross) represents the absence of ORFs within the region. Genome organization and accessory ORFs of these CoVs were confirmed except for the n2s of PHEV. All the accessory genes are group-specific and highly diverged within subgroups, particular within the S—E intergenic region. SARS-CoV has a very similar genome structure with group 3 CoVs, with two ORFs located between M and N gene, and a conserved stem-loop motif s2m at their 3’ UTR. Although the ORF 5a/5b of group 3 CoVs and ORF 5/6 of SARS-CoV are in homologous location, they do not have any significant sequence homology. FECV: feline enteric coronavirus 41., 42., 43., 44., 45.; FIPV: feline infectious peritonitis virus 41., 42., 43., 44., 45.; CCV: canine coronavirus 43., 46.; TGEV: transmissible gastroenteritis virus 41., 47., 48.; PRCV: porcine respiratory coronavirus 41., 47., 48.; PEDV: porcine epidemic diarrhea virus 49., 50.; HCV 229E: human coronavirus 229E 49., 51.; MHV: murine hepatitis virus 52., 53.; RCV: rat coronavirus (54); BCV: bovine coronavirus (55); PHEV: porcine hemagglutinating encephalomyelitis virus (56); HCV OC43: human coronavirus OC43 57., 58.; TCV: turkey coronavirus 59., 60., 61.; IBV: infectious bronchitis virus 62., 63., 64..
Based on the confirmed ORFs of the SARS-CoV described above, a comparison of all homologous accessory and essential ORFs of known coronaviruses with the novel SARS-CoV is shown in Figure 1. From the results, it does not seem that the coding regions are a consequence of a newly occurring recombination event between any of the existing known coronaviruses, similar to the conclusion made by Holmes (9). Interestingly, the SARS-CoV genome has a very similar organization to that of group 3 avian coronaviruses (IBV and TCV), with the presence of three ORFs within the M-N intergenic region, two ORFs spanning between the S and E genes (65), and a stem-loop motif s2m in 3’ UTR. The presence of s2m and the finding that the 3’ fragment of SARS-CoV RDRP clustered into group 3 in the phylogenetic analysis (39) suggest that the avian coronaviruses and the SARS-CoV might share a common ancestor which gained the s2m from a single RNA horizontal transfer event from a non-related virus family, as the astroviruses did 39., 66.. Another possibility, that a common coronavirus ancestor had once gained the motif but subsequently lost it, except the group 3 and SARS-CoV, cannot of course be excluded. Pairwise sequence homology search among the accessory ORFs at the S-E intergenic region of the SARS-CoV and all other coronaviruses shows no significant sequence homology 12., 13., 14. but they are homologous within subgroups. The ORF 5a/5b of group 3 coronaviruses and ORFs 6-8 of the SARS-CoV are in a homologous location, but they do not have any significant sequence homology. The above results imply that, although the SARS-CoV and group 3 coronaviruses have a very similar genome organization, they might have acquired these accessory genes from several RNA recombination events with different hosts or viral sources. It is observed that the accessory ORFs are group-specific but are usually truncated to a different extent within a subgroup (Figure 1). Another interesting observation is the genetic diversity at the S-E intergenic region. Usually two or three group-specific ORFs are found within this region of each subgroup, but only one confirmed ORF (ORF 3) is found in this region of the SARS-CoV genome 12., 13., 14., 16., 22.. The diversity (mainly due to truncation and deletion) of these S-E intergenic ORFs within the subgroups is higher than that of other accessory ORFs. Their sequence divergence implies their common ancestors might have acquired these ORFs by RNA recombination, which is a common phenomenon in large RNA viruses 67., 68., rather than evolved from mutations of a single ancestral RNA sequence segment (9). Typical examples are the acquirement of the HE gene from Influenza C (69) and recombination events with Berne virus at the HE-ns2 region (52).
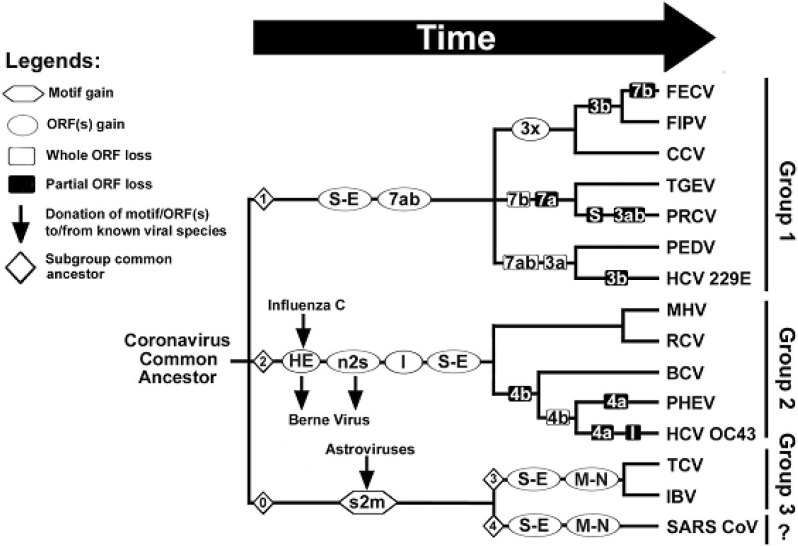
Based on the recombination and truncation events occurring within these intergenic regions, the phylogenetic relationship between the SARS-CoV and other group 3 coronaviruses has been reconstructed (Figure 2). At least four subgroup common ancestors (◊ in Figure 2) have acquired their S-E intergenic ORFs and other group-specific ORFs from several independent RNA recombination events. Moreover, there is a tendency of deletions or truncations of these ORFs when crossing the species barriers within the subgroups, e.g. ORF 4a/b in group 2 54., 55., 56., 57., 58.; ORF 3a/b and ORF 7a/b in group 1 41., 42., 47., 48., 50., 70., 71., 72.. The deletions of these redundant accessory ORFs are likely to be the result rather than the cause of crossing the host barriers, as coronavirus host range specificity and tropism have been demonstrated, at least in four studies 7., 73., 74., 75., as determined by the receptorbinding domain of the spike glycoprotein.
Fig. 2.
