Abstract
There is a large gap between the number of membrane protein (MP) sequences and that of their decoded 3D structures, especially high-resolution structures, due to difficulties in crystal preparation of MPs. However, detailed knowledge of the 3D structure is required for the fundamental understanding of the function of an MP and the interactions between the protein and its inhibitors or activators. In this paper, some computational approaches that have been used to predict MP structures are discussed and compared.
Key words: structure prediction, membrane proteins
Introduction
Membrane proteins (MPs) constitute about 30% of all the proteins encoded in the currently known genomes, and play critical roles in cell signaling, ion transport, and cell-cell communications, as well as assist the folding of other MPs (1). Because of these biological significance, MPs represent the most important class of drug targets—about 50% of current molecular targets are membrane-bound (2). However, only about 2% (518 of 25,176) of the 3D structures deposited in the Protein Data Bank (PDB; ref. 3) are for MPs. And the number of high-resolution structures (from X-ray diffraction and more recently from NMR) remains even smaller, largely because of the difficulties in crystallizing MPs. Recently, some new ideas and experimental approaches have been introduced in the area of MP crystallization (4), all of which exploit the spontaneous self-assembling properties of lipids and detergent as vesicles (vesicle-fusion method), discoidal micelles (bicelle method), and liquid crystals or mesophases (in meso or cubic-phase method). Despite these promising new methods, the current gap between need and supply of MP 3D structures makes prediction algorithms important and essential.
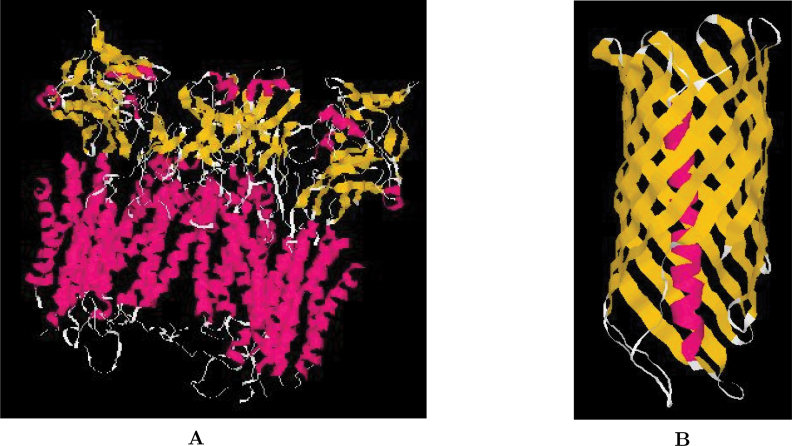
MPs come in a variety of sizes and shapes, though the available 3D structure principles are far less diverse than those of the globular proteins. From a structural point of view, there are two major groups of MPs. One is the α-helix bundle protein, in which one or several α-helices span the membrane; and the other is β-barrel protein, in which eight or more antiparallel TM β-strands form a closed barrel 5., 6.. Two recent examples 7., 8. are shown in Figure 1.
Fig. 1.
The crystal structures of two new MPs by X-ray diffraction. A. The cytochrome B6F complex of an α-helix bundle protein from Mastigocladus Laminosus, PDB Id: 1VF5(7). The red helices are the TM α-helix segments. B. The translocator domain of autotransporter nalp of a β-barrel protein from Neisseria Meningitidis, PDB Id: 1UYN(8). The yellow segment is the TM β-barrel composed of 12 membrane strands, and an N-terminal α-helix is in the center of the barrel.
Since Jähnig and Edholm in 1992 presented one of the first methods using secondary structure prediction to build suitable model structures as initial conformations for molecular dynamic studies (9), several groups have tried computational approaches to elucidate MP structures. In 1993, Milik and Skolnick presented a method based on the combination of a hydropathy scale for the prediction of trans-bilayer fragments with dynamic Monte Carlo simulation techniques (10). In 1994, Taylor et al. adapted some programs originally developed for the prediction of globular protein structures to derive a method for the prediction of integral MP structures (11). Each step in the method is fully automated, from the initial sequence data bank searches to the final construction of 3D models. The major problem of MP prediction is lack of high-resolution experimental data. Consequently, estimates for prediction accuracy are perhaps overly optimistic. Here, we summarize recent attempts within the field of computational biology and bioinformatics to predict an MP’s structure.
