Abstract
Genomic research has made a large number of sequences of novel genes or expressed sequence tags available. To investigate functions of these genes, a system for conditional control of gene expression would be a useful tool. Inducible transgene expression that uses green fluorescent protein gene (gfp) as a reporter gene has been investigated in transgenic cell lines of cotton (COT; Gossypium hirsutum L.), Fraser fir [FRA; Abies fraseri (Pursh) Poir], Nordmann fir (NOR; Abies nordmanniana Lk.), and rice (RIC; Oryza sativa L. cv. Radon). Transgenic cell lines were used to test the function of the chemical inducer dexamethasone. Inducible transgene expression was observed with fluorescence and confocal microscopy, and was confirmed by northern blot analyses. Dexamethasone at 5 mg/L induced gfp expression to the nearly highest level 48 h after treatment in COT, FRA, NOR, and RIC. Dexamethasone at 10 mg/L inhibited the growth of transgenic cells in FRA and NOR, but not COT and RIC. These results demonstrated that concentrations of inducer for optimum inducible gene expression system varied among transgenic cell lines. The inducible gene expression system described here was very effective and could be valuable in evaluating the function of novel gene.
Key words: inducible gene expression, confocal microscopy, transcriptional activator GVG, fluorescence microscopy
Introduction
Inducible gene expression systems using chemical inducers will be powerful tools for basic research in functional genomics and plant biology. Different chemical-inducible systems, based on activation and inactivation of the target gene by regulatory elements from prokaryotes, insects, and mammals, had been developed and used in a number of plant species 1., 2., 3., 4., 5.. In these systems, different inducers such as antibiotic tetracycline 6., 7., 8., glucocorticoid or steroid hormone dexamethasone and estrogen 9., 10., 11., 12., 13., 14., ethanol 15., 16., 17., 18., copper 19., 20., benzothiadiazol (21), herbicide safeners (22), and ecdysone 3., 23., 24., have been used. Although the advantages and limitations of different inducible gene expression systems to regulate the temporal and spatial expression patterns existed (3), well developed inducible gene expression systems will dramatically increase the application of transgenic technology in forestry and agriculture 5., 25., 26., 27..
Compared to regulated gene expression using constitutive promoters to transcribe a gene of interest, chemical-inducible gene expression systems offer a more general and flexible solution because chemical-inducible systems are quiescent in the presence or absence of inducers and therefore will not inhibit physiological activities 1., 28., 29., 30.. If a foreign gene product expressed in animals and plants is going to interfere with regeneration, growth or reproduction, an inducible promoter is required 1., 2.. With such a tool, plants can be regenerated while the promoter is inactive. Further analysis can then be performed after activating expression of the transgene 5., 28..
In this study, the glucocorticoid-inducible system 9., 12. was used. In this system, the chimaeric transcription factor GVG consists of the yeast Gal4 binding domain (BD), the Herpes simplex VP16 activation domain (AD), and the rat glucocorticoid receptor (GR; ref. 9., 12.). Induction is based on the property of the GR domain to localize the constitutively expressed GVG protein to the cytoplasm in the absence of the inducer (9). After treatment with glucocorticoid hormones such as dexamethasone, GVG is transferred to the nucleus and interact with GVG recognition sites cloned upstream of the targeting gene and activate transcription of the targeting gene 12., 31.. The GVG-regulated gene expression has been reported to be rapidly inducible and its control is very tight 9., 12..
Although the GVG inducible gene expression system has been investigated extensively in dicotyledonous and monocotyledonous plants such as tobacco, Arabidopsis, and rice 9., 12., 32., it was not used to compare the differential gene expression induced by inducers in angiosperms and gymnosperms. We adapted the GVG system for specific use in transgenic angiosperm and gymnosperm cell lines using green fluorescence protein gene (gfp) as a reporter gene. Inducible expression of gfp reporter gene was analyzed qualitatively and quantitatively at different induction levels. Our results suggested that the GVG inducible gene expression system could be useful in studying regulated gene expression ranging from both angiosperm and gymnosperm plant species.
Results
Transgenic cell lines
Cell cultures derived from single callus clones were used to produce transgenic cell lines. Nine cell cultures of cotton (COT; Gossypium hirsutum L.), seven cell cultures of Fraser fir [FRA; Abies fraseri (Pursh) Poir], nine cell cultures of Nordmann fir (NOR; Abies nordmanniana Lk.), and five cell cultures of rice (RIC; Oryza sativa L. cv. Radon) were infected with the Agrobacterium tumefaciens strain EHA105 containing pINDEX3-m-gfp5-ER carrying a hygromycin phosphotransferase gene, a chimaeric transcription factor GVG, and a m-gfp5-ER reporter gene (Figure 1), respectively. Two transgenic cell lines from each species were isolated following selection on medium supplemented with hygromycin. Transgenic cell lines were transferred into fresh proliferation medium weekly for 10-12 weeks to produce more cells. Eight putative transgenic cell lines with hygromycin resistance were initially tested for the presence of the m-gfp5-ER by PCR. Eight hygromycin-resistant cell lines produced the expected band (816 bp), whereas DNA from non-transformed cell cultures did not yield such a product (Figure 2). Southern hybridization of digested genomic DNA demonstrated that the T-DNA insert was integrated in this eight transgenic cell lines (data not shown). Single copies of transgene were observed from genomic DNA of four transgenic cell lines from COT, FRA, NOR, and RIC (data not shown). GFP expression could not be visualized in all eight cell lines transformed with m-gfp5-ER due to transcription of m-gfp5-ER was not activated by GVG transcription factor. Four transgenic cell lines (COT, FRA, NOR, and RIC), each with one copy of the T-DNA insert, were used for future inducible gene expression experiments.