Phylogenetic relationship of all known coronaviruses based on the putative RNA recombination events occurred at the accessory ORFs. There are at least four subgroup common ancestors (◊ no.1-4) have acquired their redundant accessory ORFs from several independent RNA recombination events. Group 3 CoVs and SARS-CoV may have a common ancestor (◊ no.0) which gained s2m from a single RNA horizontal transfer event from a non-related family of astroviruses (see text). There is a tendency of deletions or truncations of these accessory ORFs when crossing the species barriers within the subgroups. The abbreviations of the viral species are shown in the legend of Figure 1.
Recombination within certain types of viruses is a common phenomenon in various virus families (67), particularly for large RNA viruses, as a means of shedding the deleterious effects of the errors accumulated during its genome replication (68). Recombination events within the coronavirus family 70., 76., 77. or with other non-related virus families 52., 66., 69. have been reported. Apparently, the diversity of the redundant accessory genes has been accompanied by extensive genome rearrangement by heterogeneous or homogenous RNA recombination events, providing useful information for the taxonomy of the coronaviruses. From this point of view, the SARS-CoV is definitely a new and unique member of the coronavirus family. The divergence of these redundant ORFs between the SARS-CoV and other known coronaviruses suggests that the SARS-CoV might have been circulating in other animal hosts long before its emergence, and somehow crossed into a human host several months ago either by a sudden bottleneck mutation event or a RNA recombination event with unknown sources.
Animal reservoir
It has been demonstrated that the SARS-CoV possesses the ability to infect macaques, which display symptoms similar to the clinical signs of SARS patients (78), and to replicate in cats and ferrets (79). Together with the evidence implied by the phylogenetic studies, it is tempting to identify the possible animal reservoir of the coronavirus. Recent studies of domestic and wild animals in Guangdong, where the SARS epidemic was first reported, identified the existence of the SARS-CoV from several animals found in the livestock market, including Himalayan palm civets (Paguma larvata) and raccoon dogs (Nyctereutes procyonoides; ref. 80), in spite of the failure of another group to identify any SARS-CoV after the screening of more than 60 animal species (81). The genome sequences of the coronaviruses isolated from these animals are almost identical (99.8%) to that of the SARS-CoV, revealing the extremely close phylogenetic relationship between them. Another major finding from the sequence analysis highlighted a 29-bp deletion upstream the N gene, which was noted only in one Guangdong isolate available from the Gen-Bank (GD0l, accession number 278489). Such deletion leads to the fusion of the two ORFs identified in mRNA 8 into one ORF. Yet its biological significance remains to be elucidated (8). Comparison of the S gene nucleotide sequence of the animal and human SARS-CoV indicated 11 consistent nucleotide signature mutations that appeared to distinguish them. The phylogenetic analysis of the S gene sequence between human and animal SARS-CoV likely ruled out the possibility that it is a consequence of human to animal transmission, implying the infected animals may acquired the virus from a true animal source that has yet to be identified (80). This was also supported by the host-association analysis of coronaviruses based on the nucleocapsid gene (39), which pinpointed that host-shifts had played an important role in the evolution of the virus and the host. The occurrence of avian-mammal host-shift supports the hypothesis that the SARS-CoV emerged from an unknown animal coronavirus.
Reverse genetics system
The reverse genetics system, a very useful tool in studying function of viral proteins and its mutations, was firstly described by Master’s group (82) for MHV in Coronaviridae. In less than six months since the first identification of the SARS-CoV (2), Yount et al (83) developed the reverse genetic systems for this coronavirus using the full-length cDNA clone of Urbani strain, by combining six component clones spanning through the entire genome. Following in vitro transcription and the transfection of the resulting RNA transcripts, a rescued recombinant virus was found to be capable of replication in the same way as the wild type. Expected marker mutations introduced were also identified. The success of the experiment offers hope for the development of attenuated strains of live vaccine against the SARS-CoV (9).
SARS and human leukocyte antigen (HLA) system
There is considerable scientific interest in the identification of the genetic agents responsible for the unusual susceptibility of the SARS-CoV in some ethnic groups. A molecular survey of the HLA system, a common method adopted to identify autoimmune disorders and emerging infectious diseases, was conducted in Taiwan during the SARS epidemic (84). Using PCR amplification plus sequencespecific oligonucleotide probing (PCR-SSOP), researchers identified the HLA genotype of SARS patients. Healthy, unrelated Taiwanese were used as controls, and the HLA genotype of SARS patients was compared with probable cases and with high-risk, uninfected health care workers. The results indicated that a higher frequency of HLA-B*4601 allele was found in severe SARS cases, which may explain the severity of SARS in these patients. Such genotype, as stated in the report, is common in Southern Han Chinese, Singaporeans and Vietnamese, but not in indigenous Taiwanese. There was no reported SARS case within the latter ethnic group. Such findings may explain the unusual SARS epidemic in South Asia.
Diagnosis of the SARS-CoV
Work on developing a laboratory diagnosis of the SARS-CoV began immediately after the SARS outbreak, although an ideal diagnostic system is still being sought. Numerous protocols have been developed for the diagnosis of infectious viral diseases. Most of these protocols are PCR-based, and the remainder depends on measurable immune response. Several factors affect the choice of proper diagnosis techniques, including time, the availability of equipment and expertise, the biological nature of the available samples, and the requirement of data output format (Table 2; ref. 10). The presence of the virus can be detected by molecular testing such as PCR and virus isolation. Measurable immune responses basically rely on SARS-CoV specific antibodies by enzyme-linked immunosorbent assay (ELISA).
Table 2.
Summary of Properties of Different Diagnostic Methods
| Features/Methods | RT-PCR | Virus isolation | ELISA | IFA | Microarray |
|---|---|---|---|---|---|
| Specificity | High | High | Relatively lower | Relatively lower | Relatively lower |
| Sensitivity | Not very high | Low | High | High | Not very high |
| Valid duration of +ve result# | d1–d10 | d1–d10 | d21–d31 | d1−d31 | d1–d10 |
| Valid duration of −ve result# | N/A | N/A | d21–d31 | d21–d31 | N/A |
| Convenience⁎ | Not very high | Moderate | High | Not very high | Low |
| Speed | Relatively lower | Slow | High | High | High |
Result is defined to be valid after the onset of fever where d=day.
