Abstract
Although various genome projects have provided us enormous static sequence information, understanding of the sophisticated biology continues to require integrating the computational modeling, system analysis, technology development for experiments, and quantitative experiments all together to analyze the biology architecture on various levels, which is just the origin of systems biology subject. This review discusses the object, its characteristics, and research attentions in systems biology, and summarizes the analysis methods, experimental technologies, research developments, and so on in the four key fields of systems biology—systemic structures, dynamics, control methods, and design principles.
Key words: systems biology, computer simulation, genomics, dynamics
Introduction
The notion of system-level understanding in biology has a long history 1., 2., but has not received renewed main attention until recently. The cause to accelerate such a change is the expansion of genome sequencing. Since the first genome sequence of Haemophilus influenzae was published in 1995 (3), we have received many complete genome sequences, including Escherichia coli, Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, Arabidopsis thaliana, Oryza Sativa, and Homo Sapiens, which help us to obtain the whole static information of genes, regulatory regions, proteins, tRNAs, repeats and so on underlying DNA sequences—the ultimate depository. The development of other technologies also provides us a comprehensive dynamic data: DNA microarrays can practically measure mRNA expression responses of genes (4); technologies for globally and quantitatively measuring protein expression are also becoming feasible (5); and the two-hybrid system is enabling the construction of protein interaction maps (6). All of these developments make the biological research transfer from data-poor state to data-rich state, which provide us a golden opportunity to analyze the biology on the system level.
Biologists have found that many biological functions and diseases can’t be explained by the function of individual gene or protein. Instead, they should be the exhibition of an interactional network of protein-protein or proteins with other molecules 7., 8.. They also found that some particular characteristics of biological system could display perfect adaptation and homeostatic regulation despite great changes in the environment or alterations in the internal parameters of the system 9., 10.. Such robustness undoubtedly is the result of the organism’s evolution in a long time to adapt the environmental changes. To understand the magic biological functions and robustness of the system network, researchers are required to integrate the information of genome, mRNA expression, proteome, protein with protein or other molecule interactions, and so on from the system-level view, and to analyze the composition of biological system on various levels, interactions among modules, system dynamics, underlying control methods and design principles.
Systems biology is an integrated process of computational modeling, system analysis, laboratory technologies, and quantitative experiments, which analyzes systemic components, basic modules, interactions among components and modules, as well as control methods, design principles, and so on. The development in this subject will greatly accelerate our in-depth understanding to the complex biological phenomena, and significantly increase the efficiency of drug discovery 11., 12.. This review describes the object, its characteristics, and research attentions in systems biology, and summarizes the analysis methods, experimental technologies, research developments, and so on in the four key fields of systems biology—systemic structure, dynamics, control methods, and design principles, which are suggested by Hiroaki Kitano (13).
The Object of Systems Biology
The research object of systems biology is the whole biological system, including genes, RNAs, proteins, metabolics and cells in a narrow sense, and organs, organism and ecological system in a broad sense. A simple complexity pyramid (Figure 1; ref. 14) can summarize the components of biological system on different levels in a narrow sense.
Fig. 1.
The complexity pyramid of biological system by Oltvai and Barabasi (14). At the lowest level, the various molecular components of genes, RNAs, proteins, and metabolites form genetic-regulatory motifs or metabolic pathways (Level 2), which in turn are the building blocks of functional modules (Level 3). These modules are nested, generating a scale-free hierarchical architecture (Level 4).
Characteristics of Biological System
Complexity
Although Vicsek thinks that the concept of complexity is currently abused and “it’s almost an empty statement to say that a system is complex because almost every real system is inherently complicated” (15), the biological system is really complex, no matter from the diversity of the system’s components, the hierarchy of the network’s interactions, or the various biological functions displayed by the system, if we don’t emphasize the accuracy of the concept firstly. Carlson et al. compared the complexity between biological systems and their modern technological counterparts, such as central processing unit (CPU) and jet aircraft (16).
