Abstract
Human tumor necrosis factor α (hTNFα), a pleiotropic cytokine with activities ranging from host defense mechanisms in infection and injury to severe toxicity in septic shock or other related diseases, is a promising target for drug screening. Using the SELEX (systematic evolution of ligands by exponential enrichment) process, we isolated oligonucleotide ligands (aptamers) with high affinities for hTNFα. Aptamers were selected from a starting pool of 40 randomized sequences composed of about 1015 RNA molecules. Representative aptamers were truncated to the minimal length with high affinity for hTNFα and were further modified by replacement of 2′-OH with 2′-F and 2′-NH2 at all ribopurine positions. These modified RNA aptamers were resistant to nuclease. The specificity of these aptamers for hTNFα was confirmed, and their activity to inhibit the cytotoxicity of hTNFα on mouse L929 cells was determined. Results demonstrated that four 2′-NH2-modified aptamers bound to hTNFα with high affinity and blocked the binding of hTNFα to its receptor, thus protecting the L929 cells from the cytotoxicity of hTNFα. Oligonucleotide aptamers described here are potential therapeutics and diagnostics for hTNFα-related diseases.
Key words: SELEX, hTNFα, aptamer, affinity, RNA, ELONA
Introduction
Tumor necrosis factor (TNF) is a pro-inflammatory cytokine that participates in the regulation of immune defense against various pathogens and the recovery from injury. It plays a central role in the development of many inflammatory diseases. There is clear evidence that aberration in TNF production in vivo results in diseases, such as multi-organ inflammation, rheumatoid arthritis (RA), multiple sclerosis, and inflammatory bowel disease (1). It is widely accepted that TNF plays important part in pathophysiology by directing its two transmembrane receptors to deliver signals of cellular proliferation, differentiation or apoptosis. Attempts to identify biological agents that could protect the body against these pathological effects led to the discovery of soluble TNF receptors and monoclonal antibodies, which competitively inhibit the binding of TNF to cell surface receptors and thus block TNF bio-activity 2., 3.. Blockade of TNF in rheumatoid arthritis or inflammatory bowel disease, although so far impressively beneficial for the majority of patients, has led in some cases to a significant incidence of drug induced anti-dsDNA production and lupus, and manifestations of neuroinflammatory diseases 4., 5., 6.. Although these protein drugs provide good therapeutic effect, there are still disadvantages. Firstly, the process of protein preparation is complicated and expensive; secondly, the protein drugs have high molecular weight and are difficult to get to the target tissues because of its poor permeability; thirdly, the blood clearance is so low that it is hard to excrete, which results in its accumulation in the body and causes side effect; finally, due to the introduction of the exogenous proteins and the change of structure, the proteins have immunogenicity, once repeatedly used, it induces antibody formation.
Nucleic acids, as a function of their primary structure, fold into complex three-dimensional structure with great diversity of binding specificities. Using the SELEX (systematic evolution of ligands by exponential enrichment) process, oligonucleotides ligands (aptamers) may be efficiently isolated from enormous randomized libraries of RNA, DNA, or modified nucleic acids that bind with high affinity and specificity to a wide range of selected molecular targets (7). Aptamers can be highly potent antagonists of specific protein-protein interactions with following advantages. They do not depend on animals, cells, or even in vivo conditions produced by chemical synthesis with extreme accuracy and reproducibility. Once denatured, functional aptamers could be regenerated easily within minutes. They are stable to long-term storage and can be transported at ambient temperature (8). The potential utility of aptamers as therapeutic or diagnostic agents is considerably enhanced by chemical modifications that lend resistance to nuclease attack. In particular, substitution at the 2′ position of ribonucleotides with 2′-amino (2′-NH2), 2′-fluoro (2′-F), or a variety of 2′-O-alkyl moieties confers resistance to ribonucleases that utilize the 2′-OH group for cleavage of the adjacent phosphodiester bond (9). This technology has been applied to a wide range of targets including growth factors, enzymes of HIV, and inflammation-inducing enzymes. The first aptamer that has proceeded to Phase II clinical trial is NX-1838, an injectable angiogenesis inhibitor used to treat macular degeneration-induced blindness (10).
Here we report the screening of human TNFα (hTNFα) aptamers from an oligonucleotide library with 40 randomized sequences composed of about 1015 RNA molecules. Representative aptamers were truncated to minimal sequence with high binding activity and modified by replacement of 2′-OH with 2′-F and 2′-NH2 at all ribopurines. Aptamers described here represent an initial set of lead compounds in our ongoing effort toward the development of potent and specific hTNFα antagonists.