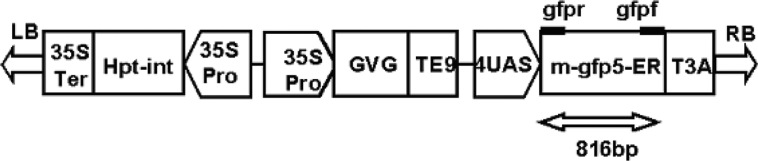
Fig. 1.
A linear plasmid map indicating the localization of the different genes, promoters, terminatiors and T-DNA borders. Genes: Hpt-int, hygromycin phosphotransferase gene with a intron from the castor bean Cat-1 gene; GVG, the chimaeric transcription factor consists of the yeast Gal4 binding domain (BD), the Herpes simplex VP16 activation domain (AD), and the rat glucocorticoid receptor (GR); m-gfp5-ER, a modified GFP protein with an endoplasmic reticulum targeting sequence. Promoters: 4UAS, the promoter containing four copies of the GAL4 UAS (upstream activation sequence) and the −46 to +1 region of the 35S promoter; 35S Pro, the cauliflower mosaic virus 35S promoter. Terminators: 35S Ter, the terminator of the CaMV 35S transcription unit; TE9, the poly(A) addition sequence of the pea ribulose biphosphate carboxylase small subunit rbcS-E9; T3A, the poly(A) addition sequence of the pea rbcS-3A. T-DNA borders: LB, left border; RB, right border. Arrows indicate gene translation orientation. The probe used in northern blot analysis of transgenic cell lines is the PCR fragment of the m-gfp5-ER gene. Binding sites of PCR primers gfpr and gfpf are shown as black rectangles, their positions are indicated immediately above the m-gfp5-ER gene, and the predicted product size (816 bp) from gfpr and gfpf is shown as a double-headed arrow.
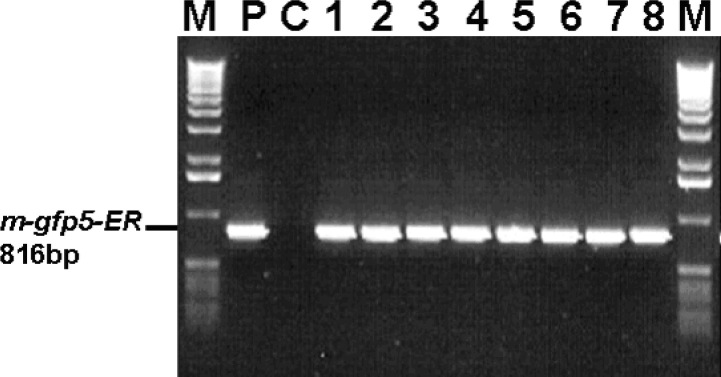
Fig. 2.
PCR analysis of DNA isolated from transgenic cell lines. Lane M: 1-Kb DNA molecular markers (Gibco-BRL); lane P: pINDEX3-m-gfp5-ER plasmid control; lane C: non-transgenic cells control; lanes 1-8: transgenic cell lines COT (lanes 1 and 2), FRA (lanes 3 and 4), NOR (lanes 5 and 6), and RIC (lanes 7 and 8), respectively. The presence of the 816-bp band amplified by primers gfpr and gfpf from templates obtained from transgenic cell lines and absence of the 816-bp band amplified by primers gfpr and gfpf from templates obtained from non-transformed control cells indicate that the T-DNA is integrated into the pine genome.
Dexamethasone-inducible gfp expression in transgenic cells
Four transgenic cell lines (COT, FRA, NOR, and RIC, each of them with a single copy of transgene) were used to test inducible gene expression by different concentrations of dexamethasone (1, 5, and 10 mg/L, respectively). After transgenic cell lines were transferred to liquid medium containing 1, 5, and 10 mg/L dexamethasone, respectively, transgenic cell lines were observed every 4 h with fluorescence microscope in blue light. Inducible gfp activity was first observed at 4 h after treated by 10 mg/L dexamethasone in COT and RIC transgenic cell lines. These transgenic cell lines produced intense green fluorescence upon blue light excitation after 24 h on medium with 1, 5 or 10 mg/L dexamethasone. Expression of m-gfp5-ER was clearly visible at three concentrations of dexamethasone tested in all four transgenic cell lines. A sample of inducible gfp expression in all four transgenic cell lines 24 h after 5 mg/L dexamethasone treatment was shown as Figure 3. Induction was also analyzed at the RNA level by northern blot analysis (Figure 4). Among samples at 24, 48, and 72 h after treatment with dexamethasone, higher transcripts were observed from all four transgenic cell lines 48 h after treatment of 5 mg/L dexamethasone (Figure 4).