Robustness
Robustness refers to the preservation of particular characteristics despite uncertainty in components or the environment (17). Among many questions in systems biology, one issue receiving considerable attention is how robustness is achieved and how it evolves within various aspects of biological systems. For example, the precision of adaptation in the chemotaxis of E. coli is robust (9); Eldar et al. found that the activation gradient of bone morphogenic protein (BMP) of Drosophila was robust to changes in gene dosage (18), and the segment polarity network of Drosophila was also a robust development module (10). The biological system uses at least the following four methods to attain the robustness aspect: a form of system control, redundancy, structure stability, and modularity, which are also applied widely in the engineering system (13).
Evolution
Evolution is just the main difference between the biological system and the engineering one: the targets’ values or statuses are received initially on the engineering system design, whereas in biological system such targets are formed by evolution (19).
Key Fields in Systems Biology
For a biological system, the system-level understanding can be derived from insight into the following four key fields—system structure, dynamics, control methods and design principles. Process in any of the four fields requires breakthroughs in our understanding of computational sciences, genomics, measurement technologies, and integration of such discoveries with existing knowledge (13). The following will describe the analysis methods and developments in the four key fields of systems biology.
System structure
The research field of system structure determines the components, their relationships, and modules of the biological system, as well as builds the whole static network of system. It is the foundation of the whole systems biology research. Only when we have got an understanding of the system structure, can we then investigate the system dynamics, underlying control methods and design principles. In fact, the current research on system dynamics is mainly focusing on the structure-known systems.
1. Classifying the genome-scale genes and distributing them to various modules based on the similarity of gene expression profile, which can accelerate the determination of the biological network’s structure.
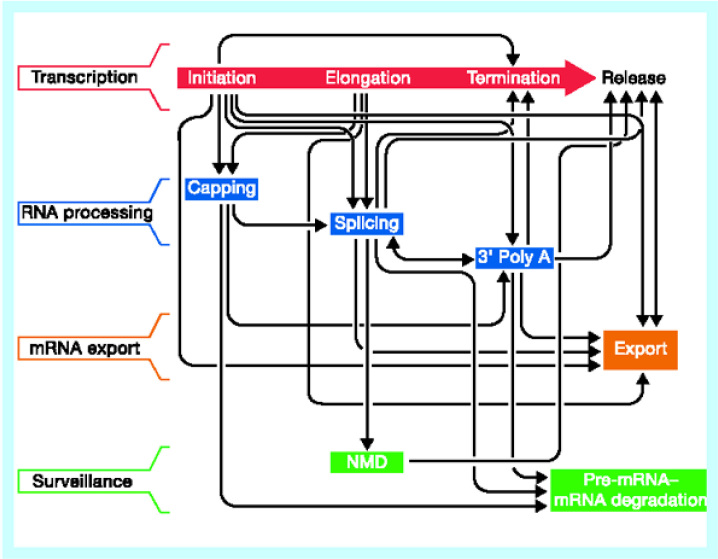
The foundation of such a method is that co-expressed genes are likely to encode proteins that participate in a common structural complex, metabolic pathway, or biological process 20., 21.. There have been many clustering algorithms to classify the large-scale expression profile data produced by DNA microarray 22., 23., 24., 25., 26., 27.. For the issues about the low accuracy and high error rate of repeat experiments in the data produced by DNA microarray 28., 29., 30., Kalir et al. measured promoter activity in the flagellar system of E. coli at high accuracy and temporal resolution by means of green fluorescent protein (GFP) reporter plasmid, and ordered the genes in a flagella pathway by analyzing expression kinetics (31). Extended research is to identify the cis-regulatory elements associated with the genes in the same group (32), and search the genome-wide distribution of such cis-regulatory elements to complement the genes in one module given by the DNA microarray profile data (33). Recent studies lead to the view that, in contrast to a simple linear assembly line, a complex and extensively coupled network has evolved to coordinate the activities of the gene expression machines (Figure 2; ref. 34).