Convenience means the requirement of expensive equipment and skilled labor.
Molecular assays
Advances have been made in molecular diagnostic techniques in recent years, and such rapid and sensitive methods allow efficient monitoring of infectious viral diseases. For SARS, the first genetic fragment of the virus was generated by reverse transcriptase-polymerase chain reaction (RTPCR; ref. 2). Two RT-PCR protocols were then developed by two WHO SARS network laboratories (http://www.who.int/csr/sars/primers/en). The sensitivities of the assay were demonstrated to be at least 50%, with the highest percentage found in throat swab specimens (85). No false positive was found in these assays.
The first rapid real-time assay was developed based on the most conserved region of the ORF1b gene sequence 86., 87.. A person will be confirmed to be infected by the SARS-CoV if viral RNA is detected by either the two PCR assays, two aliquots of specimen, or two sets of primers (http://www.cdc.gov/ncidod/sars/specimen_collection_sars2.htm). The second generation of this test protocol can detect the existence of the virus within 10 days after the onset of fever 87., 88., 89. and provides 80% sensitivity and 100% specificity in the testing of 50 NPA samples collected from SARS patients within three days after the onset of the disease (87). To further increase the sensitivity, one-step real-time RT-PCR has been recently developed (89). Specificity of the PCR can be enhanced by coupling it with the use of an additional amplification target using the virus N gene fragment (89), which is theoretically the most abundant subgenomic mRNA produced during transcription (13). The technique provides information on viral load during anti-viral treatments in real time, so that the efficacy of the therapy can be evaluated (10). However, although the PCR assays are powerful, their performance is also technically demanding and labor intensive (10).
The development of microarray technology for viral discovery was firstly described by Wang et al in 2002 (90). The capability of the rapid high throughout screening of unknown viral pathogen gives it great potential to be used as a diagnostic tool. In the identification of the SARS-CoV, Wang et al (11) employed the use of an improved microarray platform, which comprised conserved 70mers from each of the 1,000 viruses, to characterize the coronavirus genome. Four hybridizing oligonucleotides from Astroviridae which share the s2m motif and three from Coronaviridae sharing conserved ORF1ab fragment were firstly recognized in the experiment. The sequence recovered from the surface of the microarray further confirmed that it is a member of the coronavirus family. The identity of the SARS-CoV was confirmed within 24 hours, and this feat was followed by the partial sequencing of the novel virus a few days later. Such technique demonstrated a rapid and accurate means of unknown virus characterization through genetic data.
Virus isolation
Virus isolation by cell culture is used extensively as a traditional technique in virology. Coronavirus presenting in the clinical specimens of SARS patients was detected by inoculating the clinical specimens in cell cultures to allow the infection and the subsequent isolation of the virus. Fetal rhesus kidney (FRhK-4; ref. 2) and vero cells (3) were found to be susceptible to SARS-CoV infection. After the isolation procedure, the pathogen was identified as the SARS-CoV by further tests, such as electron microscopy, RT-PCR, or immunofluorescent viral antigen detection. Virus isolation is the only means to detect the existence of live virus from the tissue. The methodology is generally employed only for a preliminary identification of an unknown pathogen, as the procedure requires skillful technicians and is time consuming. The requirement of infectious viruses and that the duration of live virus existence varies add on further problems for conducting such assays, but they are nevertheless of very high specificity.
Enzyme-linked immunosorbent assay (ELISA)
The N protein is usually chosen as the antigen for anticoronavirus antibody detection assay 91., 92. as it is believed to be a predominant antigen of the SARS-CoV 35., 36.. It is also the only viral protein recognized by acute and early convalescent sera from patients recovering from SARS (29). In addition to the N protein, the S protein in the SARS-CoV was also reported as an antigen eliciting antibodies in human body (29), but at a much lower titer than that of the N protein 35., 36..
The assay based on the presence of SARS-CoV antibodies is suggested to be valid only for specimens obtained more than three weeks after the onset of fever 88., 89., although some patients have detectable SARS-CoV antibodies within 14 days of the onset of illness. Nevertheless, the negative result, i.e. absence of SARS-CoV antibodies, within the first three weeks cannot conclude that the patient is free of the virus, though the ELISA method was still defined as a good standard for rapid diagnosis of SARS (85). Seroconversion from negative to positive or a four-fold rise in antibody titer from acute to convalescent serum indicates recent infection (http://www.who.int/csr/sars/diagnostictests/en/).
Molecular Epidemiology and Evolution of SARS
The epidemiology of SARS has been extensively investigated since the outbreak of SARS in November 2002 in Guangdong (1). This traditional method was used to access the epidemiology of SARS initially. Molecular epidemiology can be used to trace the disease transmission by using phylogenetic analysis of viral nucleotide sequence, which can quickly identify and aid in monitoring the transmission (93).