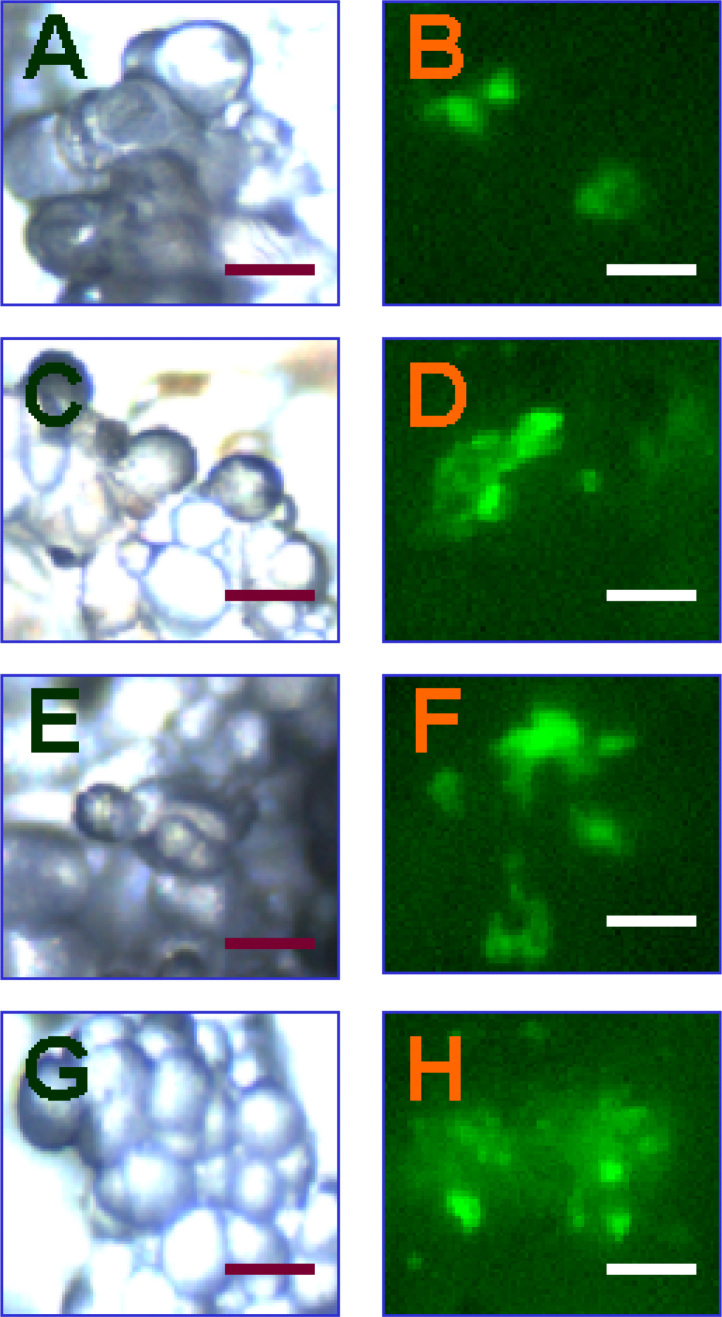
Fig. 3.
Fluorescent detection of dexamethasone-induced GFP expression in different transgenic cell lines. A-B: GFP expression induced by 5 mg/L dexamethasone from transgenic cell line COT at 0 h (A) and 24 h (B) after treatment, respectively; C-D: GFP expression induced by 5 mg/L dexamethasone from transgenic cell lines FRA at 0 h (C) and 24 h (D) after treatment, respectively; E-F: GFP expression induced by 5 mg/L dexamethasone from transgenic cell lines NOR at 0 h (E) and 24 h (F) after treatment, respectively; G-H: GFP expression induced by 5 mg/L dexamethasone from transgenic cell lines RIC at 0 h (G) and 24 h (H) after treatment, respectively. No GFP fluorescence was observed in transgenic cell lines without addition of dexamethasone and in non-transformed control (bars = 0.1 mm).
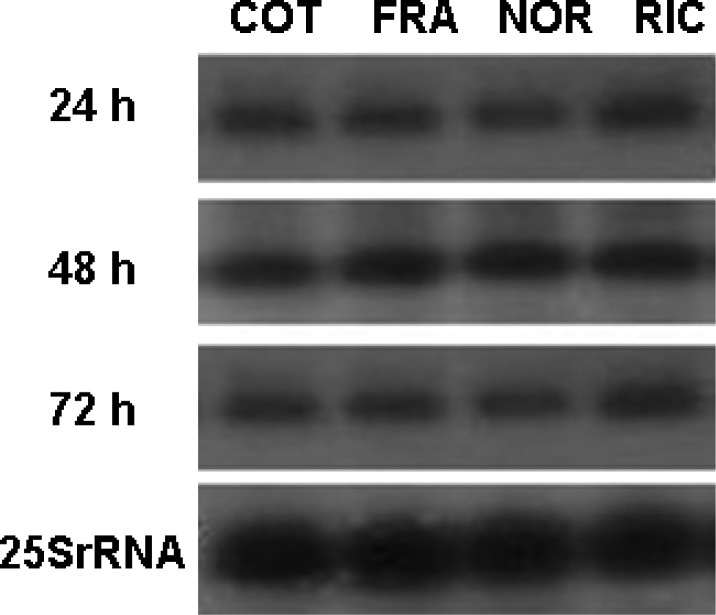
Fig. 4.
Northern blot analysis of total RNA from transgenic cell lines. RNA (15 μg) was prepared as described in the experimental protocol section from transgenic cell lines COT, FRA, NOR, and RIC at 24 h, 48 h, and 72 h after treatment of 5 mg/L dexamethasone, respectively, and were hybridized (at 65°C) with the 816-bp m-gfp5-ER probe corresponding to the m-gfp5-ER gene. The integrity and the amount of RNA applied to each lane were verified by the control of 25S rRNA (lower panel).
Effect of dexamethasone-mediated induction on cell growth was evaluated in transgenic cell lines. Transgenic cell lines (COT, FRA, NOR, and RIC) were sub-cultured weekly on liquid medium containing 0, 1, 5, and 10 mg/L, respectively. Transgenic cell cultures were harvested at 21 d after treatment with dexamethasone for measurement of fresh weight and dry weight. There are no significant differences in fresh and dry weight increase when 1 or 5 mg/L dexamethasone was applied for all four transgenic cell lines. The results demonstrated that cell growth was not inhibited by the addition of dexamethasone at 1 and 5 mg/L (Table 1, Table 2). However, 10 mg/L dexamethasone inhibited cell growth of FRA and NOR transgenic cell lines, but did not inhibit growth and development of COT and RIC transgenic cell lines (Table 1, Table 2). This reduction in growth is probably due to the fact that high levels of dexamethasone could potentially have toxic effects on transgenic cell cultures.
Table 1.
