Abstract
To obtain a primary overview of gene diversity and expression pattern in Lycoris longituba, 4,992 ESTs (Expressed Sequence Tags) from L. longituba bud were sequenced and 4,687 cleaned ESTs were used for gene expression analysis. Clustered by the PHRAP program, 967 contigs and 1,343 singlets were obtained. Blast search showed that 179 contigs and 227 singlets (totally 1,066 ESTs) had homologues in GenBank and 3,621 ESTs were novel.
Key words: Lycoris longituba, EST
Introduction
A rapid growing area of genome research is the generation of expressed sequence tags (ESTs) in which large numbers of randomly selected cDNA clones are partially sequenced. The collection of ESTs reflects the level and complexity of gene expression in the sampled tissue (1). EST provides a rapid method of gene discovery that has been widely applied to human and other species (2).
Lycoris longituba is a perennial, bulbiferous, herbaceous plant in the genus of Lycoris, Amaryllidaceae family. A unique biological characteristic of this plant is that its leaves come out in spring and die in early summer, followed by flowering. Its vegetative growth and reproduction are discrete, which is rare in angiosperm. With a vigorous and long scape, its flowers are diverse in both color and floral form, and fragrant in smell, having a high ornamental value. With a reputation for Chinese tulip, this species is ideal for ikebana. So far, there has been only a little research on floral ESTs all over the world, and there is no ESTs recorded in the genus of Lycoris in public EST databases. Therefore, studies on floral form and color of L. longituba described in this paper will establish a theoretical foundation for the isolation and cloning of genes related to these properties.
We collected more than 70 variants of this species in our laboratory with different floral colors and forms. In order to understand the reasons behind the variation, an L. longituba bud cDNA library, named LLB, was constructed for analyzing EST. To identify flowering-related genes and analyze expression pattern in L. longituba, 4,992 ESTs from this library were sequenced and analyzed.
Results
The sizes of inserts in the LLB library were 0.5–3.0 Kb. Totally 4,992 clones were randomly selected and sequenced from the 5’ end. After removing all sequences that were either contaminated or of low quality (Q20<100 bp), 4,687 cleaned EST sequences were available for further analysis. The average read-length of cleaned ESTs was 545 bp. The cleaned ESTs were assembled into 967 contigs and 1,343 singlets (clusters) by the PHRAP program. Blast search showed that 406 clusters (1,066 ESTs) had homologues in GenBank. Among them, 946 (20.18%) ESTs had functional homologues in NR, Swissprot, or unigene databases of rice, maize and Arabidopsis. In the other 3,621 (79.82%) ESTs, there were no significant homology to known sequences (Table 1).
Table 1.
General Data of ESTs from LLB
| ESTs | Number | Percentage (%) |
|---|---|---|
| Total ESTs | 4,992 | |
| Cleaned ESTs | 4,687 | 93.89 (4,687/4,992) |
| Average length (bp) | 545 | |
| Average GC content | 39.46 | |
| Average score (quality) | 39.28 | |
| Known genes matched EST | 946 | 20.18 (946/4,687) |
| Repeats | 169 | 3.61 (169/4,687) |
| rRNA | 9 | 0.19 (9/4,687) |
| Mitochondrial | 1 | 0.021 (1/4,687) |
| Novel transcripts | 3,621 | 77.26 (3,621/4,687) |
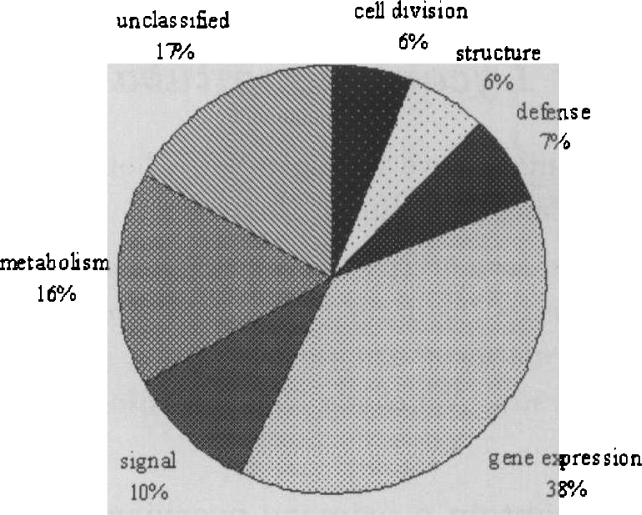
All the clusters annotated in GenBank were classified into seven groups according to their functions: signaling and communication, gene expression, cell division, structure and motility, defense and homeostasis, metabolism, and unclassified (Figure 1; ref. 3).
