Abstract
Microsatellites or simple sequence repeats (SSRs) have been found in most organisms during the last decade. Since large-scale sequences are being generated, especially those that can be used to search for microsatellites, the development of these markers is getting more convenient. Keeping SSRs in viewing the importance of the application, available CDS (coding sequences) or ESTs (expressed sequence tags) of some eukaryotic species were used to study the frequency and density of various types of microsatellites. On the basis of surveying CDS or EST sequences amounting to 66.6 Mb in silkworm, 37.2 Mb in fly, 20.8 Mb in mosquito, 60.0 Mb in mouse, 34.9 Mb in zebrafish and 33.5 Mb in Caenorhabditis elegans, the frequency of SSRs was 1/1.00 Kb in silkworm, 1/0.77 Kb in fly, 1/1.03 Kb in mosquito, 1/1.21 Kb in mouse, 1/1.25 Kb in zebrafish and 1/1.38 Kb in C. elegans. The overall average SSR frequency of these species is 1/1.07 Kb. Hexanucleotide repeats (64.5%–76.6%) are the most abundant class of SSR in the investigated species, followed by trimeric, dimeric, tetrameric, monomeric and pentameric repeats. Furthermore, the A-rich repeats are predominant in each type of SSRs, whereas G-rich repeats are rare in the coding regions.
Key words: insect, eukaryote, CDS, EST, microsatellite
Introduction
Microsatellites or simple sequence repeats (SSRs) are tandem short stretches of DNA and consist of the repeat units of 1-6 bp in length. They are ubiquitous in eukaryotic genomes and can be analyzed by PCR (polymerase chain reaction) technology. The SSR markers have been extensively used for genetic mapping and population studies (1). SSRs also provide molecular tools to understand spatial relationships among the chromosome segments, which in turn, aid in analyzing temporal relationships between species and genera (2). Furthermore, it is abundant in each organisms, for instance, in humans about 3% of the genome is occupied by SSRs, and it is becoming clear that such repeats are important in genomic organization and function and may be associated with disease conditions 3., 4.. However, their systematic analysis between species has not been much reported. Moreover, the development of SSR markers is expensive, labor intensive and time consuming, in particular if they are developed from genomic libraries.
Even though, because of the importance of microsatellites, they have been developed in a large number of species such as maize (5), rice 6., 7. and mouse (8). Thanks to the current emphasis on functional genomics, coding sequences (CDS) are fast accumulating in genomics and EST (expressed sequence tag) databases of a large number of species. These CDS or EST databases can be mined for SSRs that would be served for designing locus-specific primers. Following this procedure, SSR markers can be obtained at significantly reduced costs, as CDS-derived (including EST) SSRs are free by-product of the currently expanding CDS databases. While CDS-derived SSRs have been shown to be less polymorphic than those derived from genomic sequences (9), they have some intrinsic advantages: they are quickly obtained by electronic sorting, unbiased in their repeat type, present in gene-rich regions of the genome, and are still abundant (10). Since they represent the transcribed part of the genome, CDS-based SSR markers lead to the direct mapping of genes. Furthermore, compared to SSR markers derived from genomic DNA sequences, these CDS-based SSR markers have a higher level of transferability among related species as they are located in more conserved regions of the genome (11). Also, certain repeats are preferred and often predominant in certain genomic locations, for examplee, triplets predominate in coding regions. However, the significance of this observation is unclear (12). There is accumulating evidence to suggest that SSRs function to regulate gene expression 13., 14.. The study of repeat density and its distribution pattern in the genome, especially in CDS or EST, is expected to help in understanding their significance and controlling diseases. Comparing the density and distribution of SSRs in several eukaryotic species, consensus and different characters of SSRs in Bombyx mori can be used for breeding purposes, since the hypervariability of SSRs has been proven to be the markers of choice in genetics research. The availability of large number genome sequences of many organisms has made SSRs possible to carry out genome-wide analyses. In the study here, CDS or EST databases of three insects and other three eukaryotic species were mined for the presence of SSRs (1–6 bp) and were analyzed on their frequency and density for the development of genetic markers.