Fresh Weight Increases of Transgenic Cell Lines (mg fresh weight/liter cell cultures/day)*
| Dexamethasone (mg/L) | Fresh weight increase |
|||
|---|---|---|---|---|
| COT | FRA | NOR | RIC | |
| 0 | 7.9±1.9a | 7.5±2.4a | 8.3±3.7a | 9.6±2.9a |
| 1 | 7.8±2.1a | 7.1±2.5a | 7.9±2.5a | 9.5±2.8a |
| 5 | 7.8±2.2a | 7.0±2.2a | 7.7±3.2a | 9.3±2.5a |
| 10 | 7.6±1.8a | 5.1±1.5b | 5.3±1.7b | 9.2±2.4a |
Fresh weight increases (mg fresh weight/liter cell cultures/day) of transgenic cell lines 21 days after treatment by different concentrations of dexamethasone. Data represent the mean ± SD. Values followed by different letters are significantly different (α=0.05) by ANOVA.
Table 2.
Dry Weight Increases of Transgenic Cell Lines (mg dry weight/liter cell cultures/week)*
| Dexamethasone (mg/L) | Dry weight increase |
|||
|---|---|---|---|---|
| COT | FRA | NOR | RIC | |
| 0 | 9.1±1.6a | 8.1±1.4a | 8.3±1.5a | 9.6±1.7a |
| 1 | 8.9±1.4a | 8.0±1.5a | 8.2±1.2a | 9.5±1.8a |
| 5 | 8.8±1.2a | 8.0±1.2a | 8.1±1.2a | 9.3±1.5a |
| 10 | 8.7±1.3a | 6.6±1.3b | 6.5±1.0b | 9.2±1.4a |
Dry weight increases (mg dry weight /liter cell cultures/week) of transgenic cell lines 21 days after treatment by different concentrations of dexamethasone. Data represent the mean ± SD. Values followed by different letters are significantly different (α=0.05) by ANOVA.
Quantitative analysis of inducible gfp expression
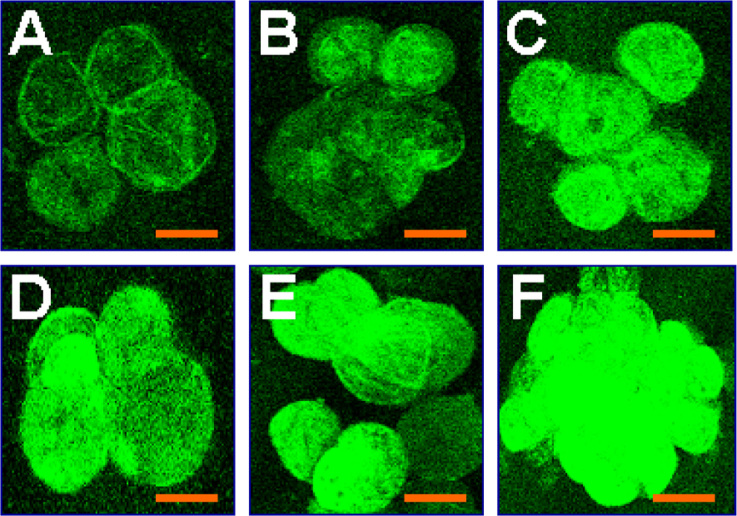
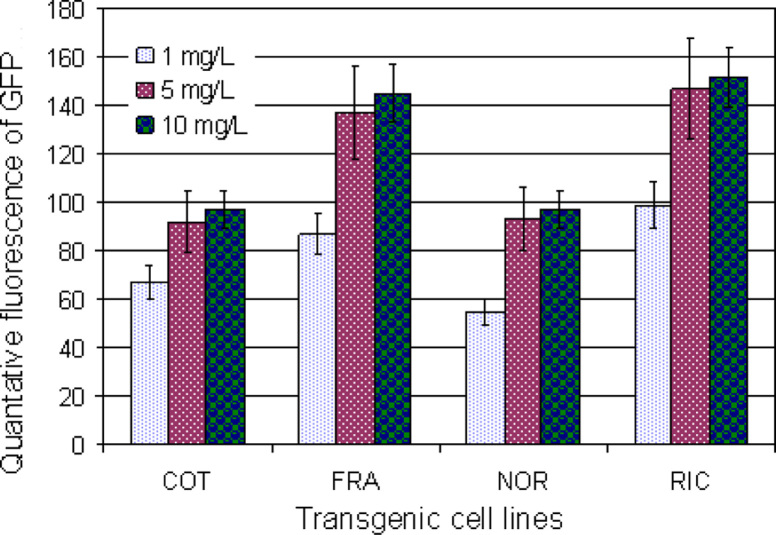
With the confocal microscopy, we have quantitatively determined dexamethasone-inducible transgene expression at different levels of inducer in COT, FRA, NOR, and RIC transgenic cell lines. Transgenic cell lines were sub-cultured on liquid medium with 1, 5, and 10 mg/L dexamethasone, respectively, for 24 h. Confocal images were taken by a LSM 510 Laser Scanning Microscope (Carl Zeiss, Inc., Thornwood, USA) at different times (0, 12, 24, 36, 48, and 72 h, respectively) after treatment of dexamethasone, and an example was shown in Figure 5A-F. Quantitative fluorescence intensities (arbitrary unit) were determined from the confocal images. As shown in Figure 6, inducible m-gfp5-ER activities ranged between 52 (NOR transgenic cells at 1 mg/L dexamethasone) and 151 (RIC transgenic cells at 10 mg/L dexamethasone) arbitrary units of fluorescence intensity, with the higher fluorescence from transgenic cell lines on medium supplemented with 10 mg/L dexamethasone in all four transgenic cell lines (Figure 6). In the absence of dexamethasone, no m-gfp5-ER activity could be detected.
Fig. 5.
Confocal images of GFP expression in transgenic cell line RIC at 0 (A), 12 (B), 24 (C), 36 (D), 48 (E), and 72 (F) h at induction levels of 5 mg/L dexamethasone. No GFP fluorescence was detected in transgenic cell line without inducer.
Fig. 6.
