Abstract
The Coronaviridae family is characterized by a nucleocapsid that is composed of the genome RNA molecule in combination with the nucleoprotein (N protein) within a virion. The most striking physiochemical feature of the N protein of SARS-CoV is that it is a typical basic protein with a high predicted pI and high hydrophilicity, which is consistent with its function of binding to the ribophosphate backbone of the RNA molecule. The predicted high extent of phosphorylation of the N protein on multiple candidate phosphorylation sites demonstrates that it would be related to important functions, such as RNA-binding and localization to the nucleolus of host cells. Subsequent study shows that there is an SR-rich region in the N protein and this region might be involved in the protein-protein interaction. The abundant antigenic sites predicted in the N protein, as well as experimental evidence with synthesized polypeptides, indicate that the N protein is one of the major antigens of the SARS-CoV. Compared with other viral structural proteins, the low variation rate of the N protein with regards to its size suggests its importance to the survival of the virus.
Key words: SARS-CoV, nucleoprotein, phosphorylation, SR-rich region, antigenic sites
Introduction
It has been established that a variant of coronaviruses, SARS-CoV, is the pathogen of SARS (1). The N protein (nucleoprotein) is one of the major structural proteins in a viral particle, playing a critical role in the transcription regulation of the genomic RNA and other viral proteins (2). It might also be involved in the virulence and virus-specific post-translational modifications (3).
In this paper, we report the predicted structure, possible functions, evolution, and the immunoassay of the N protein of SARS-CoV to confirm that the N protein is one of the major antigens with synthesized polypeptides.
Results and Discussion
The ORF of the N protein
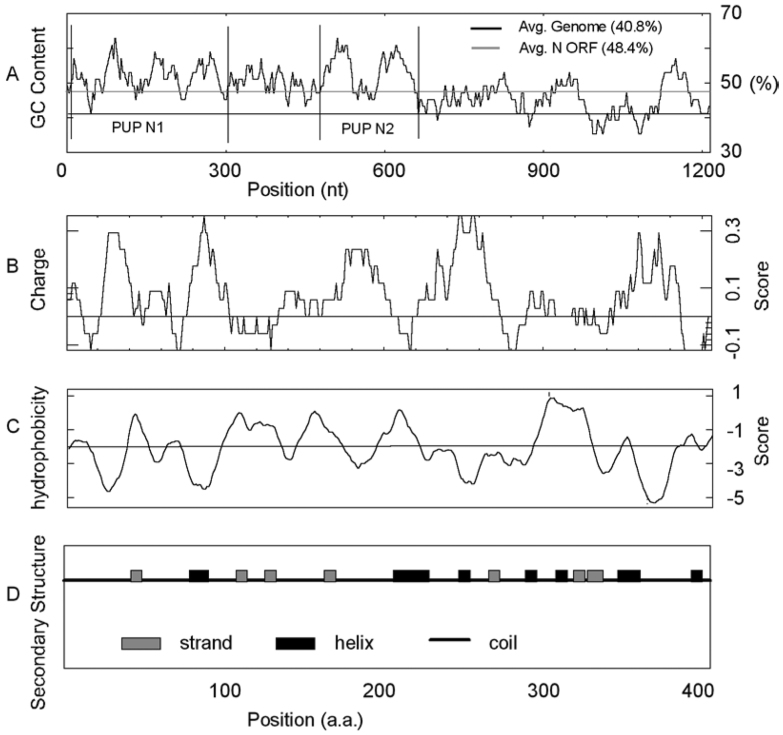
The ORF (open-reading frame) for the N protein is located at the 3′ end (nucleotide position 28,101-29,369) of the SARS-CoV genome. The ORF is 1,269 nucleotide (nt) long, accounting for 4.27% of the total genome. It has a GC content of 48.38% (A: U: C: G = 31.60: 20.02: 26.32: 22.06), which is significantly higher than the average of the complete SARS-CoV genome (40.76%). And the GC content in the left half near the 5′ end (51.63%) is obviously higher than that in the right half near the 3′ end (44.68%) (Figure 1).
Fig. 1.
The predicted distributions of GC content (A), electric charge (B), hydrophobicity (C) and secondary structure (D) in the N protein of SARS-CoV.