Fig. 2.
A complex network of coupled interactions in eukaryotic gene expression by Maniatis and Reed (34).
The genes classifying method by expression similarity still has some shortages: all the biological processes are interlocked, and proteins may play multiple cellular roles, which may produce errors in the association between genes’ expression profiles and their functions. So Ker-Chau Li presented a co-expression dynamics theory to try to solve this issue (35).
2. Determining the proteins’ relationship to build a protein-protein interaction network.
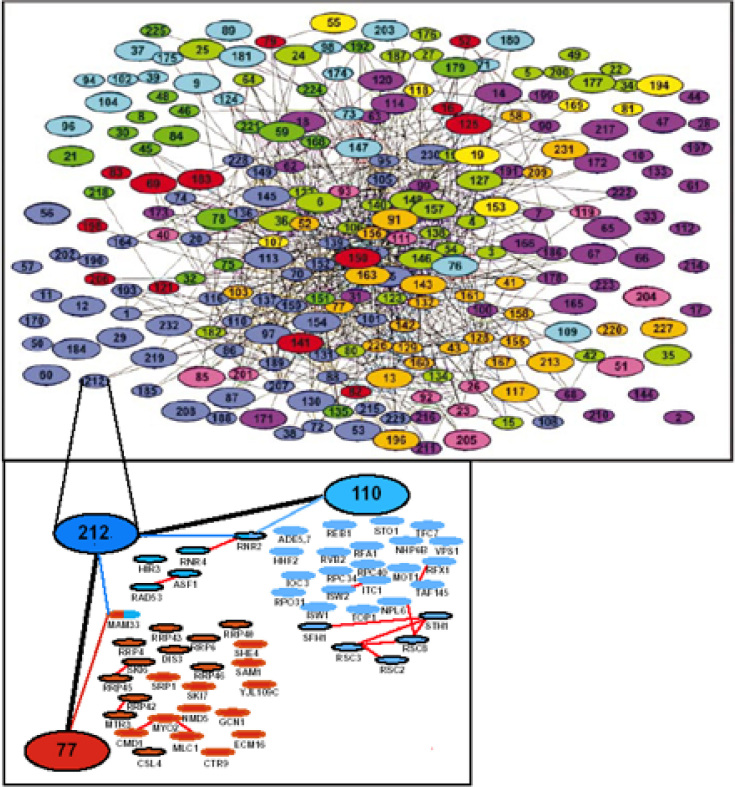
Protein interactions on a proteome-wide scale have already been analyzed in the methods of yeast two-hybrid assay 36., 37., protein chips (38), and so on. Yeast two-hybrid assay is a high-throughput method of mapping pair-wise protein interactions on a large scale, and has been applied in the construction of large-scale proteins interaction networks of S. cerevisiae 39., 40., C. elegans (41), D. melanogaster (42), etc. Gavin et al. (43) and Ho et al. (44) took a more effective approach to identify protein complexes that contain three or more proteins. They attached tags to hundreds of different proteins as bait proteins, and allowed such baits to be expressed in the cell and form physiological complexes with other proteins, then fished out the entire complex by extracting the tag on the bait protein. At the end, the proteins extracted with the tagged bait were identified using the standard mass-spectrometry method. Figure 3 represents the protein complex network built by Gavin et al. in such a way.
Fig. 3.
The yeast protein complex network, and grouping of connected complexes by Gavin et al. (43).
3. Distributing certain genes to some modules that govern the biological functions or phenotypes by measuring the phenotypic aspect of haploid deletion strains.
Jorgensen et al. obtained the five most potent genes and the corresponding complex network that govern the critical cell size in S. cerevisiae (45) by determining the cell size distributions for the complete set of ~6000 yeast gene deletion strains (46). This method is more directly than other ways to determine the system functional modules, for it directly measures the phenotypic aspects of certain gene deletion strains. Its shortages are that it can only be used to analyze the modules whose phenotypic characteristics are easy to measure, and it can’t consider the genes whose absence would lead to the cell’s death.