Quantitative analysis of GFP fluorescence from different transgenic cell lines COT, FRA, NOR, and RIC at different induction levels of dexamethasone (1, 5, and 10 mg/L), respectively. Quantitative fluorescence is expressed as intensity (arbitrary units). GFP fluorescence was detected 48 h after addition of inducer. No GFP fluorescence was detected in transgenic cell line without inducer. Experiments were repeated three times, and each replicate consisted of 30 cells. Values represent the means ± SD.
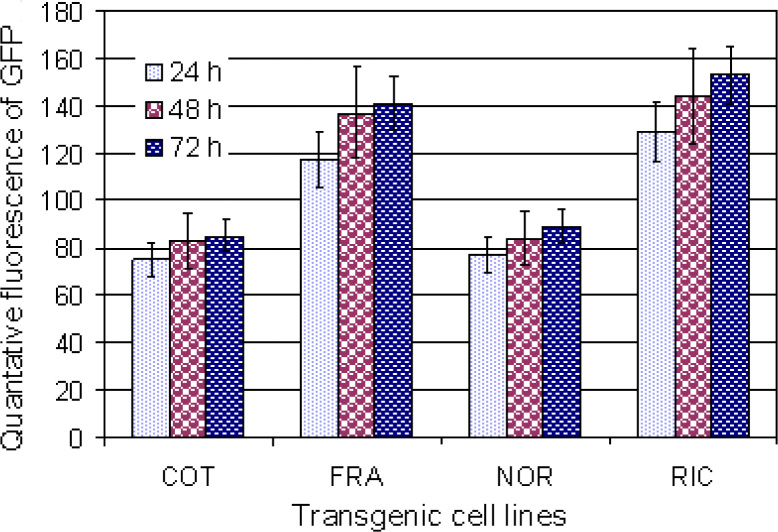
Dexamethasone-inducible gene expression was also quantitatively analyzed at different times (24, 48, and 72 h) after treatment with inducer in four transgenic cell lines, respectively. Transgenic cell lines (COT, FRA, NOR, and RIC) were transferred to medium containing 10 mg/L dexamethasone and cultured for 24, 48, and 72 h, respectively, prior to GFP fluorescence measurements. No GFP fluorescence was detected in all four transgenic cell lines cultured on medium without addition of dexamethasone. The differences in inducible gfp activity occurred among different transgenic cell lines and times of applying dexamethasone (Figure 7). The highest GFP fluorescence was observed from RIC transgenic cells on medium supplemented with 10 mg/L dexamethasone after 72 h treatment (Figure 7). Compared to transgenic cells grown at 1 and 5 mg/L dexamethasone, transgenic cells grown at 10 mg/L dexamethasone showed stronger fluorescence (Figure 6). Clearly, the concentration of inducer and time of induction required to saturate inducible gene expression will be dependent on transgenic cell lines.
Fig. 7.
Quantitative analysis of GFP fluorescence from different transgenic cell lines COT, FRA, NOR, and RIC at different times of induction (24, 48, and 72 h), respectively. Quantitative fluorescence is expressed as intensity (arbitrary units). Induction level of dexamethasone was 10 mg/L for all four transgenic cell lines. No GFP fluorescence was detected in transgenic cell line without inducer. Experiments were repeated three times, and each replicate consisted of 30 cells. Values represent the means ± SD.
Discussion
Inducible gene expression system would be useful in identifying functions of expressed sequence tags (ESTs) and novel genes produced from genomic research. Although different inducible gene expression systems are available 1., 2., 3., 27., 33., we have chosen the GVG system 9., 12. for dexamethasone-mediated transcriptional induction in four transgenic cell lines derived from different gymnosperm and angiosperm species. We used the m-gfp5-ER reporter gene (34) to test the inducible GVG system. The m-gfp5-ER reporter gene allows us to identify transgenic cells visually and to determine inducible gene expression quantitatively. In our study, four transgenic cell lines with one copy of m-gfp5-ER were used to analyze the dexamethasone-inducible gene expression. All four transgenic cell lines showed dexamethasone-inducible m-gfp5-ER activity 12 h after treatment. The fact that all cell lines were positive indicates that the organization of the T-DNA in the pINDEX3 vectors guarantees a high frequency of transgenic cell lines with an intact T-DNA insertion (12). Although different inducible gene expressions were observed among transgenic cell lines, our results demonstrated that the widely used CaMV 35S promoter is a useful alternative for dexamethasone-inducible gene expression in transgenic cell lines derived from different plant species.
Confocal microscopy of transgenic cell lines showed that the 4UAS sequence upstream of the m-gfp5-ER is not recognized by native transcriptional activators within plant cells, because no m-gfp5-ER activities were detected in the absence of dexamethasone and differential gfp expressions were observed under different concentrations of dexamethasone. Our results demonstrated that expression of m-gfp5-ER gene was controlled tightly by dexamethasone. This is in agreement with the results of others in tobacco and Arabidopsis 1., 33.. In the present investigation, we observed that 5 mg/L dexamethasone was already sufficient to obtain high levels of m-gfp5-ER induction (Figure 6). Although it has been reported that methylation of UAS sites can interfere with reporter-gene activation sites in transgenic tobacco plants (35), our results demonstrated that the use of the GVG system was not hampered by UAS methylation problems 9., 31., 33..
For determining the effects of dexamethasone on cell growth as reported in Kang et al. (32), due to the nuclear localization of GVG after dexamethasone treatment, we quantitatively analyzed the fresh weight and dry weight increase of transgenic cells under different concentrations of dexamethasone (Table 1, Table 2). Our results demonstrated that no growth retardation occurred when 1 and 5 mg/L dexamethasone were used. However, decreased cell growth was observed in two transgenic cell lines (FRA and NOR) when 10 mg/L dexamethasone was applied (Table 1, Table 2). Higher concentrations of dexamethasone caused growth retardation in FRA and NOR transgenic cell cultures, but did not influence growth of COT and RIC cell cultures. The fact that the growth retardation derived from 10 mg/L dexamethasone was depended on cell lines demonstrated that optimum inducible gene expression systems varied among plant species. Therefore, selection of transgenic cell lines with a mild GVG phenotype in combination with optimized induction conditions makes this system suitable for different plant species (12). The system can nevertheless have useful applications and GVG effects can be minimized by careful adjustment of the induction conditions. Our results also demonstrated that quantitative analysis of GFP fluorescence based on confocal images of transgenic cells provided a reliable way to identify differentially inducible gene expression in transgenic plant cells.