General physiochemical features of the N protein
The N protein is composed of 422 amino acids (a.a.) with an estimated molecular weight of 46.03 KD, the second largest of the structural viral protein. It has a low percentage (< 1.7%) of methionine, tryptophan and histidine residues, but does not contain any cysteine (Table 1).
Table 1.
The Amino Acid Composition of the SARS-CoV N Protein
| Number | Percentage (%) | |
|---|---|---|
| Non-polar, Neutral | ||
| Ala, A | 34 | 8.06 |
| Phe, F | 13 | 3.08 |
| Gly, G | 45 | 10.66 |
| Ile, I | 11 | 2.61 |
| Leu, L | 26 | 6.16 |
| Met, M | 7 | 1.66 |
| Pro, P | 31 | 7.35 |
| Val, V | 11 | 2.61 |
| Trp, W | 5 | 1.18 |
| Total | 183 | 43.36 |
| Polar, Neutral | ||
| Cys, C | 0 | 0.00 |
| Asn. N | 25 | 5.92 |
| Gln, Q | 34 | 8.06 |
| Ser, S | 35 | 8.29 |
| Thr, T | 33 | 7.82 |
| Tyr, Y | 11 | 2.61 |
| Total | 138 | 32.70 |
| Polar, Positive | ||
| His, H | 5 | 1.18 |
| Lys, K | 29 | 6.87 |
| Arg, R | 31 | 7.35 |
| Total | 65 | 15.40 |
| Polar, Negative | ||
| Asp, D | 22 | 5.21 |
| Glu, E | 14 | 3.32 |
| Total | 36 | 8.53 |
Absence of the cysteine is one of the common features in coronavirus N proteins. Cysteine is an important amino acid in zinc knuckle structure that may be involved in packaging signal recognition (4). It has been reported that the N protein of MHV (murine hepatitis virus) has no any known RNA-binding motif, such as arginine-rich motif or zinc finger (5). The lack of cysteine in the N protein of SARS-CoV indicates that the interaction of the N protein and signal packaging may occur in the absence of zinc knuckle structure.
The complete N protein is a highly basic protein. It has positively net charges, and has the highest pI (pI 10.11) among all known structural proteins in the virus, making it easier to interact with acidic genomic RNA. The charge distribution shows five positive peaks with relatively even distance (Figure 1). Positively charged amino acids, histidine, lysine and arginine, account for a big portion in each peak. In addition to high pI and positive charges, the N protein has a high hydrophilicity (54%). The middle of the N protein is relatively hydrophobic, but the two termini are hydrophilic (Figure 1). The N-terminus is basic with positive charge, while the C-terminus is acidic with negative charge. In a model for demonstrating the interaction of multiple N proteins, the two termini are supposed to be linked end to end (6).
Phosphorylation of the N protein
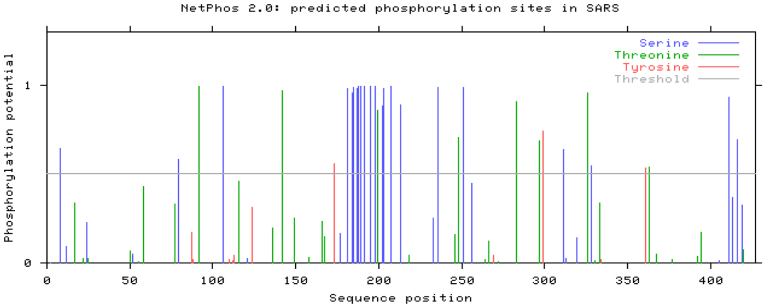
Phosphorylation of the N protein is related to RNA-binding, oligomerization and localization to nucleoli (3). We identified 33 potential phosphorylation sites in the N protein, including 22 serines, 8 threonines and 3 tyrosines. The average score of serines is the highest among the three amino acids. This demonstrates that serines are the predominant phosphorylated residues in the N protein, consistent with the previous reports (5). The phosphorylation sites concentrate in the middle of the N protein (Figure 2). However, the exact number and location of phosphoserines have not been identified by experiments yet.
Fig. 2.
The predicted phosphorylation sites on the N protein. We identified 33 potential phosphorylation sites in the N protein, including 22 serines, 8 threonines and 3 tyrosines. The average score of serines are significantly higher than that of the other two. The phosphorylation sites concentrate in the middle of the N protein.