4. Analyzing the biological interaction network to distribute the proteins into different modules with graph theory, topological structure analysis, etc.
Depending on the more reliable 11,855 interactions among 2,617 yeast proteins evaluated by von Mering et al. (47), Bu et al. identified topological structures of such proteins’ interaction network (quasi-cliques and quasi-bipartites) to classify the proteins to different groups by a spectral analysis method. They found that the proteins within the same group share similar biological functions (48). Because interacting proteins usually localize in the same subcellular compartments (47), Rives et al. integrated the high-throughput yeast protein-protein interaction data 39., 40. and the corresponding data of protein localization in cellular compartments (49) to identify modules of yeast cellular networks by a network-clustering method, which was validated by clustering known signaling-protein modules (50).
System dynamics
The research field of system dynamics is to measure the components’ variety (for example, the mRNAs’ expression profiles) in a time scale or different conditions for a system with structure mainly known, choose certain mathematical model, list corresponding equations to describe the model, determine the dynamic parameters based on the kinetic data, and at last compare the prediction data with the experimental data, and if needed, revise the model. In fact, the dynamic analysis can also give useful predictions and complements to the system structure. For example, Ideker et al. integrated the methods of DNA microarrays, proteome, database searching and so on to analyze the pathway of yeast galactose utilization, and complemented some interactions to the metabolic network (51).
1. Choosing a model.
The choice of certain mathematical model is a key step for system dynamic analysis, which depends on the availability of the corresponding biology knowledge to compare with the scope and abstraction level of the model. With unknown rate constants for particular reactions, the constraints-based models can be adopted utilizing only the network structure information 52., 53., 54.. Because of the large number of unknown kinetic parameters and computational complexity, genome-scale models of metabolism or other biological systems have only been analyzed with the constrains-based modeling philosophy (55). With some known steady-state rate constants, many modeling approaches can be used to model cellular processes and to analyze how system behavior changes and investigate dynamic aspects. These approaches include kinetic 56., 57., stochastic 58., 59., and cybernetic ones 60., 61.. Bifurcation analysis can provide a detailed illustration of dynamic behavior 62., 63..
2. Determining the kinetic parameters.
After building the model and listing the corresponding equations, the next step is to specify the kinetic parameters for various regulations. We need to measure the system elements’ varying profiles, and give the prediction profiles by model simulations, then determine the kinetic parameters by minimizing the difference between the measured profiles and the predicted ones. Ronen et al. developed a system for real-time monitoring of the transcriptional activity of operons in the SOS DNA repair system of E. coli by means of low-copy reporter plasmid, in which a promoter controls the GFP, described the subsystem by the Michaelis-Menten model, and determined the corresponding dynamic parameters (64). The model for circadian oscillations in Drosophila was described by a set of ten kinetic equations 65., 66., 67., and Leloup et al. extended the rhythm model to mammals, which was governed by sixteen kinetic equations (68).
Control methods
1. Determining the central components, modules or aspects of the system on different levels.
After the system network is constructed, the next important analysis is to address the key components in the system, which can be the candidates for the drug targets, and the control methods, such as feedback and feedforward controls, to extend our understanding to the system. The quantitative analysis in the perturbation to protein interactions of S. cerevisiae has demonstrated that the most highly connected proteins in the cell are the most important for its survival (69). Stelling et al. utilized the elementary-mode analysis to simultaneously predict the key aspects of network functionality, robustness and gene regulation of E. coli central metabolism (70).
2. Exploring the relationship between the changes of system parameters or components and the variety of system’s phenotypes and functions.