Results
Cloning and sequencing of selected oligonucleotides
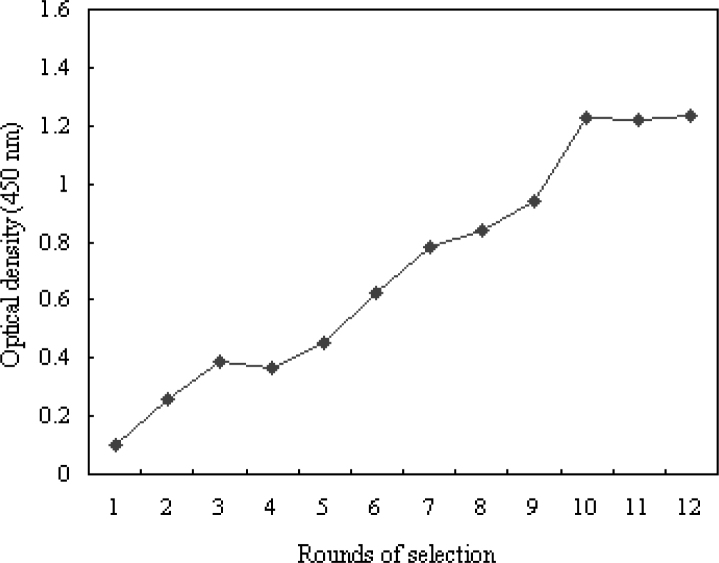
RNA aptamers for hTNFα were isolated from an oligonucleotide library with 40 randomized sequences. Twelve rounds of selection were performed, and enzyme-linked oligonucleotide assay (ELONA) was used to evaluate the binding affinity between the production of every round and hTNFα (Figure 1). As no further improvement in binding affinity was observed after Round 12, sequences in the pool of the 12 rounds were cloned respectively, and then plasmids of the 40 clones were purified. Biotin-labeled RNA aptamers were prepared by in vitro transcription. The binding affinity was evaluated by filter binding assay, and 20 clones with higher affinity for hTNFα were sequenced among the 40 clones (Table 1). All the sequences were different from each other. There was no signal in the negative control of RNA library and hTNFα, neither in the blank control of RNA aptamer 1 alone.
Fig. 1.
Binding affinity analysis of aptamers with ELONA. Oligonucleotides were labeled by biotin, and 5 μg/mL hTNFα was coated in every well. The binding affinity for hTNFα was determined by ELONA as described in Materials and Methods.
Table 1.
Sequences of 20 RNA Aptamers for hTNFα
| Family | Aptamer | Sequence of variable region* |
|---|---|---|
| I | 3.1 | CCCCGGGUUCUGUAUGAUCCGACCGGUCAGAUAAGACCAC |
| 7.5 | CGCAUCGUUUGCGUGGCGUGUCCGGGCGCCGAUUCGUAAA | |
| 12 | CUAGGCGGAUUGUUUCGAUUCUUUGCCUUGUCCCUAGUGC | |
| 14.8 | CGUAUAUACGGAUUAGGUUGUAGCUCAGACCAGUAAUGUC | |
| 16.3 | CGUGCUAGAUGCUACGAGUGGUCUCCUCACGUAGAAGGGG | |
| 18.10 | GGUCCCACAUAGGUUGGUCUUGUUGUAUGGGCUGUUUGCA | |
| II | 1 | GUGUUUUGGGAGAGAAAAGGGGGAGCCUUUACUUUGUUGG |
| 2 | GACGAUGUUAUCAGGGAGUUGGGAUCAUAUAGUCUUACAU | |
| 4 | CGCAAGAGCCGCCCUAAUGGUUCAAUGGUAACUGUAUAUG | |
| 6 | GACUUCUUGUGCCAUUAUGAAUUAUUGCUAAUCCUCUUGA | |
| 8.6 | AGGACGUACUUGGAAAAGAGGCGCGAAGAACCUGGUAUGU | |
| 9 | UAGGACGUACUUGGAAAAGAGGCGCGAAGAACCUGGUAUG | |
| 10 | UGGCCACCUUGCCACUCUUCCUUGCAUAUUUUACUCCCGC | |
| 11.7 | CAAGCCGAGGGGGAGUAUCUGAUGACAAUUCGGAGCUCCA | |
| 13.2 | UCAUGGUGUGUGAGUUAGCUCACGUGCCGUUUCGAAGGCG | |
| 17.9 | CAUGGGCUAGACCGGCAUAAAACUGCUGUAGUUGCACGCC | |
| 19 | UGGCCACCUUGCCACUCUUCCUUGCAUAUUUUACUCCCGC | |
| 20.4 | CGUUGUAGUAGUGGCUUGGGCAUAACUCAGUUAAACACUA | |
| III | 5 | GACCGCGGAAAAGGAAGGAAUUAGAUACAACGGAGAAGUG |
| 15 | GACCGCGGAAAAGGAAGGAAUUAGAUACAACGGAGAAGUG |
Aligned sequences isolated from the affinity-enriched RNA pools are shown. For each clone only the variable 40-nt region is shown.
Analysis of selected oligonucleotides
Ligands containing the primary structure motif of GGAG/AAAGU/AG were Aptamers 5, 8, 9, 15, and 20. Aptamers 5, 8, 9, 15, and 16 shared the conserved primary structure motifs CGNAGAA. Aptamers 1, 5, 8, 9, and 15 shared the conserved primary structure motifs GAG/AAAG. Ligands containing the primary structure motif of GAG/AUUA were Aptamers 5, 6, 13, 14, and 15. Aptamers 6, 8, 9, 10, 12, 18, 19, and 20 shared the same motif of CUUG. Other sequences were not obviously related to any other sequence. These motifs shared by some aptamers maybe related to the part that binds to hTNFα. However, the secondary structure is the most important for ligand binding.