Fig. 1.
Classification of ESTs annotated functions from the LLB library.
BLASTX searches of L. longituba bud ESTs revealed that 3.3% (156 of 4,687) ESTs were significant matched to plant transposable elements (TEs). Among them, 29% (45 of 156) ESTs were homologous to class-I DNA elements (Mutator, Ac/Ds, or En/Spm), and the others were homologous with polyproteins or reverse transcriptases of class-II RNA elements (copia, gypsy; ref. 4)· Rossi et al. reported a lower range of TEs among sugarcane ESTs (4); and Kuhl et al. reported a similar range of sugarcane TEs among onion ESTs (5).
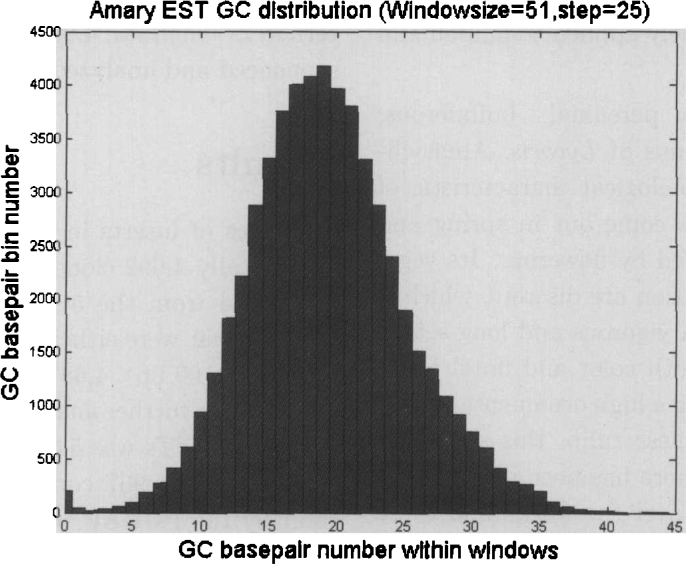
In previous studies, GC gradient was described as a property of Gramineae genes and perhaps all monocot genes, yet it has not been observed in eudicot genes so far (6). To capture the general GC characteristics of Lycoris radiate contig consensuses (total 2,310 consensuses), we used a 51-bp sliding window. The GC number within the window for each consensus segment was counted. As shown in Figure 2, the statistics basically follow a normal distribution, meanwhile a slight left shift. The peak at the x-axis at zero is likely to be fluctuated by poly-A signal.
Fig. 2.
GC distributions of Lycoris radiate contig consensuses (sliding window size 51 bp, step length 25 bp). The gradient is the slope of the trend in segment number versus average GC number within sliding window.