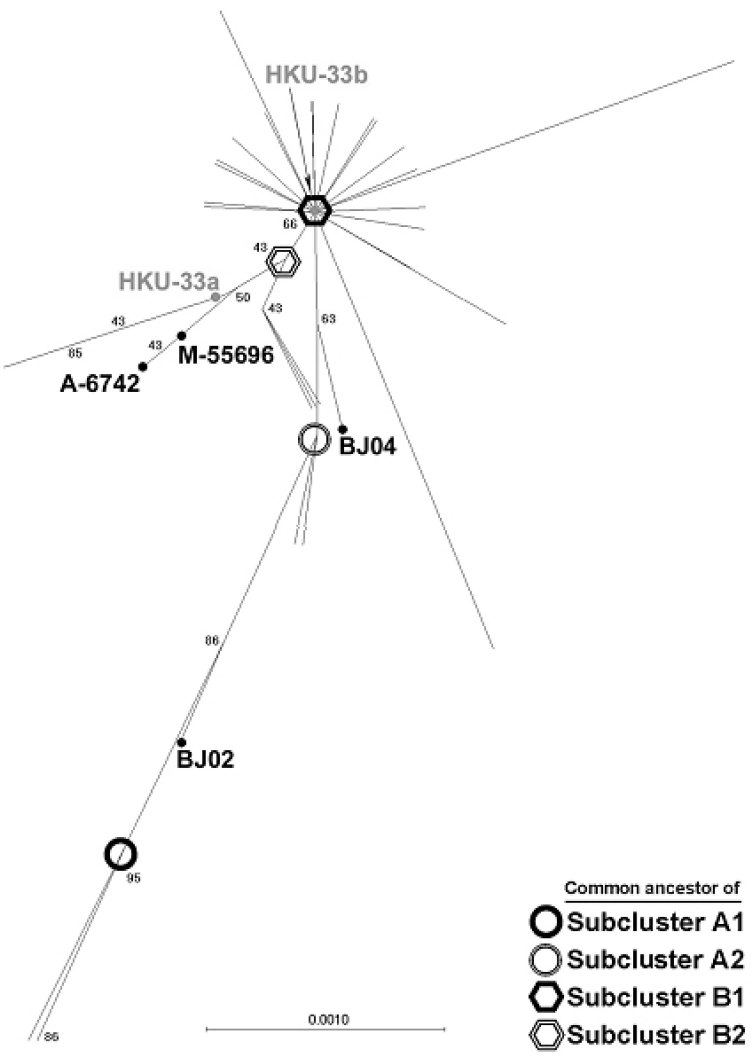
In coronavirus, variations in the spike protein can drastically affect viral entry, pathogenesis (94), antiviral immune response (29), virulence (95), cellular (6), or even species tropism (7). The S gene has been used as a target for genotyping most coronaviruses, like human coronaviruses (96) and IBV (97). Study of the N-terminal region of the SARS-CoV spike protein produced similar conclusions by conventional epidemiology methods (98). The investigation included the collection S1 gene sequences from SARS patients in Hong Kong and Guangdong during February-April 2003 mainly by direct sequencing of RT-PCR products derived from clinical specimens, and compared it phylogenetically to additional 27 other sequences available from GenBank. The majority of the Hong Kong viruses, including those from a large outbreak in a high-rise apartment block, Amoy Garden, clustered to a single index case that came from Guangdong to Hong Kong in late February (Figure 3). Most of the viruses derived from Hong Kong patients belong to the same lineage with viruses derived from the Hong Kong index case. Outbreaks in Canada, Singapore, Taiwan and Vietnam were also derived from the SARS-CoV of the same initial virus lineage as judged from the same phylogenetic analysis. A number of viruses derived from the early patients were excluded from the major lineage and formed distinct cluster, implying multiple introductions of the virus have occurred, although these viruses did not caused large-scale outbreaks. Viral sequences identified in Guangdong and Beijing are genetically more diverse 1., 98., implying that the SARS-CoV has been circulating there for a while before the introduction to Hong Kong. The Hong Kong index case that initiated the first super-spreading incident to affect 12 other patients might be simply a matter of chance or the viruses found in that patient were contagious to initiate super-spreading events, but these still need further investigations. Apart from findings that indicate the possible transmission routes, transitional isolates that possess both the characteristics of two lineages were also identified. Ruan et al (99) and Tsui et al (100) performed similar analysis based on the comparison of full genome sequences of different SARS-CoV isolates. They independently identified some of the variations, as Guan et al (98) did. Chiu et al (101) have recently identified the nucleotide substitution in the S gene that is unique to the Taiwan isolates and was linked to the Hong Kong index case. Sequence comparison of the Amoy Garden isolates revealed no significant variations within the S1 gene, or across the whole genome, implying that other non-viral factors may contribute to the abnormal transmission and clinical presentation of SARS in this cluster of high-rise apartments 98., 102.. In summary, the transmission route of the SARS-CoV in different countries and areas correlates well with the traditional epidemiological findings, implying the successful application of molecular epidemiological techniques in tracing the virus transmission history.
Fig. 3.
Phylogenetic analysis of 169 SARS-CoV spike genes. Unrooted trees were constructed based on the optimal alignment by neighbor-joining method using MEGA 2. Number at the nodes indicates boostrap values in percentage. The branch length shows the genetic distance with reference to the horizontal scale bar. All sample names were hidden for the convenience of display, except the index case isolate HKU-33 (gray) and subcluster transition isolates (dark). The locations of these isolates on the tree were pinpointed by dots besides their names. The hypothetical common ancestors of the subclusters were highlighted as described in the right bottom of the figure.
Concerning viral evolution, Zeng et al (103) have performed a linear regression analysis and tried to estimate the last appearance of the SARS-CoV common ancestor. With such effort, which has been successfully applied in timing of the ancestral sequence of human immunodeficiency virus (HIV; ref. 104), the ancestral sequence is believed to have appeared last in late 2002. These preliminary findings provide important information for tracing the origin of the SARS-CoV and monitoring its spread.
Immunity, Vaccination and Antiviral Drug Design
Current knowledge on coronavirus immunity has mainly been acquired from research on animal coronaviruses. Clinical observations have shown that humoral and cell-mediated immune responses may be both necessary against SARS-CoV infection (105). It was reported that T cell (CD3+, CD4 and CD8+) depletion was observed in early infection, but that levels returned to normal as the disease was improved (106). IgG antibody could be detected at the 7th day after the onset of symptoms and kept at high titer at least three months (107). Another report indicated that the virus was still detectable in respiratory and stool specimens by RT-PCR diagnosis but could not be cultured more than 40 days after presentation (108), implying that the antibody could be stimulated rapidly and might restrict the virus infection. However it has also been reported in fowl and feline coronaviral diseases that low-level antibody may exacerbate diseases (109). It is therefore important to conduct further investigations into the immune response to SARS patients in the future so as to benefit the vaccine development and disease control.
Concerning the candidate target for vaccine development, the S1 unit of the spike proteins has been identified as the host protective antigen and used as a vaccine candidate in other coronaviruses (110). An extensive structural analysis of the corresponding protein in SARS is thus desirable. With the identification of the SARS-CoV functional receptor (30) and the mapping of the receptor-binding domain on the spike protein (31), subunit vaccine targeting the receptorbinding domain and the preparation of killed or attenuated vaccine using ACE2 expression cell line may be promising (30).