This research is an important application field for systems biology, which explores the variety of system’s components or parameters and the organism’s phenotypes, analyses the corresponding control methods, and gives effective predictions. On the sub-cell level: Lev Bar-Or et al. modeled the oscillations of p53 and Mdm2 proteins by a mathematical model to describe the p53-Mdm2 feedback loop (71); Ideker et al. integrated genomic and proteomic methods to systematically perturb and analyze the yeast galactose utilization pathway (51); methods in computational biology contribute to clarify the molecular and dynamical bases of cellular rhythms (72); Davidson et al. gained some properties of the Strongylocentrotus purpuratus’s endomesoderm development process by analyzing the gene regulatory network, and tried to explain the developing phenotype (73). On the cell level: Jamshidi et al. developed a mathematical application package to perform dynamic simulations of the metabolic network of human red blood cell (74); the E-Cell project is developed to visualize and dynamically simulate the cell (75); and the Alliance for Cellular Signaling (AfCS) is systematically exploring the signaling networks in two types of mouse cells—B lymphocytes and cardiac muscle cells 76., 77., 78.. At the organ level, the whole organ of heart can be modeled to explore its functionality (79).
Design principles
The research field of design principles is to explore the principles of creating organisms by “God”, and is the highest level of systems biology. However, we haven’t uncovered the system-scale dynamics, let alone comprehensive control methods, so it is a long way to understand the system design principles underlying them. Presently there are the following ways applied in this field:
1. Exploring the similarity of system-level design between the engineering system and the biological one by the knowledge in our system design of modern engineering.
Csete et al. described insights from engineering theory and practice that can shed some light on biological complexity (17). Strogatz explored the topology of complex networks, such as food web, electric power grid, and regulatory network in cell (80).
2. Analyzing the system network to determine some system aspects and provide clues to clarify the system design principles in the end.
The interaction network of yeast proteins has been shown to have a nonrandom power-law distribution of node connectivity (number of interactions of each protein) and a low frequency of direct connections between high-connectivity nodes (81). Shen-Orr et al. found that much of the transcriptional regulation network of E. coli is composed of repeated appearances of three highly significant motifs, the feedforward loop, single input module and densely overlapping regulons, each of which has a specific function in determining the gene expression (82). Network motif was analyzed in complex networks, such as that of biochemistry, neurobiology, ecology, and engineering, to uncover their structural design principles (83).
Attentions to the Systems Biology Research
1. The comprehensiveness in measurements.
The high throughput, systemic and accurate measurements needed by systems biology research require the comprehensiveness in three aspects, that is, factor, time-line, and item. The factor comprehensiveness reflects almost all the mRNAs, proteins, and other components in the biological system. The time-line comprehensiveness represents the time frame within which measurements are made. And the item comprehensiveness refers to the simultaneous measurement of multiple items, such as mRNA and protein concentrations, phosphorylation, localization, and so forth (13).
2. The breakthroughs in experimental technology.
Completing the system-level analysis of biological regulation requires high throughput and exact measurements for goals that are perhaps beyond the scope of current experimental practices. Technical innovations in experimental devices, such as single-molecule measurements, femto-lasers that permit visualization of molecular interactions, and nano-technologies, are critical aspects of systems biology research (13).
3. The building of mathematical model.
When choosing certain model, it is important to firstly consider the purpose of model building: whether it is to obtain an in-depth understanding of system behavior or to predict complex behavior in response to complex stimuli, and we must first define the scope and abstraction level of the model (13).
4. The development of standard programming language to describe the biological models and the corresponding software.
It is very important to represent various biological models in a universal programming language for the information’s integration and the effective utilization of resource, which can benefit the researchers to exchange models between different model-building software, and enable them to reuse components from one model to another, thus accelerating model building. The Systems Biology Mark-up Language (SBML; ref. 84) and CellML (85) are trying to define a standard for an XML-based machine-readable model definition. The Systems Biology Workbench (SBW; ref. 86) built on SBML provides a framework of modular open-source software for systems biology research.