Structure of the N protein
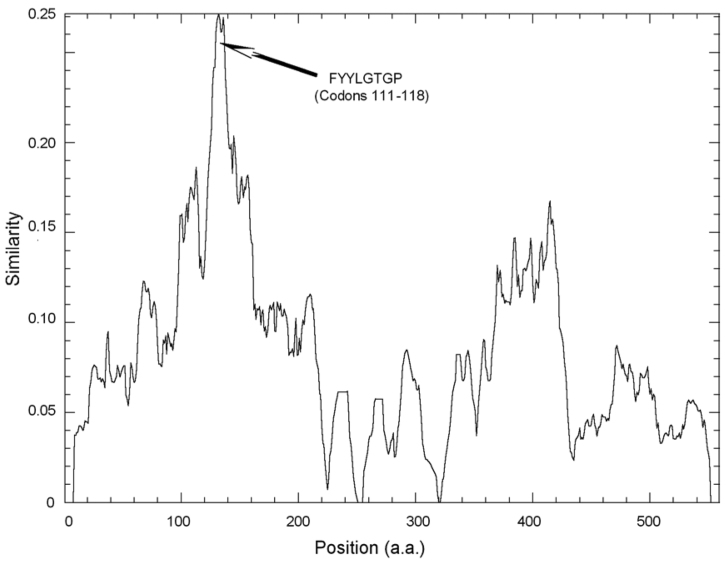
Through multi-alignment of total nineteen sequences of the coronavirus N proteins, we found two conserved structural regions at Codons 81-140 and 270-320 (a.a. positions are all referred to the N protein of SARS-CoV, Isolate BJ01), with the former more conserved (Figure 3). This result is consistent with the conserved domains that we have predicted by using CDS (Conserved Domain Search). A highly conserved domain (FYYLGTGP at Codons 111-118) was identified within the first conserved region in all coronaviruses. These conserved regions and domains could serve as potential drug targets because of their great possibility of performing critical functions. In contrast, the two termini of the N protein are more variable, particularly the C-terminus.
Fig. 3.
The similarity chart of the N protein. Based on multi-alignment of totally nineteen coronavirus N proteins, two conserved regions were found around a.a. 81-140 and a.a. 270-320 (amino acid positions are all referred to the N protein of SARS-CoV, Isolate BJ01). The arrow indicates the most conserved domain, and its sequence and amino acid position are given. In contrast, the two termini of the N protein are more variable, particularly the C- terminal. The figure was generated by Plotcon in the EMBOSS package (http://www.hgmp.mrc.ac.uk/Software/EMBOSS/).
We also predicted the secondary structure of the N protein by using PSIPRED. According to the result, the N protein is composed of coils, strands, and helices. There are totally eight helices and seven of them are distributed in the 3′ end of the N protein.
SR-rich region of the N protein
An SR-rich region (a region rich of serine and arginine) was identified in the coronavirus N protein, which has a core motif of SR{X2}SR{X2}SR (X indicates any amino acid, and subscript number indicates the number of amino acids between two SRs.). In the N protein of SARS-CoV, its SR-rich region presents a symmetric structure: SR{X6}SR{X2}SRSR{X2}SR{X6}SR (Codons 177-204).
The SR-rich region has been found in all coronavirus N proteins. However, it is not within conserved regions that we have detected this region from similarity analysis. Further study showed that this region is relatively variable. For example, in the N protein of SARS-CoV, there is a substitution in SR-rich region. Therefore, we used SR as a marker to identify this region, though amino acid change may occur in SR and cause marker to disappear. We classified all SR-rich regions of coronavirus N proteins into a few types (Table 2), and discovered that the classification of those coronaviruses by their core motifs of the SR-rich region was consistent with the phylogenic tree that we had constructed. It seems that the SR-rich region is typically common and representative, though it is outside the conserved regions and has easily-varied sequence.
Table 2.