Antiviral drugs represent an alternative anti-SARS strategy to vaccination. Inhibiting chemicals targeting the SARS-CoV replication-related proteins were considered as anti-SARS-CoV drug candidates, e.g. inhibition the enzymatic activity of 3CLPRO. An extensive structural analysis of 3CLPRO encoded from nsp5 on ORF 1a was performed 28., 111.. The 3CLPRO structure showed a considerable degree of conservation of the substrate-binding sites, with the evidence that it could retain its proteolytic activity upon TGEV (transmissible gastroenteritis virus) main proteinase (111), though another group mentioned that the inactive property of the enzyme might exist in vitro (112). From this result, these authors suggested that the use of rhinovirus 3CPRO inhibitor might be useful in anti-SARS therapy. Two months later, a research group from the US conducted a study on the interaction of two chemicals (KZ7088 and the AVLQSGFR octapeptide) with 3CLPRO (113), further highlighting the importance of the main proteinase as a target for anti-viral drug design. Fan et al (114) provided valuable additional information, and concluded that only the dimeric form of the 3CLPRO is active and that the proteinase-substrate interaction can be speeded up if more beta-sheet-like structure is involved in the substrate. Recently the crystal structure of 3CLPRO was reported by Yang et al (115). The 3CLPRO crystal underwent conformational changes under different pH conditions while complexing with the specific inhibitor at the same time. A serine-protease fold with a Cys-His at the active site was recognized. On the other hand, the modeling of the structure of 20-O-MT domain located at nsp16 was proposed by von Grotthuss et al (116) using the 3D jury system with high reliability (3D jury score >100). The conservation of the unique tetrad residues K-D-K-E of the domain assigned a proposed mRNA cap methylation function of this domain, suggesting an alternative target for anti-viral drug design. In addition to main proteases, blocking the virus entry should be considered as well. Structural analysis of the S2 domain of the SARS-CoV S protein, which plays a role in fusion of the virus with host cell, revealed a conservation of sequence motifs with the wellstudied gp41 protein of HIV-1 and other viruses with class I transmembrane domain (27). Such a structure may be another target for drug design.
Conclusion
The collaborative efforts of the global scientific community have provided invaluable insights into the molecular biology of the SARS-CoV. The development of a rapid and accurate method of diagnosis based on the molecular findings has helped to identify SARS patients at an early stage of the disease, thereby providing valuable information for national authorities to monitor the spread of the disease and take effective quarantine measures, and contributing to the understanding of the clinical presentations of the syndrome. The elucidation of the molecular biology of the SARS-CoV has provided a foundation for vaccine design and narrowed down the targets for large-scale high throughput drug screening program for anti-viral therapy. These advances helped the global community to contain the spread of SARS within four months since its first identification. However, much remains to be discovered about this novel coronavirus, and it may yet pose a serious threat. Unlike other recently identified viral diseases like Ebola and West Nile virus, it seems the transmission of SARS-CoV does not need a visible vector for spreading, and that a tiny, invisible, respiratory droplet is sufficient to infect another person (117). The nearly undetectable symptom presented by the recently confirmed SARS case in Singapore suggests that the virus may continue to circulate undetectably (65). The possibility that common domestic animals are also a virus reservoir for SARS further complicates the struggle to contain and ultimately eradicate this disease. In these aspects, sensitive, accurate and rapid diagnosis plays an extremely important role in limiting the disease spread, especially in the developing world and densely populated countries. Luckily, the aggressive quarantine measures imposed by the WHO proved to be effective in containing the outbreak, and the experience gained in the last SARS outbreak has prepared us to face another outbreak with some confidence. Nevertheless, nobody can predict exactly when an effective vaccine or anti-viral drug will be developed. All that can be said is that, based on our growing knowledge of the molecular epidemiology and evolution of the virus, the successful development of countermeasures to SARS is very possible.
Acknowledgements
We wish to thank specifically our colleagues at the Department of Microbiology inviting us to join in the SARS research effort. We feel sad about all the life loss caused by SARS and particularly the medical staffs who were infected and died in taking care of SARS patients.
This work was supported by Research Grant Council Grant HKU 7553/03M and The University of Hong Kong.
References
- 1.Zhong N.S. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peiris J.S. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drosten C. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 4.Ksiazek T.G. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1956. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 5.Peiris J.S. Severe acute respiratory syndrome (SARS) J. Clin. Virol. 2003;28:245–247. doi: 10.1016/j.jcv.2003.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casais R. Recombinant avian infectious bronchitis virus expressing a heterologous spike gene demonstrates that the spike protein is a determinant of cell tropism. J. Virol. 2003;77:9084–9089. doi: 10.1128/JVI.77.16.9084-9089.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haijema B.J. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 2003;77:4528–4538. doi: 10.1128/JVI.77.8.4528-4538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sstadler K. SARS—Beginning to understand a new virus. Annu. Rev. Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmes K.V. SARS coronavirus: a new challenge for prevention and therapy. J. Clin. Invest. 2003;111:1605–1609. doi: 10.1172/JCI18819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis J.S., Zambon M.C. Molecular diagnosis of influenza. Rev. Med. Virol. 2002;12:375–389. doi: 10.1002/rmv.370. [DOI] [PubMed] [Google Scholar]
- 11.Wang D. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:E2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marra M.A. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 13.Rota P.A. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 14.Zeng F.Y. The complete genome sequence of severe acute respiratory syndrome coronavirus (SARS-CoV) strain HKU-39498 (HK-39) Exp. Biol. Med. 2003;228:866–873. doi: 10.1177/15353702-0322807-13. [DOI] [PubMed] [Google Scholar]