Results and Discussion
Occurrence and density of microsatellites
A large set of CDS or EST data representing 66.6 Mb in silkworm, 37.2 Mb in fly, 20.8 Mb in mosquito, 60.0 Mb in mouse, 34.9 Mb in zebrafish and 33.5 Mb in Caenorhabditis elegans was procured from the public database and our silkworm EST project. The analysis of CDS or EST of these species for occurrences of various microsatellites showed in specific that the frequency of SSRs amounted to 1/1.00 Kb in silkworm, 1/0.77 Kb in fly, 1/1.03 Kb in mosquito, 1/1.21 Kb in mouse, 1/1.25 Kb in zebrafish and 1/1.38 Kb in C. elegans. The estimate of the total SSR frequency calculated here indicates the similarity across the investigated insects and other eukaryotic species, suggesting that SSRs occur at the frequency of every 0.77–1.4 Kb in the investigated insects and eukaryotic species. The overall average of SSR frequency for these species is 1/1.07 Kb, resulting in 234,982 SSRs in a total of 251 Mb of coding sequence. The frequency of SSRs in these species is not comparable to earlier results with some differences of increased frequency 11., 15.. This difference may be explained due to a variation in the quantity of the analyzed sequence data and the differences in defining the criteria for SSR mining in the CDS or EST databases.
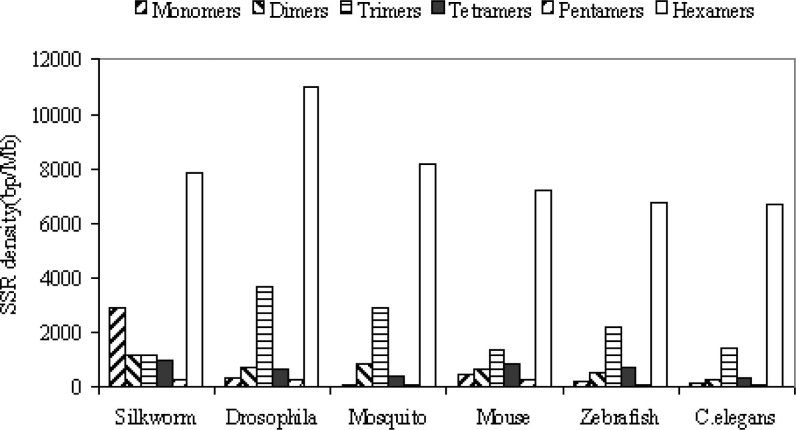
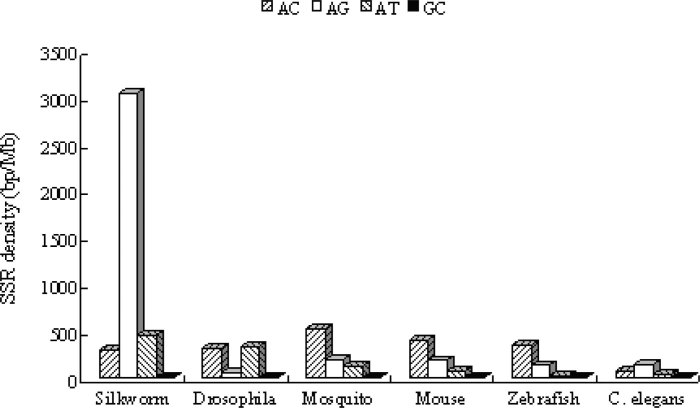
The density of SSRs of these species was also analyzed as shown in Figure 1. The results showed that there is a slight increase in SSR density of insect species compared to other three eukaryotic species. The highest SSR density was found in fly (16,632 bp/Mb), followed by silkworm (14,352 bp/Mb) and mosquito (12,473 bp/Mb), and the lowest SSR density was found in C. elegans (8,981 bp/Mb) and zebrafish (10,387 bp/Mb). Hexanucleotide repeats (6,691–10,970 bp/Mb) are the most abundant class of SSRs in all the species. This is the same as the analysis in the entire human genome (4). The trimeric, dimeric, tetrameric, monomeric and pentameric repeats are represented in decreasing proportions of 1,169–3,648 bp/Mb, 267–1,190 bp/Mb, 353–997 bp/Mb, 70–2,873 bp/Mb and 67–277 bp/Mb, respectively. It should be noted that all the SSRs densities of fly including hexamers and trimers are higher than those of other species.