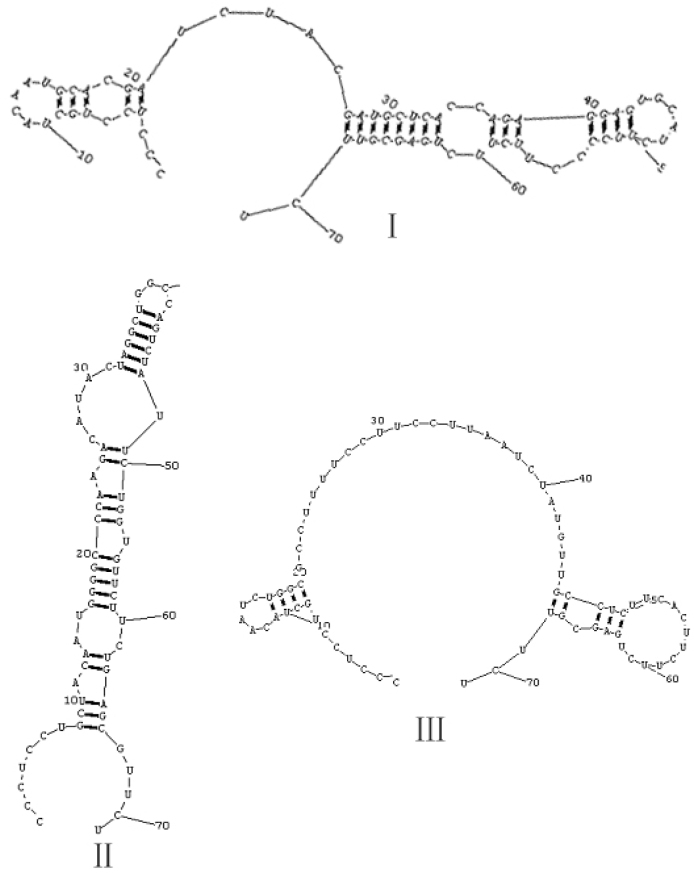
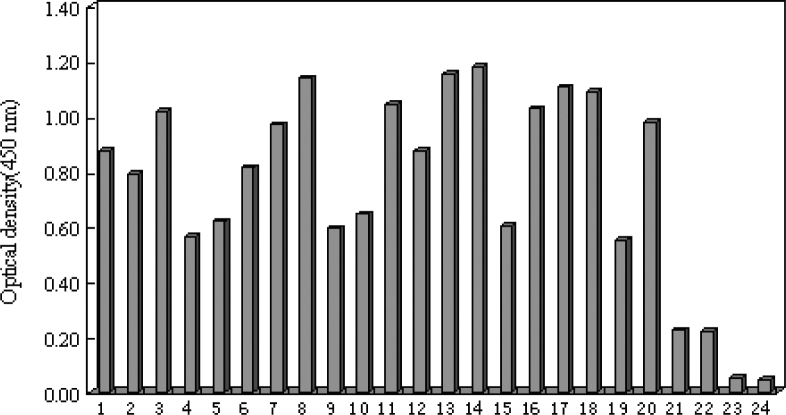
The 20 aptamers were grouped into three major families based on their secondary structures analyzed by RNA structure software (Figure 2). Ligands containing the secondary structure of a small unclosed loop in the center and two stems beside the middle loop were defined as Family I. Aptamers 3, 7, 12, 14, 16, and 18 were grouped into Family I. Ligands with 3-5 small loops bin between stems in a line were defined as Family II. Aptamers 1, 2, 4, 6, 8, 9, 10, 11, 13, 17, 19, and 20 were grouped into Family II. Ligands with a big loop in the center and 2-3 stems in the loop were defined as Family III. Aptamers 5 and 15 were grouped into Family III. We selected 10 clones with higher affinity by ELONA (Figure 3), which were Aptamers 3, 7, 8, 11, 13, 14, 16, 17, 18, and 20, belonging to Families I and II. The binding affinity of Family II is higher than that of Family I.
Fig. 2.
Three representative secondary structures of RNA stem-loops and sequences binding to hTNFα were predicted.
Fig. 3.
Binding affinity of RNA Aptamers 1–20 with hTNFα. 5 μg/mL hTNFα was coated in every well except blank control. 1–20: biotin-labeled RNA aptamers; 21: biotin-labeled RNA library; 22: tRNA; 23 and 24: blank control. Biotin-labeled RNA Aptamers 1 and 2 were added to uncoated wells.
Although possible base pairing interactions for some ligands may be identified, generally between the 5′-fixed sequence and nucleotides on the 3′-side of the conserved sequence motif, no predicted secondary structure common to most of the ligands is evident. Some of the fixed sequences in Family I were involved in the formation of the secondary structure, but whether the stem-loop structure formed by fixed sequence is pivotal for the binding activity remains unknown. It appears that some aptamers in Families I and II formed new secondary structures, but none in Family III did.
Nitrocellulose filter binding
The affinities of 10 truncated biotin-labeled aptamers for hTNF were determined by filter binding. The result indicated that ligands showed very high affinity for hTNFα. Removal of fixed sequence didn′t decrease their affinity. The fixed sequence of the 10 aptamers does not contribute to the protein-oligonucleotide binding interaction.
ELONA test
To determine the affinities of different concentration aptamers for hTNFα, ELONA was performed as described in Materials and Methods. The concentrations of biotin-labeled RNA aptamers were from 800 nM to 50 pM (Figure 4). The result indicated that the biotin-labeled RNA library had low binding affinity for hTNFα, but RNA Aptamers 3.1, 13.2, 16.3, 20.4, 7.5, 8.6, 11.7, 14.8, 17.9, and 18.10 had high binding affinity. RNA Aptamers 13.2, 7.5, 11.7, and 14.8 had higher binding affinity than the others, and 800 pM of RNA Aptamer 11.7 could get positive signals. So the RNA aptamer is available for detecting hTNFα.