The Core Motif of SR-rich Region in the Coronavirus N Protein
| Coronavirus | Core motif of SR-rich region in the N protein |
|---|---|
| SARS Coronavirus BJ01 | SRGGSQASSRSSSRSRGNSRNSTPGS SR* |
| Murine Hepatitis Virus | |
| Puffinosis Virus | |
| Rat Sialodacryoadenitis Coronavirus | SRSGSRSQSR |
| Rat Coronavirus | |
| Equine Coronavirus | |
| Bovine Coronavirus | |
| Porcine Hemagglutinating Encephalomyelitis Virus | SRSTSR[A/T]#[S/P][S/N]RA[S/P]SAGSR(SR)† |
| HCoV-OC43 | |
| Turkey Coronavirus | |
| Avian Infectious Bronchitis Virus | S(T)RAPSREG SR{Xn}‡SR |
| Porcine Respiratory Coronavirus | |
| Transmissible Gastroenteritis Virus | |
| Canine Enteric Coronavirus | SRDNSRS[R/P] SQSRS[R/Q]SRNRSQSR{Xn}SR |
| Canine Coronavirus | |
| Feline Infectious Peritonitis Virus | SRNN SRSGSQSRSVSR NRSQ{Xn}SR{Xn}SR |
| Feline Coronavirus | |
| Human Coronavirus 229E | SRAPSRSQSRSQ SR{Xn}SR{Xn}SR |
| Porcine Epidemic Diarrhea Virus | SRANSRSRSR{Xn} SR{Xn}SR{Xn}SR{Xn}SR |
Bold letter indicates the marker SR. Normal letter indicates the amino acid between two SRs.
Square brackets indicate this position may be occupied by one of the amino acids in them.
Round brackets indicate the amino acids in them may occur in some viruses while may not in other viruses.
Curly brackets indicate amino acids between SRs, X indicates any amino acid, and subscript letter n indicates the number of amino acids between SRs.
Previous study reported that the N protein of MHV could interact with the M (membrane) protein to help the envelopment of MHV nucleocapsid (7). By targeting RNA recombination, Ding, et al. found that the SR-rich region could not be transferred from MHV to BCoV (bovine coronavirus), which means that this region was unable to be substituted between various species (6). It is believed that the SR-rich region is possibly derived from the SR (or RS) domain of many RNA-binding proteins, such as SR proteins (6). The SR proteins are essential for constitutive mRNA splicing and the regulation of alternative splice site selection (8). The C-terminal SR domain of SR proteins is involved in mediating protein-protein interaction as well as nuclear localization 8., 9.. The SR-rich region might be necessary for the interaction of the N and M proteins. It also possibly contributes to the interaction of the N protein with other viral proteins, including the N protein itself.
Localization to the nucleolus is a common feature of coronavirus N proteins. This feature helps with disrupting host cell division to promote virus assembly and sequestering ribosomes for translation of viral proteins (10). It has been reported that the SR domain in SR proteins is a nuclear localization signal but not a subnuclear speckle one (8). Consequently, the SR-rich region may function only as a nuclear localization signal. Subnuclear localization of the N protein might need several additive and redundant signals. InterproScan has revealed that there is a bipartite nuclear localization signal domain (NLS-BP, IPR001472) in the N protein, which has a sequence of KKKKTDEAQPLPQRQKKQ at Codons 373-390. NLS-BP is a domain for the protein translocation from cytoplasm to nucleus.
Phosphorylation and dephosphorylation of the SR domain in SR proteins are necessary for its function. We also detected that in the N protein, the SR-rich region has eleven possible phosphoserines, which account for 50% of the total. This indicates that the SR-rich region is an important region involved in phosphorylation and this post-transcription modification is greatly required for RNA-binding, oligomerization, and localization to nucleoli of the N protein.