Materials and Methods
Plasmid constructs
The pINDEX3 binary vector (12) was transferred into NovaBlue competent cells (Novagen, Madison, USA) following the NovaBlue Competent Cells Protocol. Plasmid pINDEX3 was prepared by Wizard Plus Minipreps DNA Purification System (Promega Corporation, Madison, USA), digested by SpeI and XhoI (Promega) at 37°C, respectively, and purified by QIAquick Gel Extraction Kit (QIAGEN Inc., Valencia, USA). The m-gfp5-ER fragments were amplified from plasmid pBINm-gfp5-ER 34., 36.. The restriction enzyme SpeI and XhoI sites were introduced by PCR with Stratagene’s Pfu DNA polymerase (Stratagene, Cedar Creek, USA). For amplification of m-gfp5-ER, the forward primer 5′-aaacatgatgagctttaaagactc-3′ and the reverse primer 5′-cttcattgtttgatcaccttgcatcc-3′ (Sigma Chemical, St. Louis, USA) were used. A total of 200 ng of plasmid DNA was used as a template in a 50-μL PCR reaction mix. The PCR reaction was programmed as described in Tang et al. (37). PCR products were digested by SpeI and XhoI (Promega) at 37°C, respectively, and purified by QIAquick Gel Extraction Kit. Ligation of digested and purified insert (30 ng) and vector (90 ng) was conducted in 20 μL volume with 2.5 U T4 DNA Ligase (Roche Applied Science, Roche Diagnostics Corporation, Indianapolis, USA) at 16°C for 16 h. 1 μL of ligation products was used to transform 20 μL NovaBlue competent cells. Plasmid pINDEX3-m-gfp5-ER prepared by Wizard Plus Minipreps DNA Purification System was introduced into Agrobacterium tumefaciens EHA105 competent cells by electroporation (Bio-Rad Laboratories, Hercules, USA).
Transformation of cultured cells and dexamethasone treatments
Cell cultures were prepared as described in Tang et al. (38). Cell cultures of Fraser fir and Nordmann fir were used for Agrobacterium-mediated transformation as previously described (37). Cell cultures of cotton were used for Agrobacterium-mediated transformation as previously described by Sunilkumar and Rathore (39). Cell cultures of rice were used for Agrobacterium-mediated transformation as previously described 40., 41.. Transformation experiments were done three days after cell suspension cultures were transferred to fresh liquid medium. Agrobacterium tumefaciens EHA105 grown to an optical density (OD600 nm = 0.8−1.0) in 3 mL of YEP broth (42), were centrifuged and re-suspended in liquid medium, then used to infect plant cells for 15 min. The infected plant cells were co-cultivated for 2 d. Agrobacterium was removed from plant cell cultures by washing with 500 mg/L Timentin (ticarcillin/clavulanic acid 3:0.1, SmithKline Beecham, Philadelphia, USA) solution for 15 min. The plant cell cultures were then transferred to fresh liquid medium containing 500 mg/L Timentin and subcultured every three days for 10 times. Cells derived from transformed cell suspension cultures were incubated in 150 rpm shaker in the dark at room temperature (25°C). After five weeks, putative transformants were transferred to a fresh hygromycin-containing medium. After six weeks, the cultures were actively producing 50-100 mg of tissue in 1 L cultures each week, and they were then used to prepare DNA for PCR and Southern blot analysis.
Transgenic cell lines (COT, FRA, NOR, and RIC) confirmed by PCR and Southern blot analysis were used for inducible gene expression experiments. Transgenic cell lines were grown in liquid medium for 3 d, and were then transferred into fresh liquid medium supplemented with different concentration of dexamethasone (1, 5, and 10 mg/L, respectively) for 24 h. Dexamethasone (Sigma Chemical, St. Louis, MO, USA) was stored as 100 mM solution in dimethyl sulfoxide (DMSO) at −20°C. Cells were then washed and transferred into fresh liquid medium. At 24, 48, and 72 h after treatment of dexamethasone, cell cultures were harvested for quantitatively fluorescent microscopy, and for northern blot analysis. At the 21st day after treatment of dexamethasone, cell cultures were harvested by centrifuging at 8,000 rpm for 15 min for the measurement of fresh weight. Then cell cultures were dried at 85°C for 3 d for the measurements of dry weight (43).
Polymerase chain reaction and Southern blot analyses of transgenic cultures
Genomic DNA was isolated from 500 mg cell cultures using a Genomic DNA Isolation Kit (Sigma) following the manufacturer’s protocol. For amplification of gfp, the forward primer (gfpf) 5′-aaacatgatgagctttaaagactc-3′ and the reverse primer (gfpr) 5′-cttcattgttatatcaccttgcat-3′ were used. A total of 200 ng genomic DNA was used as a template in a 50-μL PCR reaction mix. The PCR mixture consisted of 200 μM each of dATP, dCTP, dGTP, dTTP, 35 pmol of each primer, 2.5 U Taq DNA polymerase (Promega), 1.5 mM MgCl2, and 5 μL 10× buffer [500 mM KCl, 100 mM Tris-HCl (pH 9.0 at 25°C), 1% Triton X-100, 15 mM MgCl2]. The PCR conditions were 95°C for 5 min followed by 29 cycles at 95°C for 60 s, 57°C for 40 s, and 72°C for 90 s. Cycling was followed with a final incubation of 72°C for 10 min. PCR products were separated by electrophoresis on 1.1% agarose gels in 1× TAE buffer (42) and were detected by UV light after staining with 0.1% ethidium bromide. A molecular marker of 1-Kb (Gibco-BRL, Gaithersburg, USA) was used. Southern blot analysis was conducted as previously described (37).