- 15.Leung F.C. Hong Kong SARS sequence. Science. 2003;301:309–310. doi: 10.1126/science.301.5631.309c. [DOI] [PubMed] [Google Scholar]
- 16.Thiel V. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 17.Lai M.M., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai M.M. Coronavirus: organization replication and expression of the genome. Annu. Rev. Microbiol. 1990;44:303–333. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- 19.Lai M.M. Coronavirus: how a large RNA viral genome is replicated and transcribed. Infect. Agents Dis. 1994;3:98–105. [PubMed] [Google Scholar]
- 20.Lai M.M. Coronavirus: a jumping RNA transcription. Cold Spring Harb. Symp. Quant. Biol. 1987;52:359–365. doi: 10.1101/sqb.1987.052.01.041. [DOI] [PubMed] [Google Scholar]
- 21.Lai M.M., Holmes K.V. Coronaviridae and their replication. In: Knipe D., editor. Fields’ Virology. Lippincott Williams & Wilkins; Philadelphia, USA: 2001. [Google Scholar]
- 22.Snijder E.J. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao F. Prediction of proteinase cleavage sites in polyproteins of coronaviruses and its applications in analyzing SARS-CoV genomes. FEBS Lett. 2003;553:451–456. doi: 10.1016/S0014-5793(03)01091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanner J.A. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J.X. The structural characterization and antigenicity of the S protein of SARS-CoV. Geno. Prot. Bioinfo. 2003;1:108–117. doi: 10.1016/S1672-0229(03)01015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spiga O. Molecular modelling of S1 and S2 subunits of SARS coronavirus spike glycoprotein. Biochem. Biophys. Res. Commun. 2003;310:78–83. doi: 10.1016/j.bbrc.2003.08.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosch B.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan L. Assessment of putative protein targets derived from the SARS genome. FEBS Lett. 2003;554:257–263. doi: 10.1016/S0014-5793(03)01115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krokhin O. Mass spectrometric characterization of proteins from the SARS virus. Mol. Cell Proteomics. 2003;2:346–356. doi: 10.1074/mcp.M300048-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao X. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen X. Small envelope protein E of SARS: cloning, expression, purification, CD determination, and bioinformatics analysis. Acta Pharmacol. Sin. 2003;24:505–511. [PubMed] [Google Scholar]
- 33.Wu Q.F. The E protein is a multifunctional membrane protein of SARS-CoV. Geno. Prot. Bioinfo. 2003;1:131–144. doi: 10.1016/S1672-0229(03)01017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Y.W. The M protein of SARS-CoV: basic structural and immunological properties. Geno. Prot. Bioinfo. 2003;1:118–130. doi: 10.1016/S1672-0229(03)01016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J.Q. Assessment of immunoreactive synthetic peptides from the structural proteins of severe acute respiratory syndrome coronavirus. Clin. Chem. 2003;49:1989–1996. doi: 10.1373/clinchem.2003.023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He R. Activation of AP-1 signal transduction pathway by SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2003;311:870–876. doi: 10.1016/j.bbrc.2003.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J.Q. The structure analysis and antigenicity study of the N protein of SARS-CoV. Geno. Prot. Bioinfo. 2003;1:145–156. doi: 10.1016/S1672-0229(03)01018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin Y. Identification of an epitope of SARS-coronavirus nucleocapsid protein. Cell Res. 2003;13:141–145. doi: 10.1038/sj.cr.7290158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rest J.S., Mindell D.P. SARS-associated coronavirus has a recombinant polymerase and coronaviruses have a history of host-shifting. Infect. Genet. Evol. 2003;3:219–225. doi: 10.1016/j.meegid.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai M.M., Holmes K.V. Coronaviruses. In: Knipe D., editor. Fields’ Virology. Lippincott Williams & Wilkins; Philadelphia, USA: 2001. pp. 1163–1185. [Google Scholar]
- 41.de Groot R.J. Sequence analysis of the 3′ end of the feline coronavirus FIPV 79–1146 genome: comparison with the genome of porcine coronavirus TGEV reveals large insertions. Virology. 1988;167:370–376. doi: 10.1016/0042-6822(88)90097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kennedy M. Deletions in the 7a ORF of feline coronavirus associated with an epidemic of feline infectious peritonitis. Vet. Microbiol. 2001;81:227–234. doi: 10.1016/S0378-1135(01)00354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vennema H. Genomic organization and expression of the 3′ end of the canine and feline enteric coronaviruses. Virology. 1992;191:134–140. doi: 10.1016/0042-6822(92)90174-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vennema H. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology. 1998;243:150–157. doi: 10.1006/viro.1998.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamanaka M. Nucleotide sequence of the inter-structural gene region of feline infectious peritonitis virus. Virus Genes. 1998;16:317–318. doi: 10.1023/A:1008099209942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horsburgh B.C. Analysis of a 9.6 Kb sequence from the 3′ end of canine coronavirus genomic RNA. J. Gen. Virol. 1992;73:2849–2862. doi: 10.1099/0022-1317-73-11-2849. [DOI] [PubMed] [Google Scholar]
- 47.Rasschaert D. Porcine respiratory coronavirus differs from transmissible gastroenteritis virus by a few genomic deletions. J. Gen. Virol. 1990;71:2599–2607. doi: 10.1099/0022-1317-71-11-2599. [DOI] [PubMed] [Google Scholar]
- 48.Vaughn E.M. Sequence comparison of porcine respiratory coronavirus isolates reveals heterogeneity in the S, 3, and 3-1 genes. J. Virol. 1995;69:3176–3184. doi: 10.1128/jvi.69.5.3176-3184.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duarte M. Genome organization of porcine epidemic diarrhoea virus. Adv. Exp. Med. Biol. 1993;342:55–60. doi: 10.1007/978-1-4615-2996-5_9. [DOI] [PubMed] [Google Scholar]
- 50.Duarte M. Sequence analysis of the porcine epidemic diarrhea virus genome between the nucleocapsid and spike protein genes reveals a polymorphic ORF. Virology. 1994;198:466–476. doi: 10.1006/viro.1994.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bridgen A. Sequence determination of the nucleocapsid protein gene of the porcine epidemic diarrhoea virus confirms that this virus is a coronavirus related to human coronavirus 229E and porcine transmissible gastroenteritis virus. J. Gen. Virol. 1993;74:1795–1804. doi: 10.1099/0022-1317-74-9-1795. [DOI] [PubMed] [Google Scholar]