Prediction and immunoassay confirmation of antigenic sites of the N protein
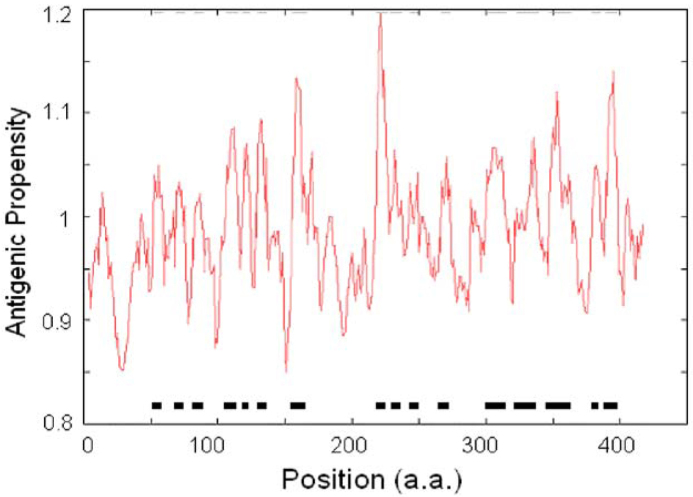
Previous experiments showed that hydrophobic residues, such as cysteine, leucine and valine, on the surface of the protein are most likely to be part of antigenic determinants. Based on a semi-empirical method through making a statistics of appearance frequency of each amino acid in known segmental epitopes (11), we have predicted 16 antigenic sites of the N protein and found they are clustering in the middle and the C-terminus (Figure 4 and Table 3). There is a strong antigenic site (TALALLLLDR) located around Codons 218-227. In addition, another three strong antigenic sites were detected around Codons 156-166 (AATVLQLPQGT), Codons 347-363 (FKDNVILLNKHIDAYKT) and Codons 389-398 (KQPTVTLLPA) (Table 3).
Fig. 4.
The possible antigenic sites of the N protein. We have predicted sixteen antigenic sites of the N protein, and found they are clustering in the middle and the C-terminal. There is a strong antigenic site (TALALLLLDR) located around Codons 218-227. In addition, another three strong antigenic sites are detected around Codons 156-166 (AATVLQLPQGT), Codons 347-363 (FKDNVILLNKHIDAYKT) and Codons 389-398 (KQPTVTLLPA).
Table 3.
The Predicted Antigenic Sites on the SARS-CoV N Protein
| No. | Start Position* | Sequence | End Position |
|---|---|---|---|
| 1 | 52 | SWFTALTQ | 59 |
| 2 | 69 | RGQGVPI | 75 |
| 3 | 83 | DQIGYYR | 89 |
| 4 | 106 | SPRWYFYYLG | 115 |
| 5 | 118 | PEASLPY | 124 |
| 6 | 130 | GIVWVAT | 136 |
| 7 | 156 | AATVLQLPQGT | 166 |
| 8 | 218 | TALALLLLDR | 227 |
| 9 | 229 | NQLESKVSG | 237 |
| 10 | 243 | QGQTVTK | 249 |
| 11 | 267 | KQYNVTQ | 273 |
| 12 | 299 | YKHWPQIAQFAPSASAF | 315 |
| 13 | 323 | MEVTPSGTWLTYHGAIK | 339 |
| 14 | 347 | FKDNVILLNKHIDAYKT | 363 |
| 15 | 379 | EAQPLPQ | 385 |
| 16 | 389 | KQPTVTLLPA | 398 |
amino acid position in the SARS-CoV N protein (BJ01)
To confirm the antigenicity of the N protein, we synthesized peptides of 20-25 amino acids covering the complete N protein. We designed fourteen peptides that are in the region of low conservativeness, excluding those of high conservativeness (Table 4). By using sera samples of nine SARS patients with ELISA (enzyme-linked immunosorbent assay), We found that two out of fourteen peptides showed strong immunogenicity and seven peptides showed medium-strong immunogenicity (Table 4).
Table 4.
The Synthesized Peptides Representing the N Protein of SARS-CoV and Their ELISA Result
| Peptides Number | Start Position* | Sequence | End Position | ELISA Result |
|---|---|---|---|---|
| N1 | 1 | MSDNGPQSNQRSAPRITFGGPTD | 23 | ++ |
| N21 | 21 | PTDSTDNNQNGGRNGARPKQRR | 42 | ++ |
| N35 | 35 | GARPKQRRPQGLPNNTASWFTA | 56 | + |
| N99 | 99 | DGKMKELSPRWYFYYLGTGPEA | 120 | - |
| N161 | 161 | QLPQGTTLPKGFYAEGSRGGSQ | 182 | +++ |
| N177 | 177 | SRGGSQASSRSSSRSRGNSRNS | 198 | ++ |
| N196 | 196 | RNSTPGSSRGNSPARMASGGGE | 217 | - |
| N215 | 215 | GGETALALLLLDRLNQLESKVSGKG | 239 | ++ |
| N245 | 245 | QTVTKKSAAEASKKPRQKRTATKQ | 268 | ++ |
| N258 | 258 | KPRQKRTATKQYNVTQAFGRRG | 279 | + |
| N355 | 355 | NKHIDAYKTFPPTEPKKDKKKK | 376 | ++ |
| N371 | 371 | KDKKKKTDEAQPLPQRQKKQ | 390 | +++ |
| N385 | 385 | QRQKKQPTVTLLPAADMDDFSRQ | 407 | ++ |
| N401 | 401 | MDDFSRQLQNSMSGASADSTQA | 422 | - |
amino acid position in the SARS-CoV N protein (BJ01)
Compared with the experimental outcome on immunogenicity of synthesized peptides, the predicted result about the antigenicity of the N protein is fairly reliable. Synthesized peptides containing the corresponding predicted antigenic sites showed strong or medium-strong immunogenicity, except for peptides N99 (Codons 99-120). This might because that its position is within the conserved region, which decreases the specificity of reaction. The different immunogenicity of peptides in different regions on the N protein is also consistent with the predicted antigenicity map.