Fig. 4.
Binding affinity of random sequences of 10 RNA aptamers for hTNFα. 1–10: biotin-labeled RNA Aptamers 3.1, 13.2, 16.3, 20.4, 7.5, 8.6, 11.7, 14.8, 17.9, and 18.10; 11: biotin-labeled RNA library. 5 μg/mL hTNFα was coated in every well. The binding affinity for hTNFα was determined by ELONA. The concentrations of biotin-labeled RNA aptamers were from 800 nM to 50 pM.
Inhibition of hTNFα-mediated cytotoxicity
As shown in Figure 5, no inhibition was observed with equimolar concentrations of RNA library for the identification of antagonists of hTNFα activities. RNA Aptamers 3.1, 13.2, 16.3, 20.4, 7.5, 8.6, 11.7, 14.8, 17.9, and 18.10 with high affinity for hTNFα showed a clear inhibitory effect on hTNFα-induced cytotoxicity, and the cytotoxicity in vitro is in a dose-dependent fashion. RNA Aptamers 13.2, 7.5, 11.7, and 14.8 showed higher inhibition effect on hTNFα-induced cytotoxicity. 1 nM aptamer is enough to inhibit the cytotoxicity. We found that there is no similarity between the RNA aptamer sequences and the gene of hTNFα. This suggests that the identified sequence is likely to bind to hTNFα and inhibit the binding of hTNFα to its receptor.
Fig. 5.
Protection of RNA aptamers to L929 cell destructed by TNF. 1–10: RNA Aptamers 3.1, 13.2, 16.3, 20.4, 7.5, 8.6, 11.7, 14.8, 17.9, and 18.1; 11: RNA library. The cytotoxic effect of hTNFα was assayed on L929 cell line. The concentrations of biotin-labeled RNA aptamers were from 800 nM to 800 pM.
Size minimization
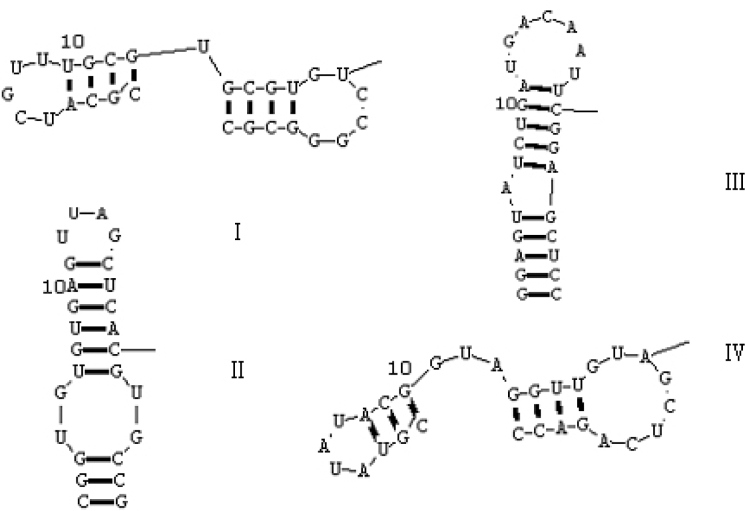
To prepare an aptamer for in vivo experimentation, we made three alterations: size minimization, further stabilization against nuclease activity, and incorporation of a bioconjugation handle for addition of biotin. Ideally, such changes maintain the affinity and specificity of the aptamer. Reducing the size of the RNA resulted in a few changes to the sequence compared with the initial clone; the stem was shortened and changes were made in the stem to optimize both transcription and stability. To identify the minimal sequence elements that confer high binding affinity for hTNFα, we used predictions based on conserved secondary structure motifs to derive truncated aptamers with high affinity from the oligo, and totally got four such aptamers (Table 2). The secondary structure showed that there were two types of the aptamer structures (Figure 6). The truncated four clones T1.7.5, T2.13.2, T3.11.7, and T4.14.8 came individually from Aptamers 7.5, 13.2, 11.7, and 14.8. Their binding affinity and biological effect changed after truncation. The sequence of the 30-nt fragment with high affinity, named T1.7.5, encompassed the conserved sequence motif, and an oligonucleotide corresponding to the selected fragment (10 nucleotides of 3′-end truncated aptamer derived from oligonucleotide 7.5) showed an approximately 3-fold loss in binding affinity for hTNFα relative to the full-length ligand. Further truncation at the 3′- or 5′-end of this molecule caused a precipitous loss in affinity. The resultant 30-nt aptamer T2.13.2 included all of the consensus secondary structure motifs. In contrast, up to 9 additional nucleotides could be removed from the 5′-end with little or no effect on binding affinity. The resultant 28-nt aptamer T3.11.7, with 11 nucleotides of 5′- and 3′-end truncated aptamer, derived from clone 11.7. They not only have the same secondary structure but also the same affinity as the initial full-length oligonucleotides. Deletion of 11 nucleotides from 5′-end and 1 nucleotide from 3′-end of aptamer reduced the length of the putative stem. For the 29-nt aptamer T4.14.8, removal of an additional base pair from the base of the proposed stem (6 nucleotides of 5′-end and 5 of 3′-end derived from clone 14) did not affect the binding affinity.