Fig. 1.
Comparison of densities of each SSR across the eukaryotic genomes. The density is expressed in base-pairs of SSRs sequence per megabase-pairs of dataset sequences.
Effect of non-redundant EST database on distribution and abundance of SSR
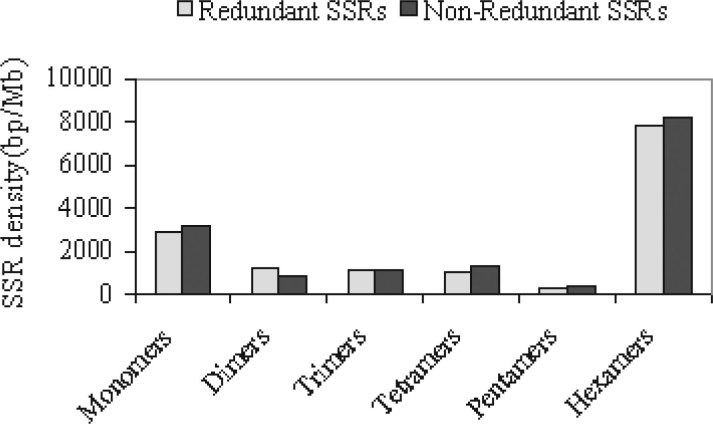
Latter-mentioned SSR density of silkworm is based on a redundant set of ESTs. Thus it may not provide a true picture on the frequency and density of SSRs in the expressed portion of the genome. However, for the silkworm ESTs, we compared the distribution and density of microsatellites in redundant and non-redundant SSRs (Figure 2). In both cases, the density of different types of microsatellites is comparable. In the silkworm genome, we have demonstrated that the frequency of non-redundant SSRs is 1/ 0.95 Kb in the expressed portion of the silkworm genome on the basis of the identification of 14,930 non-redundant SSRs in a set of 17,661 assembled ESTs (representing 13.7 Mb). The detailed analysis suggests that, apart from minor deviations, there is no significant difference in the distribution and density of microsatellites between the redundant and non-redundant sets of silkworm ESTs. The results also demonstrated that it is reliable to develop the SSR markers for a given redundant set of ESTs (11).
Fig. 2.
Comparative analysis of redundant and non-redundant SSRs in silkworm EST set. Redundant SSRs were identified in total set of ESTs (116,914), and non-redundant SSRs were identified in non-redundant set of ESTs (14,081) and were selected after assembling.
Distribution of microsatellite classes
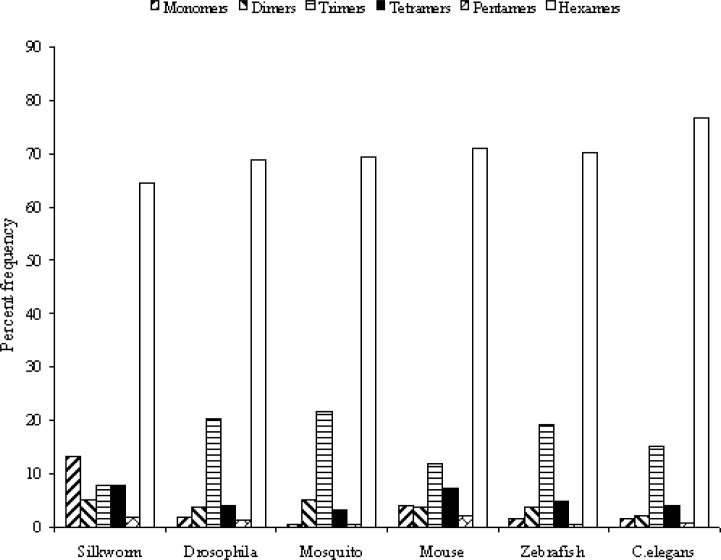
The proportion of the various classes of SSRs (that is, mono-, di-, tri-, tetra-, penta- and hexameric repeats) was not evenly distributed in all the species. The hexameric repeats in the range of 64.5%-76.6% of the total SSRs are the most abundant class of microsatellites in all the species (Figure 3). The trimeric, dimeric, tetrameric, and monomeric repeats are represented in decreasing proportions of 7.6%-21.5%, 2.2%-5.2%, 3.1%-7.8% and 0.5%-13.1%, respectively.