- 52.Snijder E.J. Comparison of the genome organization of toro- and coronaviruses: evidence for two nonhomologous RNA recombination events during Berne virus evolution. Virology. 1991;180:448–452. doi: 10.1016/0042-6822(91)90056-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss S.R. The ns 4 gene of mouse hepatitis virus (MHV), strain A 59 contains two ORFs and thus differs from ns 4 of the JHM and S strains. Arch. Virol. 1993;129:301–309. doi: 10.1007/BF01316905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoo D. Primary structure of the sialodacryoadenitis virus genome: sequence of the structural-protein region and its application for differential diagnosis. Clin. Diagn. Lab. Immunol. 2000;7:568–573. doi: 10.1128/cdli.7.4.568-573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abraham S. Sequence and expression analysis of potential nonstructural proteins of 4.9, 4.8, 12.7, and 9.5 kDa encoded between the spike and membrane protein genes of the bovine coronavirus. Virology. 1990;177:488–495. doi: 10.1016/0042-6822(90)90513-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sasseville A.M. Sequence of the 3’-terminal end (8.1 Kb) of the genome of porcine haemagglutinating encephalomyelitis virus: comparison with other haemagglutinating coronaviruses. J. Gen. Virol. 2002;83:2411–2416. doi: 10.1099/0022-1317-83-10-2411. [DOI] [PubMed] [Google Scholar]
- 57.Mounir S., Talbot P.J. Human coronavirus OC43 RNA 4 lacks two open reading frames located downstream of the S gene of bovine coronavirus. Virololgy. 1993;192:355–360. doi: 10.1006/viro.1993.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vieler E. The region between the M and S genes of porcine haemagglutinating encephalomyelitis virus is highly similar to human coronavirus OC43. J. Gen. Virol. 1996;77:1443–1447. doi: 10.1099/0022-1317-77-7-1443. [DOI] [PubMed] [Google Scholar]
- 59.Breslin J.J. Sequence analysis of the turkey coronavirus nucleocapsid protein gene and 3ʹ untranslated region identifies the virus as a close relative of infectious bronchitis virus. Virus Res. 1999;65:187–193. doi: 10.1016/S0168-1702(99)00117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Breslin J.J. Sequence analysis of the matrix/nucleocapsid gene region of turkey coronavirus. Intervirology. 1999;42:22–29. doi: 10.1159/000024956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verbeek A., Tijssen P. Sequence analysis of the turkey enteric coronavirus nucleocapsid and membrane protein genes: a close genomic relationship with bovine coronavirus. J. Gen. Virol. 1991;72:1659–1666. doi: 10.1099/0022-1317-72-7-1659. [DOI] [PubMed] [Google Scholar]
- 62.Boursnell M.E. Sequencing of coronavirus IBV genomic RNA: three open reading frames in the 5′ ‘unique’ region of mRNA D. J. Gen. Virol. 1985;66:2253–2258. doi: 10.1099/0022-1317-66-10-2253. [DOI] [PubMed] [Google Scholar]
- 63.Cavanagh D., Davis P.J. Evolution of avian coronavirus IBV: sequence of the matrix glycoprotein gene and intergenic region of several serotypes. J. Virol. 1988;69:621–629. doi: 10.1099/0022-1317-69-3-621. [DOI] [PubMed] [Google Scholar]
- 64.Jia W.N., Naqi S.A. Sequence analysis of gene 3, gene 4 and gene 5 of avian infectious bronchitis virus strain CU-T2. Gene. 1997;189:189–193. doi: 10.1016/S0378-1119(96)00847-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lai M.M. SARS virus: the beginning of the unraveling of a new coronavirus. J. Biomed. Sci. 2003;10:664–675. doi: 10.1007/BF02256318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jonassen C.M. A common RNA motif in the 3′ end of the genomes of astroviruses, avian infectious bronchitis virus and an equine rhinovirus. J. Gen. Virol. 1998;79:715–718. doi: 10.1099/0022-1317-79-4-715. [DOI] [PubMed] [Google Scholar]
- 67.Lai M.M. RNA recombination in animal and plant viruses. Microbiol. Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lai M.M. Recombination in large RNA viruses: coronaviruses. Semin. Virol. 1996;7:381–388. doi: 10.1006/smvy.1996.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luytjes W. Sequence of mouse hepatitis virus A59 mRNA 2: indications for RNA recombination between coronaviruses and influenza C virus. Virology. 1988;166:415–422. doi: 10.1016/0042-6822(88)90512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Herrewegh A.A. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 1998;72:4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Connor J.B., Brain D.A. The major product of porcine transmissible gastroenteritis coronavirus gene 3b is an integral membrane glycoprotein of 31 kDa. Virology. 1999;256:152–161. doi: 10.1006/viro.1999.9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vennema H. Genetic drift and genetic shift during feline coronavirus evolution. Vet. Microbiol. 1999;69:139–141. doi: 10.1016/S0378-1135(99)00102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baric R.S. Episodic evolution mediates interspecies transfer of a murine coronavirus. J. Virol. 1997;71:1946–1955. doi: 10.1128/jvi.71.3.1946-1955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuo L. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 2000;74:1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sanchez C.M. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 1999;73:7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee C.W., Jackwood M.W. Evidence of genetic diversity generated by recombination among avian coronavirus IBV. Arch. Virol. 2000;145:2135–2148. doi: 10.1007/s007050070044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rowe C.L. Quasispecies development by high frequency RNA recombination during MHV persistence. Adv. Exp. Med. Biol. 1998;440:759–765. doi: 10.1007/978-1-4615-5331-1_98. [DOI] [PubMed] [Google Scholar]
- 78.Fouchier R.A. Aetiology: Koch’s postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martina B.E. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425:915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guan Y. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 81.Normile D., Enserink M. SARS in China. Tracking the roots of a killer. Science. 2003;301:297–299. doi: 10.1126/science.301.5631.297. [DOI] [PubMed] [Google Scholar]