It was reported that in the coronavirus-infected cells, the N protein was a more abundant antigen than the S (spike) protein (12). We also performed the same experiments on the S protein of SARS-CoV (see the article about the S protein in this issue). The comparison of both results demonstrates that the percentage of strongly positive results in the N protein (64%) is significantly higher than that in the S protein (28%). One of the explanations might be that the N protein is the most abundant protein produced throughout infection, because its template mRNA is the smallest and it has the most abundant sgRNA (subgenome RNA) during transcription (2). When the infected cells broke up and released the inside content, the N protein showed the strongest antigenicity. Combined with our prediction and experimental results, we believe that the N protein is a predominant antigen of SARS-CoV.
Evolution and substitution of the N protein
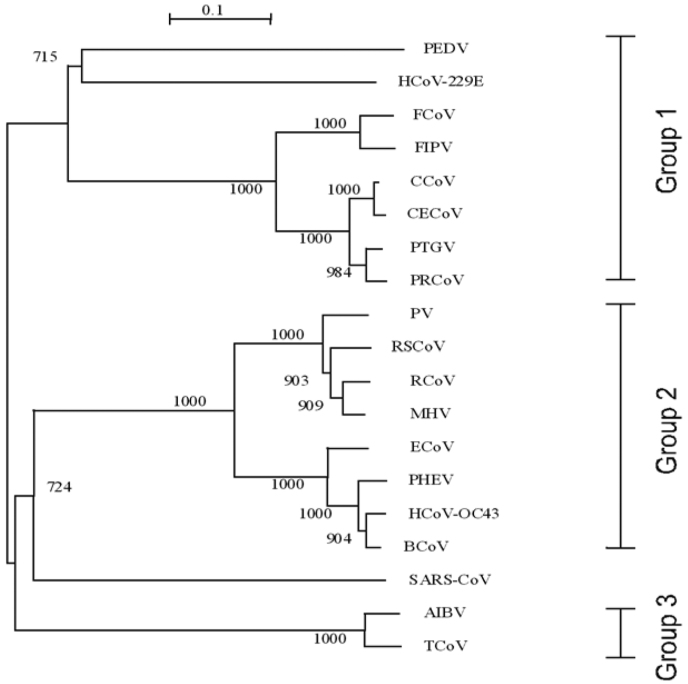
Based on the nineteen coronavirus N proteins, we constructed an evolutionary tree (Figure 5). It reveals that SARS-CoV is closer to Group 2 than to Groups 1 and 3. We also performed a global pair-wise alignment of these nineteen sequences (Table S1). The N protein of SARS-CoV is highly similar to that of Equine coronavirus (49.8% similarity, 34.3% identity), with the lowest similarity to that of human coronavirus 229E strain (32.1% similarity, 21.2% identity).
Fig. 5.
The phylogenetic tree based on nineteen coronavirus N proteins. It reveals that SARS-CoV is closer to Group 2 than to Groups 1 and 3. Abbreviations: AIBV: avian infectious bronchitis virus; BCoV: bovine coronavirus; CCoV: canine coronavirus; CECoV: canine enteric coronavirus; ECoV: equine coronavirus; FCoV: feline coronavirus; FIPV: feline infectious peritonitis virus; HCoV-OC43: human coronavirus strain OC43; HCoV-229E: human coronavirus strain 229E; MHV: murine hepatitis virus; PEDV: porcine epidemic diarrhea virus; PHEV: porcine hemagglutmating encephalomyelitis virus; PRCoV: porcine respiratory coronavirus; PTGV: transmissible gastroenteritis virus; PV: puffinosis virus; RCoV: rat coronavirus; RSCoV: Rat sialodacryoadenitis coronavirus; SARS-CoV: human severe acute respiratory syndrome-associated coronavirus isolate BJ01; TCoV: turkey coronavirus.