RNA isolation and northern blot analysis
Total RNA was isolated from 2 g transgenic cell cultures using RNeasy Plant Mini Kit (QIAGEN). Electrophoresis and northern blotting of RNAs were performed as described by Tang and Tian (44). Baked blots were pre-hybridized in 1 M NaCl, 1% SDS, 10% dextran sulphate and 50 μg/mL denatured herring sperm DNA at 64°C, washed with 0.1× SSPE (1× SSPE is 180 mM NaCl, 10 mM NaH2PO4, 1 mM EDTA, pH 6.5), 0.5% SDS at 45°C, and graphed. Digoxigenin (DIG)-Labelling m-gfp5-ER DNA (816 pb; Roche) was used as hybridization probes. Equal loading of RNA samples was verified on the control of 25S rRNA.
Qualitative and quantitative analysis of inducible gfp expression
Transgenic cells was observed with a stereo dissecting microscope equipped with a fluorescence module consisting of a 100-W mercury lamp and GFP Plus excitation and emission filters (Leica, Heerbrugg, Switzerland). This system (excitation filter 480 nm; dichroic mirror 505 nm LP; barrier filter 510 nm LP) permits visualization of GFP following excitation by blue light. Transgenic cells treated with different concentrations of dexamethasone and at different times of induction were observed. For quantitative fluorescence determinations of m-gfp5-ER activity, transgenic cells were examined with a LSM 510 Laser Scanning Microscope (Carl Zeiss, Inc., Thornwood, USA) using excitation with the 488-nm Argon laser line and detection of emitted light of 520 nm. The confocal images of m-gfp5-ER expression cells were created in the Expert Mode. Fluorescence intensities (arbitrary unit) of different samples were calculated from confocal images with the Zeiss LSM Image Examiner software. The fluorescence level was quantified separately for the whole cell by circumscribing the respective area as a region of interest. Background correction was applied by deducing fluorescence levels in a neighboring non-transgenic cell. Fifty to 100 cells were used for each sample.
Acknowledgements
The authors are grateful to Dr. P. B. F. Ouwerkerk and Dr. A. H. Meijer (Leiden University, The Netherlands) for the gift of the vector pINDEX3, to Dr. C. N. Stewart and Dr. J. Haseloff for providing us with the m-gfp5-ER constructs, and to Dr. D. Weidner (The Flow Cytometry-Confocal Microscopy Core Facility, the Brody School of Medicine, East Carolina University, NC 27858, USA) for technical assistance with confocal microscopy for imaging and quantitative analysis of green fluorescence. We thank K. S. Rathore for providing us with the cotton and rice calluses.
References
- 1.Aoyama T. Glucocorticoid-inducible gene expression in plants. In: Reynolds P.H.S., editor. Inducible gene expression in plants. CAB International; Wallingford, UK: 1999. pp. 43–59. [Google Scholar]
- 2.Gatz C. Use of the Tn10-encoded tetracycline repressor to control gene expression. In: Reynolds P.H.S., editor. Inducible gene expression in plants. CAB International; Wallingford, UK: 1999. pp. 11–22. [Google Scholar]
- 3.Padidam M. Chemically regulated gene expression in plants. Curr. Opin. Plant Biol. 2003;6:169–177. doi: 10.1016/s1369-5266(03)00005-0. [DOI] [PubMed] [Google Scholar]
- 4.Reynolds P.H.S., editor. Inducible gene expression in plants. CAB International; Wallingford, UK: 1999. [Google Scholar]
- 5.Zuo J., Chua N.H. Chemical-inducible systems for regulated expression of plant genes. Curr. Opi. Biotechnol. 2000;11:146–151. doi: 10.1016/s0958-1669(00)00073-2. [DOI] [PubMed] [Google Scholar]
- 6.Bohner S. Transcriptional activator TGV mediates dexamethasone-inducible and tetracycline-inactivatable gene expression. Plant J. 1999;19:87–95. doi: 10.1046/j.1365-313x.1999.00503.x. [DOI] [PubMed] [Google Scholar]
- 7.Love J. Differential Top10 promoter regulation by six tetracycline analogues in plant cells. J. Exp. Bot. 2002;53:1871–1877. doi: 10.1093/jxb/erf050. [DOI] [PubMed] [Google Scholar]
- 8.Weinmann P. Achimeric transactivator allows tetracycline-responsive gene expression in whole plants. Plant J. 1994;5:559–569. doi: 10.1046/j.1365-313x.1994.5040559.x. [DOI] [PubMed] [Google Scholar]
- 9.Aoyama T., Chua N.H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- 10.Bruce W. Expression profiling of the maize flavonoid pathway genes controlled by estradiol-inducible transcription factors CRC and P. Plant Cell. 2000;12:65–80. doi: 10.1105/tpc.12.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore I. A transcription activation system for regulated gene expression in transgenic plants. Proc. Natl. Acad. Sci. USA. 1998;95:376–381. doi: 10.1073/pnas.95.1.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouwerkerk P.B.F. Glucocorticoid-inducible gene expression in rice. Planta. 2001;213:370–378. doi: 10.1007/s004250100583. [DOI] [PubMed] [Google Scholar]
- 13.Schena M. A steroid-inducible gene expression system for plant cells. Proc. Natl. Acad. Sci. USA. 1991;88:10421–10425. doi: 10.1073/pnas.88.23.10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuo J. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
- 15.Caddick M.X. An ethanol inducible gene switch for plants used to manipulate carbon metabolism. Nat. Biotechnol. 1998;16:177–180. doi: 10.1038/nbt0298-177. [DOI] [PubMed] [Google Scholar]