5. The development of corresponding databases.
There have built some databases concerned with biological pathways, interactions and so on, such as the Kyoto Encyclopedia of Genes and Genomes (KEGG; ref. 87), Alliance for Cellular Signaling (AfCS; ref. 76), Munich Information Center for Protein Sequences (MIPS; ref. 88), and Signal Transduction Knowledge Environment (STKE; ref. 89).
Conclusion
In the past tens of years, biologists have always tried to locate, clone, and express various genes, and to associate individual gene’s role with some biological functions or phenotypes, and have explained some system behavior by the characteristics of components, which in fact belong to the reductionism. These methods have produced many achievements and are still important, whereas during the progress of our research and the accumulation of genomic, proteomic and dynamic measuring data nowadays, we should exceed reductionism (90) and focus on the system-level research of biology, which includes the above fields of system structure, dynamics, control methods and design principles. Because the biological system is not just a simple assembly of genes, proteins and other molecules, many of its properties cannot be fully understood by merely listing the whole components of the system and drawing maps of their interactions. If molecular biology, genomics, and proteomics have provided us some knowledge of individual components’ roles and system behavior, and listed for us most components of the system, now it is time to put them back to the system, and analyze the systemic hierarchy, dynamics, control methods and design principles.
Acknowledgements
The author thanks the discussion with Prof. Bailin Hao in Institute of Theoretical Physics, China, the suggestions of Prof. Uri Alon in Weizmann Institute of Science, Israel, and the precious comments of the reviewers.
References
- 1.von Bertalanffy L. Oxford Univ. Press; Oxford, United Kingdom: 1933. Modern Theories of Development: An Introduction to Theoretical Biology. [Google Scholar]
- 2.Weiner N. MIT Press; Cambridge, United States: 1948. Cybernetics or Control and Communication in the Animal and the Machine. [Google Scholar]
- 3.Fleischmann R.D. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 4.Lander E.S. Array of hope. Nat. Genet. 1999;21:3–4. doi: 10.1038/4427. [DOI] [PubMed] [Google Scholar]
- 5.Gygi S.P. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 6.Schwikowski B. A network of protein-protein interactions in yeast. Nat. Biotechnol. 2000;18:1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- 7.Hartwell L.H. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 8.Eisenberg D. Protein function in the post-genomic era. Nature. 2000;405:823–826. doi: 10.1038/35015694. [DOI] [PubMed] [Google Scholar]
- 9.Alon U. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 10.von Dassow G. The segment polarity network is a robust developmental module. Nature. 2000;406:188–192. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- 11.Bailey J.E. Lessons from metabolic engineering for functional genomics and drug discovery. Nat. Biotechnol. 1999;17:616–618. doi: 10.1038/10794. [DOI] [PubMed] [Google Scholar]
- 12.Cascante M. Metabolic control analysis in drug discovery and disease. Nat. Biotechnol. 2002;20:243–249. doi: 10.1038/nbt0302-243. [DOI] [PubMed] [Google Scholar]
- 13.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 14.Oltvai Z.N., Barabasi A.L. Systems biology. Life’s complexity pyramid. Science. 2002;298:763–764. doi: 10.1126/science.1078563. [DOI] [PubMed] [Google Scholar]
- 15.Vicsek T. The bigger picture. Nature. 2002;418:131. doi: 10.1038/418131a. [DOI] [PubMed] [Google Scholar]
- 16.Carlson J.M., Doyle J. Complexity and robustness. Proc. Natl. Acad. Sci. USA. 2002;99:2538–2545. doi: 10.1073/pnas.012582499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Csete M.E., Doyle J.C. Reverse engineering of biological complexity. Science. 2002;295:1664–1669. doi: 10.1126/science.1069981. [DOI] [PubMed] [Google Scholar]
- 18.Eldar A. Robustness of the BMP morphogen gradient in Drosophila embryonic patterning. Nature. 2002;419:304–308. doi: 10.1038/nature01061. [DOI] [PubMed] [Google Scholar]