Secondary Structure Prediction and Transmembrane Segments Topology Prediction
Most current methods of theoretical MP structure prediction do not actually deal with predicting the 3D structure, but rather try to predict the most likely topology of the protein, that is to say, the in/out location of the N and C termini relative to the membrane, and the number and position of transmembrane (TM) segments. A high-quality model of secondary structure and topology is a prerequisite for experimental structure-function studies, and can be a starting point for attempts to model the 3D structure before molecular dynamics or simulated annealing simulations. In recent years, various accurate methods have been applied to the topology prediction of TM α-helices and β-strands, respectively. Table 1 shows the main methods of TM segment topology prediction. Because the number of high-resolution structures of β-barrel proteins is less than that of the α-helix proteins, the neural network has been more frequently adopted in the β-strand topology prediction. The details of some methods based on Hidden Markov Models (HMMs) are listed in Table 2.
Table 1.
The Main Methods of Transmembrane Segments Topology Prediction
| Segment type | Method | Approach | Self-proclaimed accuracy (segments) | Self-proclaimed accuracy (proteins) |
|---|---|---|---|---|
| Transmembrane α-helices |
TMHMM | HMM | 97%-98% | 77%-78% |
| HMMTOP | HMM | >98% | 85% | |
| MEMSAT | HMM | 92% | 77% | |
| PHDhtm | homologous & neural network | 98% | 89% | |
| TopPred | hydrophobicity analysis & positive-inside rule | – | 96% | |
| DAS-TMfilter | dense alignment surface | – | 95% | |
| ConPred_elite | consensus approach | – | 95%-98% | |
| Membrane β-strands | Gromiha’s | based on the conformational parameters and surrounding hydrophobicities | – | 82% |
| Diederichs’s | neural network | – | – | |
| Jacoboni’s | neural network | 93% | 78% | |
| Martelli’s | HMM | – | 84% | |
Table 2.
Several Methods Based on Hidden Markov Model
| Method | Number of states | Type of states |
|---|---|---|
| TMHMM | 7 | helix core, helix caps on either side of the membrane, short loop on cytoplasmic side/inside, short and long loop on noncytoplasmic side/outside, and a globular domain state |
| HMMTOP | 5 | inside loop, inside helix tail, helix, outside helix tail, and outside loop |
| MEMSAT | 5 | inside loop, inside helix tail, helix, outside helix tail, and outside loop |
| Martelli’s | 6 | 2 β-strand cores and 1 β-strand cap on either side of the membrane; 1 inner loop, 1 outer loop, and 1 globular domain state in the middle of each loop |
Many secondary structure prediction methods are based on statistical methods, physicochemical methods, sequence pattern maching, and evolutionary conservation (12). The main methods for identifing TM helices are on the basis of their hydrophobicity and known minimum length (at least 15 residues; ref. 13). Membrane propensities were defined by a statistical analysis carried out on a set of 640 TM helices, belonging to 133 MPs extracted from SWISS-PROT (14) that have experimentally defined topologies.
The five widely used prediction methods for predicting the topology of α-helix bundle MPs are TMHMM (15), HMMTOP (16), MEMSAT (17), PHDhtm (18), and TopPred (19). TMHMM, HMMTOP, and MEMSAT are all based on HMMs with 5~7 types of structural states. PHDhtm is designed to use information from homologous proteins. TopPred was the first topology prediction method that combined hydrophobicity analysis and the positive-inside rule. Generally, these sequence-based methods for predicting the number and approximate location of TM helices within MPs have about 85% accuracy. In 2003, Karin Melén et al. tried to construct useful reliability scores for these methods (20). They estimated an overall topology prediction accuracy of 55%-60% when entire proteomes are analyzed. The DAS (dense alignment surface; ref. 21) algorithm can provide a solution to the problem that non-transmembrane query sequences may give false positive hits (20%-30%) in the prediction process. The upgraded and modified version of the DAS-prediction method, DAS-TMfilter algorithm, has been distributed (22). The new algorithm is designed to make distinction between protein sequences with and without TM helices at a reasonably low rate of false positive prediction (~1 among 100 unrelated queries) while the high efficiency of the original algorithm locating TM segments in queries is preserved (sensitivity of ~95% among documented proteins with helical TM regions). In 2003, Xia and colleagues presented a new approach, ConPred_elite (23), that can predict the whole topology with accuracies of 98% for prokaryotic and 95% for eukaryotic proteins as they reported.
Besides the TM helix, another TM segments type is β-barrel, which consists of several TM strands. Unlike α-helical MPs, there are no simple low-resolution experiments that yield large amounts of data for β-barrel MPs. This has constrained the ability to develop prediction methods. All early attempts to predict membrane strands employed the amphipacity and hydrophobicity of β-strands. Unfortunately, membrane strands have no long stretch of consecutive hydrophobic residues. In fact, the overall hydrophobicity for β-barrel MPs is similar to that of soluble proteins (13).