Fig. 3.
Distribution of SSRs in eukaryotic genomes. In all species hexameric repeats are the most frequent followed by trimeric repeats, while penatameric repeats are under 2% of the total number of SSRs.
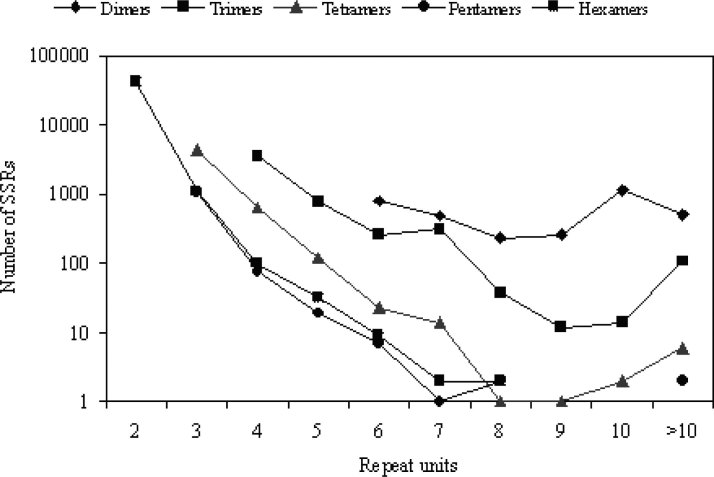
The pentameric repeats were the least frequent (always <2%). These findings are in consistency with previous observations about differences in abundance of SSR unit sizes classes (7). It can be concluded that the hexameric and trimeric SSRs are highly abundant in the coding region sequences. This dominance of hexameric and trimeric SSRs over mono-, di-, tetra-, and pentameric ones may be explained on the basis of the suppression of non-trimeric-times SSRs in coding regions due to the risk of frameshift mutations that may occur when those microsatellites alternate in size of one unit (16). We have also confirmed previously aspects that for all the investigated species and every class of microsatellites, the frequency of microsatellites decreases with increasing repeat length (7). In silkworm, for instance, the single category of SSRs consisting of four repeat units represents 69.7% of the total number of trimeric SSRs, and among the tetrameric SSRs, the category with three repeat units shares as much as 84.3% of the total class (Figure 4). If all microsatellites of different types are classified into two categories of <10 and >10 repeat units, we observe that the category of >10 repeat units contributes only as much as 25% to the total number of microsatellites (data not shown). In a few cases, especially in the tetrameric, pentameric and hexameric microsatellites, all the microsatellites (100%) fall into the category of <10 repeat units.
Fig. 4.
An example of the distribution of microsatellites of different repeat units in ESTs of silkworm. The number of SSRs in a particular class decreases with the increasing number of repeat units.
Between the two types of monomer repeats, poly(A) or poly(T) was far more abundant than poly(C) or poly(G) in all the species. These findings are in consistency with previous observations about differences in abundance for monomer repeats 4., 17..