Table 2.
Sequences of Four Truncated RNA Aptamers for hTNFα
| Aptamer | Sequence* |
|---|---|
| T1.7.5 | CGCAUCGUUUGCGUGGCGUGUCCGGGCGCCGAUUCGUAAA |
| T2.13.2 | UCAUGGUGUGUGAGUUAGCUCACGUGCCGUUUCGAAGGCG |
| T3.11.7 | CAAGCCGAGGGGGAGUAUCUGAUGACAAUUCGGAGCUCCA |
| T4.14.8 | CGUAUAUACGGAUUAGGUUGUAGCUCAGACCAGUAAUGUC |
The truncated aptamers are shown in black and the deleted are in gray.
Fig. 6.
Predicted secondary structures of the truncated aptamers, T1.7.5, T2.13.2, T3.11.7, and T4.14.8, which individually came from 7.5, 13.2, 11.7, and 14.8.
Further nuclease stabilization
For RNA aptamers, a 2′-NH2 or 2′-F group replaced the ribose 2′-OH group of pyrimidines. After that the binding affinity assay was carried out. The results indicated that the binding affinity of modified RNA aptamer decreased (Figure 7). The binding affinity of truncated aptamer T1.7.5 modified by 2′-NH2-pyrimidine was higher than that of 2′-F-pyrimidine. The half-life of these modified aptamers in serum was increased, compared to the typical half-life of 5 min or less for natural RNA molecules. Increased stability of 2′-NH2-modified and 2′-F-modified RNAs was observed with selected aptamers for hTNFα. The stability of RNA aptamer T3.11.7 was examined in mouse serum at 37°C from 0 to 16 h. Samples were taken at 0, 1, 2, 4, 8, and 16 h. ODtest/ODundigest was calculated to describe the stability of oligonucleotides (Figure 8). After 1 h, 80% of unmodified T3.11.7 were degraded. The results indicated that the half-life of unmodified RNA aptamer was several minutes, while the half-life of modified RNA aptamer was prolonged to 8 h.
Fig. 7.
Binding affinity of modified RNA aptamers. Truncated aptamers T3.11.7 and T2.13.2 were modified by 2′-F-pyrimidine and 2′-NH2-pyrimidine. Modified aptamers were labeled by biotin, and 5 μg/mL hTNFα was coated in every well. The binding affinity for hTNFα was determined by ELONA. The concentrations of biotin-labeled RNA aptamers were from 800 nM to 50 pM.
Fig. 8.
Comparision of the stability of unmodified and 2′-NH2-pyrimidine modified RNA in human serum. Unmodified RNA T3.11.7 and 2′-NH2-pyrimidine modified RNA corresponding to it in sequence were prepared by in vitro transcription from the same synthetic DNA tamplate. Both RNAs were internally biotin-labeled and incubated at a concentration of 50 nM 85% human serum supplemented with 10 mM phosphate buffer (pH 7.4). Aliquots from unmodified and 2′-NH2-pyrimidine modified RNA incubation mixtures were withdrawn at indicated time intervals, frozen in dry ice until the end of the experiment, and loaded onto 20% denaturing polyacrylamide gels to resolve labeled degradation products. Absorbance of purified RNA was detected at 260 nm, and ODtest/Odundigest(%) were calculated.
Inhibition of hTNFα-mediated cytotoxicity by minimized and modified aptamers
The original and NH2-modified RNA aptamers T3.11.7 were transcribed in vitro by T7 polymerase and used to test their ability to inhibit hTNFα-induced cytotoxicity. Both RNA aptamer T3.11.7 and NH2-modified aptamer T3.11.7 showed a clear inhibitory effect on hTNFα-mediated cytotoxicity in L929 cells, whereas no inhibition was observed when equal concentrations of RNA library were tested (Figure 9A). Both T3.11.7 aptamers inhibited the cytotoxicity in a dose-dependent fashion. The inhibition rate of original T3.11.7 is 50%-100%, while that of NH2-modified T3.11.7 is 20%-90%. The result showed that the biological activity decreased after modification.
Fig. 9.
A. Inhibition of hTNFα-induced cytotoxicity in L929 cell by RNA aptamer T3.11.7 and 2′-NH2-modified aptamer T3.11.7. The concentrations of RNA aptamers were from 300 nM to 1 nM. Each concentration of hTNFα and aptamer mixture was tested in triplicate. B. Inhibition of 1 or 10 U hTNFα-induced cytotoxicity in L929 cell by RNA aptamer T3.11.7. The cytotoxic effect of the hTNFα was assayed oh L929 cell line. The concentrations of RNA aptamers were from 800 nM to 800 pM. Each concentration of hTNFα and aptamer mixture was tested in triplicate.
Serially diluted modified RNA aptamer T3.11.7 was preincubated with 1 U and 10 U hTNFα (Figure 9B). The results indicated that the levels of inhibition by modified T3.11.7 depend on the concentrations of used hTNFα. It can inhibit 1 U hTNFαeffectively, but for 10 U hTNFα, the inhibitory decreased.
The aptamers selected in this study could inhibit hTNFα-mediated cytotoxicity in vitro on mouse cell lines. As the binding of hTNFα to its receptors is a prerequisite for its biological activities, the ability of the aptamers to inhibit hTNFα-induced cytotoxicity suggests that it mimics the binding site of hTNFα membrane receptor.