- 16.Junker B.H. In plants the alc gene expression system responds more rapidly following induction with acetaldehyde than with ethanol. FEBS Lett. 2003;535:136–140. doi: 10.1016/s0014-5793(02)03889-9. [DOI] [PubMed] [Google Scholar]
- 17.Salter M.G. Characterisation of the ethanol-inducible alc gene expression system for transgenic plants. Plant J. 1998;16:127–132. [Google Scholar]
- 18.Sweetman J.P. Ethanol vapor is an efficient inducer of the alc gene expression system in model and crop plant species. Plant Physiol. 2002;129:943–948. doi: 10.1104/pp.010937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenzie M.J. Controlled cytokinin production in transgenic tobacco using a copper-inducible promoter. Plant Physiol. 1998;116:969–977. doi: 10.1104/pp.116.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mett V.L. Copper-controllable gene-expression system for whole plants. Proc. Natl. Acad. Sci. USA. 1993;90:4567–4571. doi: 10.1073/pnas.90.10.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedrich L. Abenzothiadiazole derivative induces systemic acquired resistance in tobacco. Plant J. 1996;10:61–70. [Google Scholar]
- 22.DeVeylder L. Herbicide safener-inducible gene expression in Arabidopsis thaliana. Plant Cell Physiol. 1997;38:568–577. doi: 10.1093/oxfordjournals.pcp.a029206. [DOI] [PubMed] [Google Scholar]
- 23.Martinez A. Creation of ecdysone receptor chimeras in plants for controlled regulation of gene expression. Mol. Gen. Genet. 1999;261:546–552. doi: 10.1007/s004380050999. [DOI] [PubMed] [Google Scholar]
- 24.Martinez A. Ecdysone agonist inducible transcription in transgenic tobacco plants. Plant J. 1999;19:97–106. doi: 10.1046/j.1365-313x.1999.00504.x. [DOI] [PubMed] [Google Scholar]
- 25.Gatz C., Lenk I. Promoters that respond to chemical inducers. Trends Plant Sci. 1998;3:352–358. [Google Scholar]
- 26.Guo H.S. A chemical-regulated inducible RNAi system in plants. Plant J. 2003;34:383–392. doi: 10.1046/j.1365-313x.2003.01723.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang R.H. Chemically regulated expression systems and their applications in transgenic plants. Transgenic Res. 2003;12:529–540. doi: 10.1023/a:1025852307127. [DOI] [PubMed] [Google Scholar]
- 28.Gatz C. Chemical control of gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:89–108. doi: 10.1146/annurev.arplant.48.1.89. [DOI] [PubMed] [Google Scholar]
- 29.Gossen M. Transcriptional activation by tetracycline in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 30.Uknes S. Regulation of pathogenesis-related protein-1a gene expression in tobacco. Plant Cell. 1993;5:159–169. doi: 10.1105/tpc.5.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNellis T.W. Glucocorticoid-inducible expression of a bacterial avirulence gene in transgenic Arabidopsis induces hypersensitive cell death. Plant J. 1998;14:247–257. doi: 10.1046/j.1365-313x.1998.00106.x. [DOI] [PubMed] [Google Scholar]
- 32.Kang H.G. A glucocorticoid-inducible transcription system causes severe growth defects in Arabidopsis and induces defense-related genes. Plant J. 1999;20:127–133. doi: 10.1046/j.1365-313x.1999.00575.x. [DOI] [PubMed] [Google Scholar]
- 33.Kunkel T. Inducible isopentenyl transferase as a high-efficiency marker for plant transformation. Nat. Biotechnol. 1999;17:916–919. doi: 10.1038/12914. [DOI] [PubMed] [Google Scholar]
- 34.Haseloff J. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gälweiler L. The DNA-binding activity of Gal4 is inhibited by methylation of the Gal4 binding site in plant chromatin. Plant J. 2000;23:143–157. doi: 10.1046/j.1365-313x.2000.00805.x. [DOI] [PubMed] [Google Scholar]
- 36.Stewart C.N. The utility of green fluorescent protein in transgenic plants. Plant Cell Rep. 2001;20:376–382. doi: 10.1007/s002990100346. [DOI] [PubMed] [Google Scholar]
- 37.Tang W. Regeneration of transgenic loblolly pine (Pinus taeda L.) from zygotic embryos transformed with Agrobacterium tumefaciens. Planta. 2001;213:981–989. doi: 10.1007/s004250100566. [DOI] [PubMed] [Google Scholar]
- 38.Tang W. Plant regeneration from embryogenic cultures initiated from mature loblolly pine zygotic embryos. In Vitro Cell. Dev. Biol.–Plant. 2001;37:558–563. [Google Scholar]
- 39.Sunilkumar G., Rathore K.S. Transgenic cotton: factors influencing Agrobacterium-mediated transformation and regeneration. Mol. Breed. 2001;8:37–52. [Google Scholar]
- 40.Hiei Y. Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol. Biol. 1997;35:205–218. [PubMed] [Google Scholar]
- 41.Hiei Y. Efficient transformation of rice (Oryza-sativa L.) mediated by Agrobacterium and sequence-analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J. 2nd edition. Cold Spring Harbor Laboratory Press; New York, USA: 1989. Molecular cloning: a laboratory manual. [Google Scholar]
- 43.Tang W. High-frequency plant regeneration via somatic embryogenesis and organogenesis and in vitro flowering of regenerated plantlets in Panax ginseng. Plant Cell Rep. 2000;19:727–732. doi: 10.1007/s002999900170. [DOI] [PubMed] [Google Scholar]
- 44.Tang W., Tian Y.C. Transgenic loblolly pine (Pinus taeda L.) plants expressing a modified delta-endotoxin gene of Bacillus thuringiensis with enhanced resistance to Dendrolimus punctatus Walker and Crypyothelea formosicola Staud. J. Exp. Bot. 2003;54:835–844. doi: 10.1093/jxb/erg071. [DOI] [PubMed] [Google Scholar]