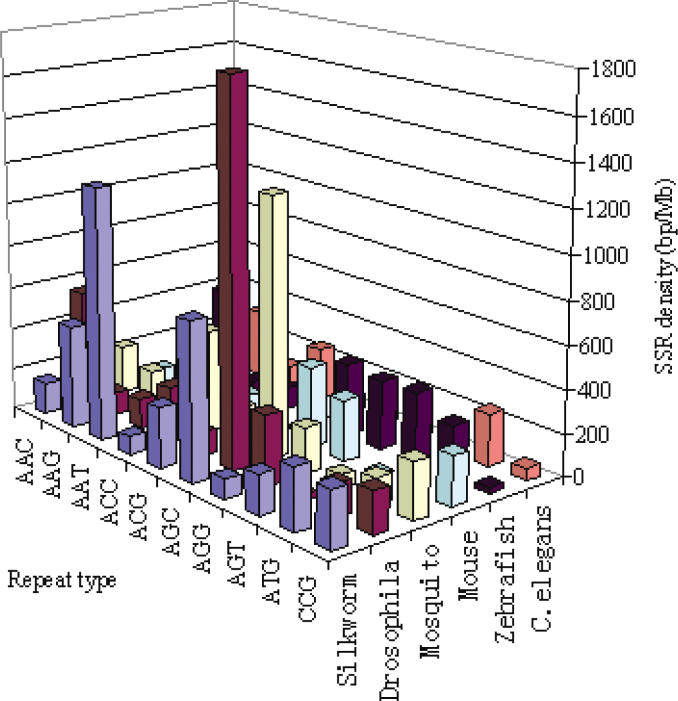
All dimeric repeat combinations excluding homomeric dimers can be grouped into four unique classes, namely, (AT)n, (AG)n, (AC)n, and (CG)n. It is evident that in silkworm, AG and AT repeats are more frequent, followed by AT and AG repeats, respectively. In contrast, other species contain more AC repeats, followed by AG and AT repeats. However, in C. elegans genome, AG repeats seem to be like those of silkworm, but just take only a slight predominant compared with other dimeric repeats. Interestingly, CG dimeric repeats are not only extremely rare in CDS or EST of the genomes studied (Figure 5), but also rare in the entire genome of many species (18). Lower frequency of CpG dinucleotides in vertebrate genomes has been attributed to methylation of cytosine, which, in turn, increases its chances of mutation to thymine by deamination (19). However, CpG suppression by this mechanism cannot explain the rarity of (CG)n dinucleotide repeats in invertebrate, since they do not show cytosine methylation (17).
Fig. 5.
Density of different dimeric repeats per million base pairs of CDS or ESTs sequences in different genomes.
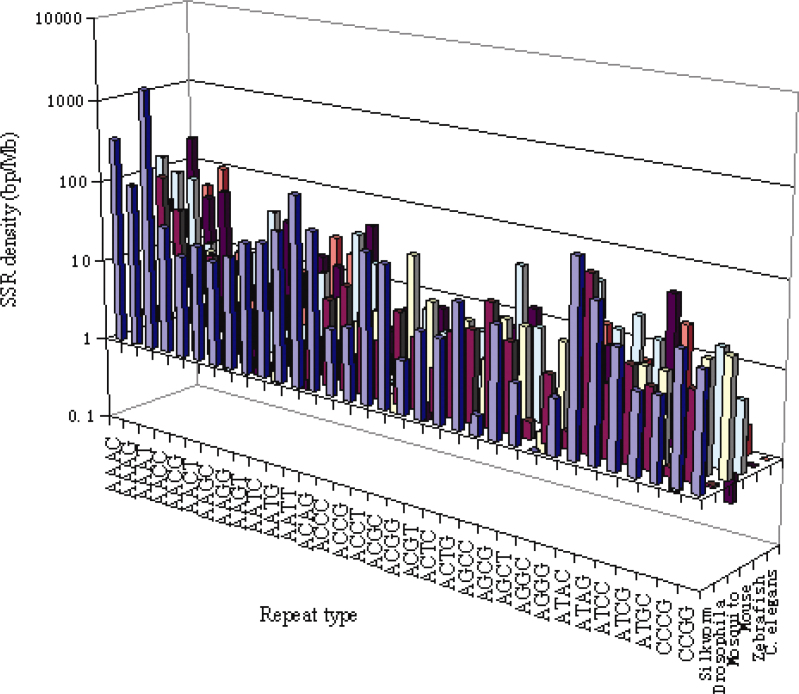
Among the trimeric repeats, the motifs AAT are the most common in silkworm, followed by AGC and AAG repeats, respectively. Drosophila melalogaster, mosquito and mouse genome have comparatively higher frequency of AGC trimeric repeats, followed by AAC, ACC and AGG motifs, respectively. In contrast, the zebrafish contains more AAG, AGC, AGG, AGT, and ATG repeats. However, C. elegans contains more AAG, ACC, and ATG repeats (Figure 6). It should be noted that densities of trinucleotide repeats in the coding regions could be partially limited by selection at the protein level (17). However, the different abundance and density of different trimeric repeats were also reported in previous investigations for different species 4., 7., 17.. This suggests that in addition to alternative DNA structures formed by repeat motifs, species-specific cellular factors interact with trimeric repeats, which are likely to play an important role in the genesis of repeats (18).