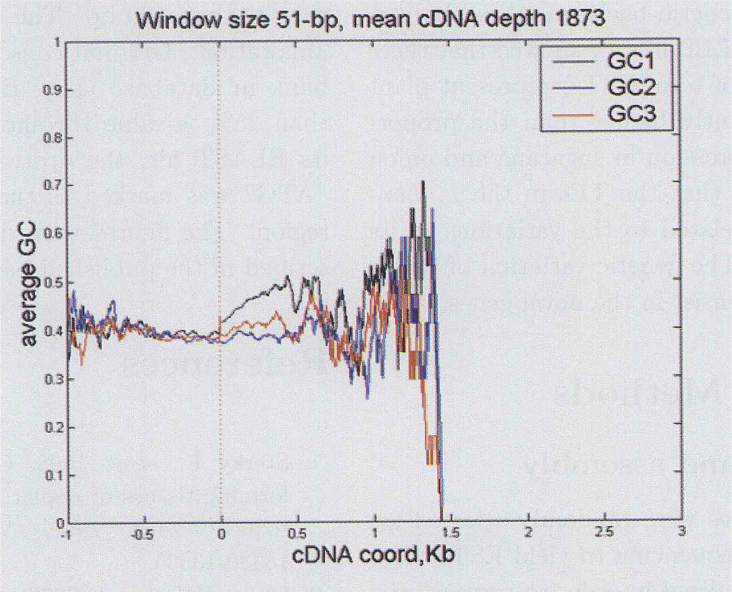
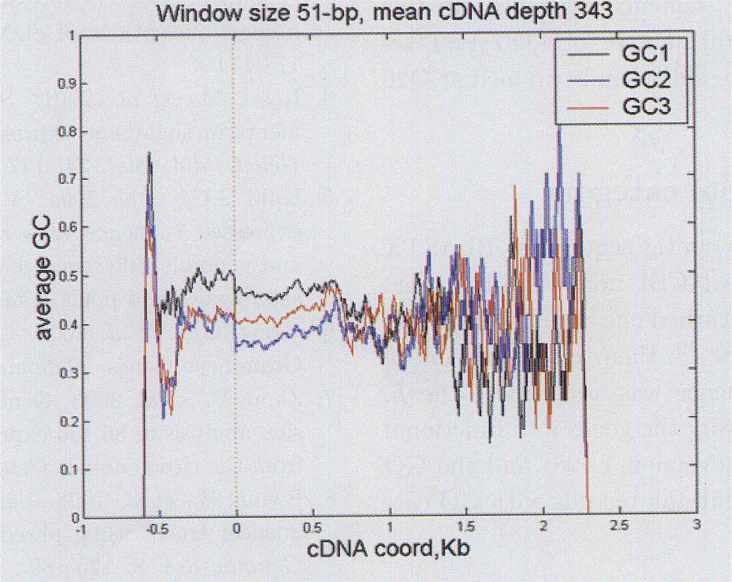
To present the GC gradient property of Lycoris radiate EST, two approaches predicting the most possible coding regions of EST contig consensuses were used (See Materials and Methods) before applying the published process (6). Totally 1,873 and 343 consensuses were retrieved respectively using the ORF (open reading frame) and annotation methods. Fig. 3, Fig. 4 depict how much effect is attributable to each of the three codon positions: GC1, GC2 and GC3. Differently from expectation, the GC gradient is not remarkable. By contraries, the distribution is slightly skewed toward positive GC content gradients. This phenomena is more obvious in Figure 3 than in Figure 4. Besides, the attribution to each of the three codon positions is also different. Interestingly, previous studies indicated that the relative magnitudes for the Gramineae genes were GC3>GC1>GC2, whereas in our study they were GC1>GC3>GC2.
Fig. 3.
GC1, GC2 and GC3 content as a function of cDNA position, relative to the start of the coding region (predicted by EMBOSS getorf, see Materials and Methods), and averaged over all cDNAs with a 51-bp sliding widow. Phase information is extended into the 5’ UTR.
Fig. 4.
GC1, GC2 and GC3 content as a function of cDNA position, relative to the start of the coding region (predicted by nr annotation, see Materials and Methods), and averaged over all cDNAs with a 51-bp sliding widow. Phase information is extended into the 5’ UTR.