Gromiha and colleagues combined amino acid preferences for β-strands with the surrounding hydrophobicity of the respective residues to predict β-strands (24). They reproduced about 82% of the residues in structure-known membrane regions. Diederichs and colleagues proposed to use a neural network to predict the topology of the bacterial outer membrane β-strand proteins and to locate residues along the axes of the pores (25). Jacoboni and colleagues applied a method combining neural networks and dynamic programming to predict the location of membrane strands (26). The authors estimated that their system correctly predicted about 93% of all known membrane strands. More recently, Martelli et al. developed a sequence-profile-based HMM model that can predict the topology of β-barrel MPs cyclicing with 6 types of states (27). They reported that the accuracy of per residue of the model was about 83%.
Lately the following protocol starting from secondary structure prediction and TM segments topology prediction are often used. Secondary structure prediction followed by TM segments identification along with prediction of loops connecting the segments, and molecular dynamics or simulated annealing simulations, may be finally used to refine these primal models. During the last refinement step, the protein is often inserted into a water/lipid bilayer/water or a water/n-octane/water environment to take into account the presence of the cell membrane. CHARMM, GROMOS, Amber, and cvff-insight are some widely used force fields in molecular dynamics calculation. The slow dynamics of lipid molecules in the bilayer might bring the difficulties in equilibrating the system (28).
The Direct Prediction of Whole 3D Structures
For globular proteins, the major successful methods for structure prediction include homology modeling, threading, and ab initio folding. Along with lucubrating the mechanism of MP folding and increasing the number of high-resolution MP structures, these methods will been applied to the direct prediction of whole MP 3D structures.
The question of how the controlled integration of an MP into the lipid bilayer takes place is still not fully worked out, and there are certainly aspects of MP structures that will probably not be fully appreciated until this step has been accomplished. Some pursuers educed the viewpoint that the prediction of MP structures from amino acid sequences was, in large measure, a problem of physicochemistry (29). Physical influences that shape MP structures include interactions of the polypeptide chains with water, bilayer hydrocarbon core, bilayer interfaces, and cofactors. Studies on the mechanism of insertion and folding of MPs into membranes are relatively rare and have been mostly performed with two model proteins: bacteriorhodopsin (BR; ref. 30) of Halobium salinarium and outer MP A (OmpA; ref. 31) of Escherichia coli. While BR is a representative α-helical bundle protein, OmpA belongs to the class of β-barrel protein.
Homology modeling constructs structures (targets) that are homologous to other protein(s) whose 3D structure is known (templates). It bases mainly on the conservation of protein folds rather than primary sequences homology. Because few high-resolution MP 3D structures are available to be used as templates, and the modeling can be unreliable when the sequence identity between the template and target proteins falls below 20%-30%, the applicability of homology modeling is limited. The same difficulties must been envisaged for threading methods.
In 2003, an ab initio method was presented (32), whose knowledge-based technique added a membrane potential to the energy terms (pairwise, solvation, steric, and hydrogen bonding). The method is based on the assembly of supersecondary structural fragments taken from a library of highly resolved protein structures using a standard simulated annealing algorithm. Results obtained by applying the method to small MPs of known 3D structures showed that the method is able to predict, at a reasonable accurate level, both the helix topology and the conformations of these proteins.
Conclusion
The structure prediction of membrane proteins still remains an interesting scientific problem. Because of the physical difference between MPs and GPs (globular proteins), more efforts have been put upon TM segment topology prediction for MPs. Current segment accuracy of reported algorithms are pretty high (above 90%), while the overall accuracy are still around 50%-60%, which gives birth to hand-raising methods to combine the reports from several other algorithms. The lack of both high-resolution and low-resolution experimental data of MP structures makes the algorithm development and their evaluation difficult, but the fact that most MP sequences are used as space blocks to get through the membrane bilayer that has predefined thickness makes the structure prediction of MPs simple on functional aspects. New algorithms will emerge and reported algorithms will be refined to give a better answer to this problem.