Fig. 6.
Density of different trimeric repeats per million base pairs of CDS or ESTs sequences in different genomes.
Analysis of density of each tetrameric repeat type revealed that AAAT, AAAG, AAAC, AATT, and ATAC were the predominant types across all the species. The overall densities of tetrameric repeats such as AATC, AATG, AACC, AACG, AAGG, ATAG, ATCC, ATCG, ACAG, ACGC and AGGG were shown in Figure 7. Surprisingly, within one class of repeats there may be a lot of difference in the abundance of a particular sequence repeat. In the case of monomeric repeats, the density of poly(A) or poly(T) is far more than that of poly(G) or poly(C). Similarly, in the case of dimeric repeats, AG, AT and AC are more abundant and CG is the least abundant. Furthermore, predominant repeats in the other various classes are AAT, AAC and AAG among trimers, while AGC is predominant too, AAAT, AAAC and AAAG in the case of tetramers, AAAAT, AAAAC and AAAAG in the case of pentamers and AAAAAT, AAAAAC and AAAAAG among hexamers. This case also existed in human, fungi and embryophytes genomes 4., 28.. It is possible that during SSR evolution the poly(A) stretches present in the genome might have been mutated to produce the A-rich repeats. It is also possible that the abundance of repeats is influenced by their secondary structures and the effect on DNA replication. If a repeat sequence is selected during evolution for transcriptional regulation or as the target of a binding protein for one or more nuclear processes (such as chromatin organization, DNA replication, transcription, and recombination), its abundance and distribution are expected to be controlled (4).
Fig. 7.
Density of different tetrameric repeats per million base pairs of CDS or ESTs sequences in different genomes.
For the abundance and distribution of SSRs in silkworm, Reddy et al. researched it successfully in the partial genome for the first time (20). They used end-labeled oligonucleotides (GT)10 and (CT)10 as probes to hybridize in partial genome library and then analyzed 28 microsatellites loci. The obtained conclusions were i) (GT)n and (CT)n were abundant in the silkworm genome; and ii) (GT)n was more abundant than (CT)n. In the present analysis, (GT)10 and (CT)10 belong to AC and AG types, respectively. Indeed, AC and AG repeats are abundant, but AG is much more abundant than AC (Figure 5). Analysis on the abundance and distribution of SSRs on the whole genome might be more accurate.
The study of SSRs in these species is just the first step towards understanding the biology of the coding DNA, and it may help us understand numerous aspects of genome organization and function. Furthermore, using this method, EST or CDS databases can be systematically searched for SSRs for the development of microsatellite markers, which are associated with transcribed genes. This approach saves both costs and time, gives a sufficient amount of available EST sequences, and can be a powerful approach to accelerate the molecular analysis of genetics, evolution, genome organization and function, and so on.
Materials and Methods
Sequence data sources
The CDS sequences that are available in the public domain of different species, fly (Drosophila melalogaster), mosquito (Anopheles gambiae), mouse (Mus muscles), zebrafish (Danio rerio) and worm (Caenorhabditis elegans), were downloaded in FASTA format from ftp://ftp.ensembl.org/pub/ in June 2003. In addition, the data of the silkworm EST-database from our lab containing 81,635 ESTs presently (80,475 entrys of which has been submitted to NCBI, accession number: CK484630-CK565104) and SilkBase (http://www.ab.a.u-tokyo.ac.jp/silkbase/) containing 35,300 ESTs from a variety of different tissues were also used for the analysis of SSRs in this paper.
Searching microsatellites
EST sequences less than 100 bp in length were not included in the analysis here. The identification and localization of microsatellites were carried out by a Perl5 script, which is capable to identify perfect microsatellites. While classifying the microsatellites into different repeat types or categories, sequence complementary was also considered, for example, repeat motifs AG, GA, TC and CT were put in the same class. For searching SSRs by the Perl5 script, microsatellites were considered to contain motifs that are in size ranged from 1 to 6 nucleotides. All theoretically possible 501 SSR types (21) were analyzed for their abundance and density per Mb. The distribution of perfect repeats with the length ≥ 12 bp was as a rule to analyze here. Thus, for a 12-bp SSR, one occurrence may comprise a repeat of 12 monomers, or six dimmers, or four trimers, or three tetramers (or pentamers), or two hexamers. The rationale for choosing the small cutoff value was that the SSRs are often disrupted by single base substitution (4).