Discussion
Previous work has shown that the approach to inhibit the binding of hTNFα and its receptor can reduce the cytotoxicity of hTNFα (11). So our studies have focused on the identification of RNA aptamers binding to hTNFα. In order to identify a physiologically active, nuclease-stabilized aptamer that inhibited the function of hTNFα in vitro, we got 20 oligonucleotides that were truncated to the minimal sequence capable of their binding affinity for hTNFα (28–31 nt) and were further modified by 2′-F-pyrimidine and 2′-NH2-pyrimidine where the substitution was tolerated. Furthermore, one 2′-NH2-modified aptamer is able to significantly inhibit hTNFα-mediated cytotoxicity in mouse fibroblast cell line L929. Results demonstrate that the aptamers potently inhibit the binding of hTNFα to the receptors.
The 20 oligonucleotides with high affinity for hTNFα were sequenced and no identical sequence was found. Their secondary structure was analyzed by RNA structure software. The data indicated that the secondary structure is the most important for the ligand binding. Molecular recognition of nucleic acids can arise by virtue of binding specificity at the level of primary and secondary structure; however, it is at the level of tertiary structure that the highest level of selectivity with non-nucleotide ligands should be achieved. Single strand of RNA that are folded into complex secondary and tertiary conformations, including local regions of duplex structure, may be distorted as a result of base mismatches, bulges, pseudoknots, and hairpins. The binding affinity of oligonucleotides is based on the secondary and tertiary structures.
In this report, biotin-labeled UTPs were incorporated by in vitro transcription with the ratio of UTPs to biotin-UTPs as 2:1. The result of ELONA was calculated according to the number of UTPs. Since almost all the oligonucleotides have the same number of UTPs, we can use them to determine the binding affinity of aptamers for hTNFα. Aptamers promise to be smaller in size and less complex than antibodies, easy to manufacture and modify, chemically consistent between lots, compatible with most current assay formats, and stable during storage (12).
A cloned aptamer sequence is typically 70–80 nt in length. Because reduced size may lead to increased tissue penetration rates (13), make these aptamers clear from the blood more rapidly than the full-length aptamer, which is an advantage for in vivo imaging, and will lead to more efficient chemical synthesis, we focused on size reduction. It is desirable to identify the minimal high-affinity aptamer sequence. Generally, the 5′-fixed and 3′-fixed sequences have no predicted secondary structure common to most of the ligands. The results of ELONA and SPR demonstrated that truncated oligonucleotides bound to hTNFα with the same affinity as originals.
To be applicable in cell culture studies, we generated nuclease-resistant aptamers in which all pyrimidine residues were substituted by 2′-NH2- and 2′-F-modified monomers. This modification is known to protect the majority of serum nucleases so that such aptamers can be applied in the presence of biological materials in which normal RNA would rapidly degrade (14). Four truncated aptamers were modified by 2′-F and 2′-NH2, followed by affinity analysis. We found affinity reductions in all modified oligonucleotides. The affinity of the 2′-NH2-pyrimidine hTNFα aptamers was indeed higher than that of most 2′-F-pyrimidine ligands. The stability of RNA aptamers in mouse serum was enhanced.
In pathological conditions caused by excessive production of hTNFα, such as in rheumatoid arthritis or septic shock, the neutralization of high-level hTNFα using anti-hTNFα monoclonal antibodies (mAb) or recombinant forms of soluble hTNFα receptors might be the strategy of choice. However, the use of these approaches has several limitations: chimerical or humanized mAb, after repeated administrations, might induce a neutralizing antibody response resulting in less effective treatment. hTNFα is a trimer molecule, and for its effective inhibition, at least two molecules of the hTNFα receptor, expressed as fusions with the immunoglobulin G molecule, are required (15). In the case of recombinant forms of soluble hTNFα receptors, these molecules can be unstable in vivo, immunogenic, and could exert an agonistic effect by acting as hTNFα carriers. The use of low-molecular-weight synthetic peptides as antagonistic molecules is another approach, but they have only low affinity and biological effect. So aptamer might be a suitable alternative for the in vivo blockade of hTNFα. In vitro, the modified hTNFα aptamers inhibit the binding of hTNFα to cell surface hTNFα receptors expressed on L929 cells. Thus, the aptamers that specifically inhibit hTNFα may have great utility in combating a variety of hTNFα human diseases.
Materials and Methods
Reagents
Recombinant hTNFα was prepared by our laboratory. The protein was resuspended in phosphate-buffered saline (PBS) to a concentration of 1 μg/μL and stored at −20°C in small aliquots before use. Aliquots were stored at 4°C for up to 4 weeks after thawing. 2′-F- and 2′-NH2-pyrimidine nucleotriphosphates were purchased from Ambion (Austin, USA). T7 Riboprobe kit was purchased from Promega (San Luis Obispo, USA). HATF membrane was purchased from Millipore (Bedford, USA). Purification system was purchased from Qiagen (Valencia, USA). DNA oligonucleotide template libraries 5′-GGGAGGACGATGTTA(N)40AAGAAGACTCGCAAGA-3′ (N means any nucleotide) were prepared by chemical synthesis. T7 RNA polymerase promoter sequence localized at the 5′-end of each template. Oligonucleotide primers (Primer 1: 5′-GGGAGGACGATGTTA-3′; Primer 2: 5′-TCTTGCGAGTCTTCTT-3′; Primer 3: 5′-TAATACGACTCACTATA-3′; Primer 4: 5′-TAATACGACTCACTATAGGGAGGACGATGTTA-3′) were synthesized by Takara (Dalian, China) for template amplification and reverse transcription. Double-stranded DNA templates were prepared by annealing Primers 1 and 2 to the libraries and extending the primers using MLV virus reverse transcriptase (Biolabs, Beverly, USA) at 37°C. 1 nmol of RNA library was synthesized by in vitro T7 transcription from the PCR-amplified double-stranded DNA pool. RNAs were purified from denaturing (7 M urea) polyacrylamide gels by excising and crushing the gel slice containing the RNA and soaking it for several hours or overnight in 2 mM EDTA. Approximately 5 nmol of RNA was obtained from each transcription.