References
- 1.White S.H., Wimley W.C. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 2.Drews J. Drug discovery: a historical perspective. Science. 2000;287:1960–1964. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- 3.Berman H.M. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caffrey M. Membrane protein crystallization. J. Struct. Biol. 2003;142:108–132. doi: 10.1016/s1047-8477(03)00043-1. [DOI] [PubMed] [Google Scholar]
- 5.Henderson R., Unwin P.N. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- 6.Koebnik R. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 2000;37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 7.Kurisu G. Structure of the cytochrome B6F complex of oxygenic photosynthesis: tuning the cavity. Science. 2003;302:1009–1014. doi: 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- 8.Oomen C.J. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 2004;23:1257–1266. doi: 10.1038/sj.emboj.7600148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jähnig F., Edholm O. Modeling of the structure of bacteriorhodopsin: a molecular dynamics study. J. Mol. Biol. 1992;226:837–850. doi: 10.1016/0022-2836(92)90635-w. [DOI] [PubMed] [Google Scholar]
- 10.Milik M., Skolnick J. Insertion of peptide chains into lipid membranes: an off-lattice Monte Carlo dynamics model. Proteins. 1993;15:10–25. doi: 10.1002/prot.340150104. [DOI] [PubMed] [Google Scholar]
- 11.Taylor W.R. A method for alpha-helical integral membrane protein fold prediction. Proteins. 1994;18:281–294. doi: 10.1002/prot.340180309. [DOI] [PubMed] [Google Scholar]
- 12.Rost B. Protein secondary structure predication continues to rise. J. Strct. Biol. 2001;134:204–218. doi: 10.1006/jsbi.2001.4336. [DOI] [PubMed] [Google Scholar]
- 13.Chen C.P., Rost B. State-of-the-art in membrane protein prediction. Appl. Bioinformatics. 2002;1:21–35. [PubMed] [Google Scholar]
- 14.Bairoch A., Apweiler R. The SWISS-PROT protein sequence database: its relevance to human molecular medical research. J. Mol. Med. 1997;75:312–316. [PubMed] [Google Scholar]
- 15.Krogh A. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 16.Tusnady G.E., Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 17.Jones D.T. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- 18.Rost B. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;5:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Heijne G. Membrane protein structure prediction—hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 20.Karin M. Reliability measures for membrane protein topology prediction algorithms. J. Mol. Biol. 2003;327:735–744. doi: 10.1016/s0022-2836(03)00182-7. [DOI] [PubMed] [Google Scholar]
- 21.Cserzo M. On filtering false positive transmembrane protein predictions. Protein Engin. 2002;15:745–752. doi: 10.1093/protein/15.9.745. [DOI] [PubMed] [Google Scholar]
- 22.Cserzo M. TM or not TM: transmembrane protein prediction with low false positive rate using DAS-TMfilter. Bioinformatics. 2004;20:136–137. doi: 10.1093/bioinformatics/btg394. [DOI] [PubMed] [Google Scholar]
- 23.Xia J.X. ConPred_elite: a highly reliable approach to transmembrane topology prediction. Comput. Biol. Chem. 2004;28:51–60. doi: 10.1016/j.compbiolchem.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Gromiha M.M. Identification of membrane spanning beta strands in bacterial porins. Protein Engin. 1997;10:497–500. doi: 10.1093/protein/10.5.497. [DOI] [PubMed] [Google Scholar]
- 25.Diederichs K. Prediction by a neural network of outer membrane beta-strand protein topology. Protein Sci. 1998;7:2413–2420. doi: 10.1002/pro.5560071119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacoboni I. Prediction of the transmembrane regions of beta-barrel membrane proteins with a neural network-based predictor. Protein Sci. 2001;10:779–787. doi: 10.1110/ps.37201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martelli P.L. A sequence-profile-based HMM for predicting and discriminating beta-barrel membrane proteins. Bioinformatics. 2002;18:46–53. doi: 10.1093/bioinformatics/18.suppl_1.s46. [DOI] [PubMed] [Google Scholar]
- 28.Faraldo-Gomez J.D. Setting up and optimization of membrane protein simulations. Eur. Biophys. J. 2002;31:217–227. doi: 10.1007/s00249-002-0207-5. [DOI] [PubMed] [Google Scholar]
- 29.White S.H. Translocons, thermodynamics, and the folding of membrane proteins. FEBS Letters. 2003;555:116–121. doi: 10.1016/s0014-5793(03)01153-0. [DOI] [PubMed] [Google Scholar]
- 30.Booth P.J., Curran A.R. Membrane protein folding. Curr. Opin. Struct. Biol. 1999;9:115–121. doi: 10.1016/s0959-440x(99)80015-3. [DOI] [PubMed] [Google Scholar]
- 31.Kleinschmidt J.H. Outer membrane protein A of E. coli inserts and folds into lipid bilayers by a concerted mechanism. Biochemistry. 1999;38:5006–5016. doi: 10.1021/bi982465w. [DOI] [PubMed] [Google Scholar]
- 32.Pellegrini-Calace M. Folding in lipid membranes (FILM): a novel method for the prediction of small membrane protein 3D structures. Proteins. 2003;50:537–545. doi: 10.1002/prot.10304. [DOI] [PubMed] [Google Scholar]