Acknowledgements
We wish to express our thanks to Jifeng Qian, Chen Ye, Guoqing Li, Ruiqiang Li, Heng Li and Wei Tong for trimming, cleaning and managing the set of sequences.
This work was supported by the Hi-Tech Research and Development Program of China (863 Program) and the National Natural Science Foundation of China (No. 30300262).
Contributor Information
Qingyou Xia, Email: xiaqy@swau.cq.cn.
Cheng Lu, Email: lucheng@swau.cq.cn.
References
- 1.Dib C. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:149–152. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 2.Kashi Y. Simple sequence repeats as a source of quantitative genetic variation. Trends Genet. 1997;13:74–78. doi: 10.1016/s0168-9525(97)01008-1. [DOI] [PubMed] [Google Scholar]
- 3.International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Subramanian S. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu J. Inconsistency between SSR groupings and genetic backgrounds of white corn inbreds. Maydica. 2001;46:133–139. [Google Scholar]
- 6.Temnykh S. Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.) Theor. Appl. Genet. 2000;100:697–712. [Google Scholar]
- 7.Temnykh S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001;11:1441–1452. doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhodes M. A high-resolution microsatellite map of the mouse genome. Genome Res. 1998;8:531–542. doi: 10.1101/gr.8.5.531. [DOI] [PubMed] [Google Scholar]
- 9.Thiel T. Technische Universität Dresden; Dresden, Germany: 2001. Identifizierung, Kartierung und Chjarakterisierung cDNA basierter Mikrosatelliten-Marker zur Diversitätsanalyse bei gerste (Hordeum vulgare L.) Diploma thesis. [Google Scholar]
- 10.Scott K.D. Microsatellite derived from ESTs, and their comparison with those derived by other methods. In: Henry R.J., editor. Plant Genotyping: The DNA Fingerprinting of Plants. CABI Publishing; Oxon, UK: 2001. pp. 225–237. [Google Scholar]
- 11.Varshney R.K. In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol. Biol. Lett. 2002;7:537–546. [PubMed] [Google Scholar]
- 12.Borstnik B., Pumpernik D. Tandem repeats in protein coding regions of primate genes. Genome Res. 2002;12:909–915. doi: 10.1101/gr.138802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunzler P. Pathological, physiological, and evolutionary aspects of short unstable DNA repeats in the human genome. Biol. Chem. Hoppe. Seyler. 1995;4:201–211. doi: 10.1515/bchm3.1995.376.4.201. [DOI] [PubMed] [Google Scholar]
- 14.Moxon E.R., Wills C. DNA microsatellites: agents of evolution? Sci. Am. 1999;280:94–99. doi: 10.1038/scientificamerican0199-94. [DOI] [PubMed] [Google Scholar]
- 15.Jurka J., Pethiyagoda C. Simple repetitive DNA sequences from primates: compilation and analysis. J. Mol. Evol. 1995;40:120–126. doi: 10.1007/BF00167107. [DOI] [PubMed] [Google Scholar]
- 16.Cardle L. Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics. 2000;156:847–854. doi: 10.1093/genetics/156.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metzgar D. Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res. 2000;10:72–80. [PMC free article] [PubMed] [Google Scholar]
- 28.Mukund V. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol. Biol. Evol. 2001;18:1161–1167. doi: 10.1093/oxfordjournals.molbev.a003903. [DOI] [PubMed] [Google Scholar]
- 19.Toth G. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 2000;10:967–981. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schorderet D.F., Gartler S.M. Analysis of CpG suppression in methylated and nonmethylated species. Proc. Natl. Acad. Sci. USA. 1992;89:957–961. doi: 10.1073/pnas.89.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy K.D. Microsatellites in the silkworm, Bombyx mori: abundance, polymorphism, and strain characterization. Genome. 1999;42:1057–1065. [PubMed] [Google Scholar]