The SELEX protocol
The SELEX process of affinity selection followed by amplification of the selected pool has been described in detail (16). In brief, one round of selection and amplification was performed as follows. hTNFα was mixed with RNA in binding buffer of PBS with 1 mM MgCl2 and 10 mM DTT. After incubation at 37°C for 30 min, the mixture was passed through 0.45-μm Type HATF nitrocellulose filters to collect complexes of hTNFα with RNA. Then RNAs were eluted from the filters by eluting buffer (binding buffer with 10 mM EDTA) and incubated in a 2:1 mixture of phenol (pH 7) and 7 M urea. After precipitation from the aqueous phase, RNAs were annealed to Primer 2 and reverse transcribed using MLV reverse transcriptase. The resultant cDNAs were amplified with 15 cycles of PCR using Primers 2, 3 and 4. Transcription of the PCR product that yielded a new library enriched for sequences with affinity for hTNFα.
Cloning and analysis of oligonucleotides
After 12 rounds of selection and amplification, individual molecules in the selected pools were cloned with pGEM-T Vector (Promega). Standard transformation was performed. Colonies grown on agar plates were picked randomly for culture. The plasmid in each colony was prepared using QIAwell Plasmid Purification System and sequenced by Takara. The primary structure sequences were analyzed by Macaw 2.05 software and conservative sequences were found. The secondary structure was predicted by RNA structure 3.5 software, which is based on the theory of the smallest free energy, Zuker algorithm. Sequences were grouped into families according to their different secondary structures.
Nitrocellulose filter binding
Nitrocellulose filter binding was used to assess aptamer affinity for hTNFα. Biotin-labeled UTPs were incorporated by in vitro transcription with the ratio of UTPs to biotin-UTPs as 2:1. The results of filter binding assay and ELONA were calculated according to the number of UTPs. Then we incubated the labeled aptamers in low concentrations (typically less than 70 pM) with various concentrations of hTNFα or without proteins in binding buffer at 37°C for 30 min. Samples were dotted on prewashed 0.45-μm nitrocellulose filters followed by a 5-10 mL wash with binding buffer. Filters were incubated with HRP-Streptavidin after blocked with 1% BSA (bovine serum albumin). The fraction of RNA that bounds to protein was measured by OPD substrate.
ELONA test
Microtiter plate was coated overnight at 4°C with 50 μL/well of a 5 μg/mL solution of the hTNFα dissolved in 0.1 M NaHCO3, pH 9.0. After 3 times washing with PBS/Tween, the plate was blocked for 2 h at 37°C with 100 μL PBS containing 1% BSA in each well. Following 3 times washing with PBS/Tween, the diluted RNA aptamers were added to wells and incubated at 37°C for 30 min. After 3 times washing with PBS/Tween, 50 μL diluted Horseradish Peroxidase conjugated streptavidin was added to each well and the incubation was washed. Then TMB and H2O2 were added. The plate was developed at room temperature for 10 min and read at 450 nm wave length.
Optimization of high affinity RNA aptamers
Aptamers that come out of a SELEX experiment are full-length sequences containing the fixed sequences that were included to aid the amplification process. These full-length aptamers are generally 70–80 nt long and could be truncated to eliminate nucleotide stretches that are not important for direct interaction with the target or folding into the structure that facilitates target binding. The identification of truncated aptamers restricted to the minimal target-binding domain requires more effort, while it has been successfully carried out to obtain functional aptamers less than 40 nt long (17).
In the majority of cases, the fixed sequence regions used for primer binding are unimportant for aptamer function and can be eliminated, thereby producing short aptamer sequences. Aptamers could be truncated and modified with high affinity. The DNA template of truncated RNA aptamer was chemically synthesized and transcribed by T7 RNA polymerase, then the binding affinity was detected. Whether further truncation should be done was decided by what the result was. At last, we got the truncated oligonucleotide that retained an affinity for hTNFα equivalent to that of the full-length one.
Stability of aptamers in mouse plasma in vitro
The stabilities of RNA-based aptamers in vitro were examined in rat serum at 37°C. Fresh serum was obtained from a Balb/c mouse′s eyes and was treated with 20 mmol/L sodium phosphate buffer. Test ligands were added to the serum to the final concentration of 300 nmol/L. The final serum concentration was 85% as a result of the addition of buffers and aptamers. From the original 100 μL incubation mixture, 20 μL aliquots were withdrawn at various periods and added to 2 μL of 500 mmol/L EDTA (pH 8.0), respectively. The mixtures were then frozen in dry ice and stored at −20°C until the end of the experiment. After purification, half of the sample was analyzed by PAGE and stained by AgNO3, the rest was detected at 260 nm wavelength. ODtest/ODundigest were calculated to describe the stability of oligonucleotides.