- 19.Kitano H. Computational systems biology. Nature. 2002;420:206–210. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 20.Eisen M.B. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcotte E.M. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 22.Alon U. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl. Acad. Sci. USA. 1999;96:6745–6750. doi: 10.1073/pnas.96.12.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ihmels J. Revealing modular organization in the yeast transcriptional network. Nat. Genet. 2002;31:370–377. doi: 10.1038/ng941. [DOI] [PubMed] [Google Scholar]
- 24.Tavazoie S. Systematic determination of genetic network architecture. Nat. Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 25.Hughes T.R. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 26.Alter O. Singular value decomposition for genome-wide expression data processing and modeling. Proc. Natl. Acad. Sci. USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown M.P. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc. Natl. Acad. Sci. USA. 2000;97:262–267. doi: 10.1073/pnas.97.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spellman P.T. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao R. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 30.Chu S. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 31.Kalir S. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science. 2001;292:2080–2083. doi: 10.1126/science.1058758. [DOI] [PubMed] [Google Scholar]
- 32.Hughes J.D. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- 33.Kurdistani S.K. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 2002;31:248–254. doi: 10.1038/ng907. [DOI] [PubMed] [Google Scholar]
- 34.Maniatis T., Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 35.Li K.C. Genome-wide coexpression dynamics: theory and application. Proc. Natl. Acad. Sci. USA. 2002;99:16875–16880. doi: 10.1073/pnas.252466999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fromont-Racine M. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 37.Phizicky E. Protein analysis on a proteomic scale. Nature. 2003;422:208–215. doi: 10.1038/nature01512. [DOI] [PubMed] [Google Scholar]
- 38.Zhu H. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 39.Uetz P. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 40.Ito T. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giot L. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 43.Gavin A.C. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 44.Ho Y. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 45.Jorgensen P. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- 46.Winzeler E.A. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 47.von Mering C. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 48.Bu D. Topological structure analysis of the protein-protein interaction network in budding yeast. Nucleic Acids Res. 2003;31:2443–2450. doi: 10.1093/nar/gkg340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar A. Subcellular localization of the yeast proteome. Genes Dev. 2002;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rives A.W., Galitski T. Modular organization of cellular networks. Proc. Natl. Acad. Sci. USA. 2003;100:1128–1133. doi: 10.1073/pnas.0237338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ideker T. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 52.Bailey J.E. Complex biology with no parameters. Nat. Biotechnol. 2001;19:503–504. doi: 10.1038/89204. [DOI] [PubMed] [Google Scholar]
- 53.Edwards J.S. Metabolic modelling of microbes: the flux-balance approach. Environ. Microbiol. 2002;4:133–140. doi: 10.1046/j.1462-2920.2002.00282.x. [DOI] [PubMed] [Google Scholar]
- 54.Edwards J.S. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat. Biotechnol. 2001;19:125–130. doi: 10.1038/84379. [DOI] [PubMed] [Google Scholar]
- 55.Price N.D. Genome-scale microbial in silico models: the constraints-based approach. Trends Biotechnol. 2003;21:162–169. doi: 10.1016/S0167-7799(03)00030-1. [DOI] [PubMed] [Google Scholar]