Discussion
In this research, a cDNA library of L. Longituba bud was constructed, and 4,687 ESTs were collected after quality assessment (trimmed at Q20, Phred scords; Table 1; ref. 7). The mean length was 545 bp and the mean Phred score was 39.28. The content of rRNA and mitochondrial mRNA was very low, indicating that the LLB library and the ESTs sequenced from it were of high quality.
The ESTs analysis will help us to uncover the reason behind the variation in the flower of L. Longituba in condition that its genetic background is obscure. Gene expression of the LLB library showed that there were 156 ESTs (3.3% of total ESTs) represent plant TEs. This is significantly higher than the proportion (0.8%) of TEs expression in sugarane and onion (4., 5.). It is suggested that the TEs in the L. Longituba bud might be related to the variations in its floral color and form. The genetic variation of flower are also supposed to consist in the novel genes.
Materials and Methods
EST sequencing and assembly
Clones from the libraries were randomly selected for single-pass, mostly 5’ sequencing to yield ESTs. The libraries were not normalized in order to preserve the random nature of the original expression patterns for quantitative analysis. We used the Phred program for base calling (8., 9.), Cross_match for vector sequences masking, and Phrap for sequence assembly. In this study we collected a total number of 4,687 EST sequences after quality assessment and trimmed at Q20 (Phred scores).
Assigning functional category
To assign annotation to contig sequences, BLASTX was used to search the NCBI non-redundant (nr) database. If BLASTX returned one or more sequences with Ε-value less than 10−10, then the annotation of the highest scoring sequence was assigned to the L. longituba contig. To classify the genes into functional categories, we used GO (Version 1.408) and the GO indices that associate GenBank records with GO categories.
GC content
To capture the general GC characteristics of Lycoris radiate contig consensuses (total 2,310 consensuses), we used a 51-bp sliding window. The GC number within the window for each consensus segment was counted. To present the GC gradient property of Lycoris radiate EST, two approaches predicting the most possible coding regions of EST contig consensuses were used. One is to extract the ORFs by EMBOSS getorf. We picked the longest ORF that begins with a start codon and ends with a stop codon, and then excluded sequences whose ORFs’ lengths are shorter than 100 bp. The other approach is by gene annotation. Original consensus was aligned to Genbank nr database using BLASTX (alignment great than 25%, e-value thresholds le-10). According to its BLAST hit, the upstream in frame start codon “ATG” was marked as the starting point of coding region. The figures were drawn by the methods described in the published paper (6).
Acknowledgments
The work was supported by Hi-tech Research & Development Program of China (863 Program, No. 2002AA241051) and Science & Technology Program for Agriculture Development of Shanghai.
References
- 1.Sterky F. Gene discovery in the wood-forming tissues of poplar: analysis of 5,692 expressed sequence tags. Proc. Natl. Acad. Sci. USA. 1998;95:13330–13335. doi: 10.1073/pnas.95.22.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y.J. Gene expression profiling in porcine fetal thymus. Geno. Prot. Bioinfo. 2003;1:171–172. doi: 10.1016/S1672-0229(03)01021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams M.D. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature. 1995;377:173–174. [PubMed] [Google Scholar]
- 4.Rossi M. Survey of transposable elements in sugarcane expressed sequence tags (ESTs) Genet. Mol. Biol. 2001;24:147–154. [Google Scholar]
- 5.Kuhl J.C. A unique set of 11,008 onion expressed sequence tags reveals expressed sequence and genomic differences between the monocot orders asparagales and poals. Plant Cell. 2004;16:114–125. doi: 10.1105/tpc.017202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong G.K. Compositional Gradients in Gramineae Genes. Genome Res. 2002;12:851–856. doi: 10.1101/gr.189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y. Gene identification and expression analysis of 86,136 expressed sequence tags (EST) from the rice genome. Geno. Prot. Bioinfo. 2003;1:26–42. doi: 10.1016/S1672-0229(03)01005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewing В. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genoms Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 9.Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]