- 82.Koetzner C.A. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J. Virol. 1992;66:1841–1848. doi: 10.1128/jvi.66.4.1841-1848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yount B. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2003;100:12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin M. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med. Genet. 2003;12:9. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yam W.C. Evaluation of reverse transcription-PCR assays for rapid diagnosis of severe acute respiratory syndrome associated with a novel coronavirus. J. Clin. Microbiol. 2003;41:4521–4524. doi: 10.1128/JCM.41.10.4521-4524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Poon L.L. Rapid diagnosis of a coronavirus associated with severe acute respiratory syndrome (SARS) Clin. Chem. 2003;49:953–955. doi: 10.1373/49.6.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Poon L.L. Early diagnosis of SARS coronavirus infection by real time RT-PCR. J. Clin. Virol. 2003;28:233–238. doi: 10.1016/j.jcv.2003.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shaila M.S. Severe acute respiratory syndrome (SARS): an old virus jumping into a new host or a new creation. J. Biosci. 2003;28:359–360. doi: 10.1007/BF02705108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ng E.K. Quantitative analysis and prognostic implication of SARS coronavirus RNA in the plasma and serum of patients with severe acute respiratory syndrome. Clin. Chem. 2003;49:1976–1980. doi: 10.1373/clinchem.2003.024125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang D. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. USA. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Y.S. Analysis of false-positive associated with antibody tests for SARS-CoV in SLE patients. Acta Biologiae Exerimentalis Sinica. 2003;36:314–317. Chinese. [PubMed] [Google Scholar]
- 92.Che X.Y. Antibody response of patients with severe acute respiratory syndrome (SARS) to nucleocapsid antigen of SARS-associated coronavirus. J. First Military Medical Univ. 2003;23:637–639. Chinese. [PubMed] [Google Scholar]
- 93.Holmes E.C. Molecular epidemiology and evolution of emerging infectious diseases. Br. Med. Bull. 1998;54:533–543. doi: 10.1093/oxfordjournals.bmb.a011708. [DOI] [PubMed] [Google Scholar]
- 94.Gallagher T.M., Buchmeier M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279:371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Phillips J.J. Murine coronavirus spike glycoprotein mediates degree of viral spread, inflammation, and virus-induced immunopathology in the central nervous system. Virology. 2002;301:109–120. doi: 10.1006/viro.2002.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hays J.P., Myint S.H. PCR sequencing of the spike genes of geographically and chronologically distinct human coronaviruses 229E. J. Viral. Methods. 1998;75:179–193. doi: 10.1016/S0166-0934(98)00116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee C.W., Jackwood M.W. Origin and evolution of Georgia 98 (GA98), a new serotype of avian infectious bronchitis virus. Virus Res. 2001;80:33–39. doi: 10.1016/s0168-1702(01)00345-8. [DOI] [PubMed] [Google Scholar]
- 98.Guan Y. Molecular epidemiology of the novel coronavirus causing severe acute respiratory syndrome (SARS) Lancet. 2004;363:99–104. doi: 10.1016/S0140-6736(03)15259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ruan Y.J. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tsui S.K. Coronavirus genomic-sequence variations and the epidemiology of the severe acute respiratory syndrome. N. Engl. J. Med. 2003;349:187–188. doi: 10.1056/NEJM200307103490216. [DOI] [PubMed] [Google Scholar]
- 101.Chiu R.W. Molecular epidemiology of SARS—from Amoy Gardens to Taiwan. N. Engl. J. Med. 2003;349:1875–1876. doi: 10.1056/NEJM200311063491923. [DOI] [PubMed] [Google Scholar]
- 102.Chim S.S. Genomic characterisation of the severe acute respiratory syndrome coronavirus of Amoy Gardens outbreak in Hong Kong. Lancet. 2003;362:1807–1808. doi: 10.1016/S0140-6736(03)14901-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zeng F.Y. Estimation of the last common ancestor of severe acute respiratory syndrome (SARS) coronavirus. N. Engl. J. Med. 2003;349:2469–2470. doi: 10.1056/NEJM200312183492523. [DOI] [PubMed] [Google Scholar]
- 104.Korber B. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 105.Tang X. Measurement of subgroups of peripheral blood T lymphocytes in patients with severe acute respiratory syndrome and its clinical significance. Chin. Med. J. 2003;116:827–830. [PubMed] [Google Scholar]
- 106.Li G. Profile of specific antibodies to the SARS-associated coronavirus. N. Engl. J. Med. 2003;349:508–509. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- 107.Chen K.H. Emerg. Infect. Dis. 2003. Determination of SARS coronavirus in patients with suspected SARS. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.de Herdt P. Infectious bronchitis serology in broilers and broiler breeders: corrections between antibody titers and performance in vaccinated flocks. Avian Dis. 2001;45:612–619. [PubMed] [Google Scholar]
- 109.Cavanagh D. Coronavirus IBV: virus retaining spike glycopolypeptide S2 but not S1 is unable to induce virus-neutralizing or haemagglutination-inhibiting antibody, or induce chicken tracheal protection. J. Gen. Virol. 1986;67:1435–1442. doi: 10.1099/0022-1317-67-7-1435. [DOI] [PubMed] [Google Scholar]
- 110.de Groot A.S. How the SARS vaccine effort can learn from HIV-speeding towards the future, learning from the past. Vaccine. 2003;21:4095–4104. doi: 10.1016/S0264-410X(03)00489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Anand K. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 112.Campanacci V. Structural genomics of the SARS coronavirus: cloning, expression, crystallization and preliminary crystallographic study of the Nsp9 protein. Acta Crystallogr. D. Biol. Crystallogr. 2003;59:1628–1631. doi: 10.1107/S0907444903016779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chou K.C. Binding mechanism of coronavirus main proteinase with ligands and its implication to drug design against SARS. Biochem. Biophys. Res. Commun. 2003;308:148–151. doi: 10.1016/S0006-291X(03)01342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fan K. Biosynthesis, purification and substrate specificity of SARS coronavirus 3C-like proteinase. J. Biol. Chem. 2004:279. doi: 10.1074/jbc.M310875200. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang H. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. USA. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.von Grotthuss M. mRNA cap-1 methyltransferase in the SARS genome. Cell. 2003;113:701–702. doi: 10.1016/S0092-8674(03)00424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Knudsen T.B. Severe acute respiratory syndrome—a new coronavirus from the Chinese dragon’s lair. Scand. J. Immunol. 2003;58:277–284. doi: 10.1046/j.1365-3083.2003.01302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]