To date there are seventeen isolates of SARS-CoV that have complete sequences. Only four substitutions have been identified in the N protein (Table 5). The substitution rate of the N protein is 0.32%, lower than the average of SARS-CoV genome (0.46%). We suggest that the N protein is much more conserved than other structural proteins for its substitution rate is the lowest among all known proteins. These four substitutions resulted in the change of three codons. There are two substitutions juxtaposed within the same codon of leucine. One is at the second nucleotide of Codon 140 (nt position 28,519), leading to amino acid change from leucine to tryptophan, and the other is at the third nucleotide of Codon 140 (nt position 28,520) as a synonymous substitution.
Table 5.
The Four Substitutions in the N Protein of SARS-CoV
| Nt Position in BJ01 | a.a. Position in the ORF | a.a. (Ratio) | Synonymous Substitution |
|---|---|---|---|
| 28,519 | 140 | L(16)/W(1) | No |
| 28,520 | 140 | L(17) | Yes |
| 28,560 | 154 | N(16)/Y(1) | No |
| 28,677 | 193 | G(16)/C(1) | No |
Methods and Materials
Samples and sequences
The SARS patients, from whom the genome sequences of Isolates BJ01-BJ04 were extracted, were diagnosed according to WHO guidelines (http://www.who.int/csr/sars/guidelines/en/) in February and March 2003 in Beijing, China. The processing of tissue samples and viral RNA, RT-PCR, cloning, and sequencing were performed according to standard protocols at Beijing Genomics Institute (BGI) and Center of Disease Control and Prevention of China (13).
The updated complete genome sequences of the BJ Group (BJ01–BJ04) have been deposited by BGI in GenBank (accession numbers: AY278488, AY278487, AY278490, and AY279354) (http://www.genomics.org.cn/bgi/news/zhongxin/news030416-2_fasta.htm). All the sequences and experimental materials are available freely.
Thirteen other full-length sequences of SARS-CoV strains, which have been published by BGI or other laboratories since March 2003, have been used in this study (accession numbers: AY278554, AY297028, AY274119, AY291451, AY283798, AY283797, AY283796, AY283795, AY283794, AY282752, AY278741, AY278491, and AY278489). Nineteen coronavirus N protein sequences were downloaded from GenBank (accession numbers: AAP30037, NP_045302, CAD67607, AAF97743, AAG39339, NP_150083, AAL80036, P33469, AAD33104, AAF23872, NP_040838, P33463, NP_058428, Q04700, BAC65328, BAC01157, BAC01161, NP_073556, and NP_598314). The nucleotide positions of SARS-CoV are all referred to the complete genome sequence of Isolate BJ01 (14).
Structure and function analysis
ORF Finder (http://ww.ncbi.nlm.nih.gov/gorf/gorf.html) was used to determine ORFs, EMBOSS package (http://www.hgmp.mrc.ac.uk/Software/EMBOSS/) to characterize proteins, NetPhos (http://www.cbs.dtu.dk/services/NetPhos/) and PhosphoBase (http://www.cbs.dtu.dk/databases/PhosphoBase/) to predict phosphorylation sites, TopPred2 (http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html) and ProtScale (http://us.expasy.org/cgi-bin/protscale.pl) to identify the hydrophobic region, ClustalW (http://www-igbmc.u-strasbg.fr/BioInfo/ClustalW/) to perform multiple-alignment and phylogenetic analysis, PSIPRED (http://bioinf.cs.ucl.ac.uk/psiform.html) to analyze secondary structures, InterproScan (http://www.ebi.ac.uk/interpro/) and CDS (Conserved Domain Search, http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) to predict conserved protein domains. All the analyses were accomplished on supercomputers DOWNING 2000/3000 (DOWNING Computers Inc., Beijing, China), SUN E10K (SUN Microsystems Inc., California, USA), SGI Origin 3800 (Silicon Graphics, Inc., California, USA), and IBM P690 (IBM Corp., New York, USA).