Inhibition of hTNFα-mediated cytotoxicity
The mouse fibroblast cell line L929 were cultured in RPMI 1640 medium (Gibco, New York, USA) supplemented with 10% (V/V) heat inactivated FCS (foetal calf serum) and antibiotics (100 U/mL penicillin, 100 μg/mL streptomycin). The cytotoxic effect of the hTNFα was assayed on L929 cell line. Cells were plated into 96-well flat bottomed microtitre plates (Nunc, Roskilde, Denmark) at 2×105/mL of the culture medium at 37°C in the presence of 5% CO2 incubator for 24 h. Serially diluted RNA aptamers in the same culture medium were preincubated with a constant amount of hTNFα at 37°C in the presence of 5% CO2 incubator for 1 h. The mixture was plated in triplicate in the cells in the presence of 2 μg/mL actinomycin D and incubated at 37°C for 24 h in the presence of 5% CO2 incubator. Viable cells were stained after incubation with 500 μg/mL MTT (thiazolyl-blue) at 37°C for 4 h. Then, 1% SDS (pH 4.5) was added. Absorbance of each well was read at 570 nm wave length. Inhibition rate was calculated by the formula: Inhibition(%) = [(ODtest − ODhTNFα)/(ODcontrol − ODhTNFα)] × 100.
References
- 1.Tracey K.J., Cerami A. Tumor necrosis factor, other cytokines and disease. Annu. Rev. Cell Biol. 1993;9:317–343. doi: 10.1146/annurev.cb.09.110193.001533. [DOI] [PubMed] [Google Scholar]
- 2.Gravestein L.A., Borst J. Tumor necrosis factor receptor family members in the immune system. Semin. Immunol. 1998;10:423–434. doi: 10.1006/smim.1998.0144. [DOI] [PubMed] [Google Scholar]
- 3.Afeltra A. Treatment of rheumatoid arthritis: new therapeutic approaches with biological agents. Curr. Drug Targets Immune Endocr. Metabol. Disord. 2001;1:45–65. doi: 10.2174/1568008013341677. [DOI] [PubMed] [Google Scholar]
- 4.Alldred A. Etanercept in rheumatoid arthritis. Expert Opin. Pharmacother. 2001;2:1137–1148. doi: 10.1517/14656566.2.7.1137. [DOI] [PubMed] [Google Scholar]
- 5.Mease P. Psoriatic arthritis: the role of TNF inhibition and the effect of its inhibition with etanercept. Clin. Exp. Rheumatol. 2002;20:S116–S121. [PubMed] [Google Scholar]
- 6.Weisman M.H. What are the risks of biologic therapy in rheumatoid arthritis? An update on safety. J. Rheumatol. Suppl. 2002;65:33–38. [PubMed] [Google Scholar]
- 7.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA Polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 8.Drolet D.W. An enzyme-linked oligonucleotide assay. Nat. Biotechnol. 1996;14:1021–1025. doi: 10.1038/nbt0896-1021. [DOI] [PubMed] [Google Scholar]
- 9.Jellinek D. Potent 2′-amino-2′-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry. 1995;34:11363–11372. doi: 10.1021/bi00036a009. [DOI] [PubMed] [Google Scholar]
- 10.Eyetech Study Group Anti-vascular endothelial growth factor therapy for subfoveal choroidal neovascularization secondary to age-related macular degeneration: phase II study results. Ophthalmology. 2003;110:979–986. doi: 10.1016/S0161-6420(03)00085-X. [DOI] [PubMed] [Google Scholar]
- 11.Akassoglou K. Oligodendrocyte apoptosis and primary demyelination induced by local TNF/p55TNF receptor signaling in the central nervous system of transgenic mice: models for multiple sclerosis with primary oligodendrogliopathy. Am. J. Pathol. 1998;153:801–813. doi: 10.1016/S0002-9440(10)65622-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubik M.F. High-affinity RNA ligands to human alpha-thrombin. Nucleic Acids Res. 1994;22:2619–2626. doi: 10.1093/nar/22.13.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hicke B.J., Stephens A.W. Escort aptamers: a delivery service for diagnosis and therapy. J. Clin. Invest. 2000;106:923–928. doi: 10.1172/JCI11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruckman J. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998;273:20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 15.Chan F.K., Lenardo M.J. Tumor necrosis factor family ligands and receptors in the immune system: targets for future pharmaceuticals. Drug News Perspect. 2002;15:483–490. doi: 10.1358/dnp.2002.15.8.840068. [DOI] [PubMed] [Google Scholar]
- 16.Pieken W.A. Kinetic characterization of ribonuclease-resistant 2′-modified hammerhead ribozymes. Science. 1991;253:314–317. doi: 10.1126/science.1857967. [DOI] [PubMed] [Google Scholar]
- 17.Eyetech Study Group Preclinical and phase 1A clinical evaluation of an anti-VEGF pegylated aptamer (EYE001) for the treatment of exudative age-related macular degeneration. Retina. 2002;22:143–152. doi: 10.1097/00006982-200204000-00002. [DOI] [PubMed] [Google Scholar]