Design and synthesis of peptides
The peptides were designed on the basis of the combined analyses of the proteins. The peptides were synthesized by Hangzhou Zhongtai Inc.(Hangzhou, China).
ELISA test
Blood samples of two normal controls and nine SARS patients were provided by Beijing Plastic Surgery Hospital, Beijing Peoples’ Hospital, and Beijing Tiantan Hospital. Peroxidase-conjugated mouse anti-human IgG and peroxidase-HRP (P6782) were purchased from Sigma (New Jersey, USA). Peptides (1 μg/mL, in 0.5 M carbonate buffer, pH 9.6) were dispensed into a 96-well microplate (100 μL/well) and then incubated at 4°C overnight. After being washed with PBS containing 0.5 M Tween-20 (PBS-T), BSA (2 mg/mL) was added up and the plates were incubated at 37°C for 1 h for blocking. The patient serum sample (10 μL), diluted with 100 μL of sample buffer, was added into each well and incubated at 37°C for 30 min. After being further washed with PBS-T, 100 μL mouse anti-human IgG was added and incubated at 37°C for 20 min. Finally, the wells were washed with PBS-T. The reaction was observed by adding the TMB solution as substrate, after inculation at 37°C for 10 min. The reaction was stopped by adding 50 μL 4 M sulphuric acid, and optical density at 450 nm (ref. 630 nm) was measured with an automatic ELISA reader (Multiskan Ascent, Finland).
Acknowledgements
The authors thank the Ministry of Science and Technology of China, Chinese Academy of Sciences, and National Natural Science Foundation of China for financial support. We are indebted to collaborators and clinicians from National Center of Disease Control of China, Beijing Plastic Surgery Hospital, Beijing Peoples’ Hospital, Beijing Tiantan Hospital, the Provincial Government of Zhejiang, the Provincial Government of Inner Mongolia, the Municipal Governments of Beijing and Hangzhou, and the Library of Chinese Academy of Science. Special gratitude is expressed here to the patients and their families for their devotion and cooperation. We appreciate the comments of Dr. Gwendolyn Zahner, visiting professor at BGI, Dr. Qimin You, Dr. Lin Hu and other colleagues on drafts of this manuscript.
Supporting Online Material
http://www.gpbjournal.org/journal/pdf/GPBl(2)-07.htm
Table S1
References
- 1.Ksiazek T.G. A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 2.Hiscox J.A. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 2001;75:10506–10512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wootton S.K. Phosphorylation of the porcine reproductive and respiratory syndrome virus nucleocapsid protein. J. Virol. 2002;76:10569–10576. doi: 10.1128/JVI.76.20.10569-10576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laity J.H. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 5.Molenkamp R., Spaan W.J.M. Identification of a specific interaction between the coronavirus mouse hepatitis virus A59 nucleocapsid protein and packaging signal. Virology. 1997;239:78–86. doi: 10.1006/viro.1997.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding P. Construction of murine coronavirus mutants containing interspecies chimeric nucleocapsid proteins. J. Virol. 1995;69:5475–5484. doi: 10.1128/jvi.69.9.5475-5484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narayahan K. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cáceres J.F. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuyama S., Bruzik J.P. Multiple roles for SR proteins in trans splicing. Mol. Cell. Biol. 2002;22:5337–5346. doi: 10.1128/MCB.22.15.5337-5346.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wurm T. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J. Virol. 2001;75:9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolaskar A.S., Tongaonkar P.C. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276:172–174. doi: 10.1016/0014-5793(90)80535-q. [DOI] [PubMed] [Google Scholar]
- 12.Daginakatte C.G. Production, characterization, and uses of monoclonal antibodies against recombinant nucleoprotein of Elk Coronavirus. Clin. Diagn. Lab. Immunol. 1999;6:341–344. doi: 10.1128/cdli.6.3.341-344.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Q.Y. Isolation and Identification of a Novel Coronavirus from Patients with SARS. Chin. J. Biotechnol. 2003:4. [Google Scholar]
- 14.Qin E.D. A complete sequence and comparative analysis of SARS-associated virus (Isolate BJ01) Chin. Sci. Bull. 2003;48:941–948. doi: 10.1007/BF03184203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1