- 56.Tomita M. E-CELL: software environment for whole-cell simulation. Bioinformatics. 1999;15:72–84. doi: 10.1093/bioinformatics/15.1.72. [DOI] [PubMed] [Google Scholar]
- 57.Loew L.M., Schaff J.C. The Virtual Cell: a software environment for computational cell biology. Trends Biotechnol. 2001;19:401–406. doi: 10.1016/S0167-7799(01)01740-1. [DOI] [PubMed] [Google Scholar]
- 58.Arkin A. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics. 1998;149:1633–1648. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McAdams H.H., Arkin A. Simulation of prokaryotic genetic circuits. Annu. Rev. Biophys. Biomol. Struct. 1998;27:199–224. doi: 10.1146/annurev.biophys.27.1.199. [DOI] [PubMed] [Google Scholar]
- 60.Varner J., Ramkrishna D. Mathematical models of metabolic pathways. Curr. Opin. Biotechnol. 1999;10:146–150. doi: 10.1016/s0958-1669(99)80025-1. [DOI] [PubMed] [Google Scholar]
- 61.Guardia M.J. Cybernetic modeling and regulation of metabolic pathways in multiple steady states of hybridoma cells. Biotechnol. Prog. 2000;16:847–853. doi: 10.1021/bp000069a. [DOI] [PubMed] [Google Scholar]
- 62.Borisuk M.T., Tyson J.J. Bifurcation analysis of a model of mitotic control in frog eggs. J. Theor. Biol. 1998;195:69–85. doi: 10.1006/jtbi.1998.0781. [DOI] [PubMed] [Google Scholar]
- 63.Chen K.C. Kinetic analysis of a molecular model of the budding yeast cell cycle. Mol. Biol. Cell. 2000;11:369–391. doi: 10.1091/mbc.11.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ronen M. Assigning numbers to the arrows: parameterizing a gene regulation network by using accurate expression kinetics. Proc. Natl. Acad. Sci. USA. 2002;99:10555–10560. doi: 10.1073/pnas.152046799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leloup J.C., Goldbeter A. A model for circadian rhythms in Drosophila incorporating the formation of a complex between the PER and TIM proteins. J. Biol. Rhythms. 1998;13:70–87. doi: 10.1177/074873098128999934. [DOI] [PubMed] [Google Scholar]
- 66.Ueda H.R. Robust oscillations within the interlocked feedback model of Drosophila circadian rhythm. J. Theor. Biol. 2001;210:401–406. doi: 10.1006/jtbi.2000.2226. [DOI] [PubMed] [Google Scholar]
- 67.Smolen P. Modeling circadian oscillations with interlocking positive and negative feedback loops. J. Neurosci. 2001;21:6644–6656. doi: 10.1523/JNEUROSCI.21-17-06644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leloup J.C., Goldbeter A. Toward a detailed computational model for the mammalian circadian clock. Proc. Natl. Acad. Sci. USA. 2003;100:7051–7056. doi: 10.1073/pnas.1132112100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jeong H. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 70.Stelling J. Metabolic network structure determines key aspects of functionality and regulation. Nature. 2002;420:190–193. doi: 10.1038/nature01166. [DOI] [PubMed] [Google Scholar]
- 71.Lev Bar-Or R. Generation of oscillations by the p53-Mdm2 feedback loop: a theoretical and experimental study. Proc. Natl. Acad. Sci. USA. 2000;97:11250–11255. doi: 10.1073/pnas.210171597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goldbeter A. Computational approaches to cellular rhythms. Nature. 2002;420:238–245. doi: 10.1038/nature01259. [DOI] [PubMed] [Google Scholar]
- 73.Davidson E.H. Regulatory gene networks and the properties of the developmental process. Proc. Natl. Acad. Sci. USA. 2003;100:1475–1480. doi: 10.1073/pnas.0437746100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jamshidi N. Dynamic simulation of the human red blood cell metabolic network. Bioinformatics. 2001;17:286–287. doi: 10.1093/bioinformatics/17.3.286. [DOI] [PubMed] [Google Scholar]
- 75.Takahashi K. Computational challenges in cell simulation: a software engineering approach. IEEE Intelligent Systems. 2002;17:64–71. [Google Scholar]
- 76.Gilman A.G. Overview of the Alliance for Cellular Signaling. Nature. 2002;420:703–706. doi: 10.1038/nature01304. [DOI] [PubMed] [Google Scholar]
- 77.Sambrano G.R. Navigating the signalling network in mouse cardiac myocytes. Nature. 2002;420:712–714. doi: 10.1038/nature01306. [DOI] [PubMed] [Google Scholar]
- 78.Sambrano G.R. Unravelling the signaltransduction network in B lymphocytes. Nature. 2002;420:708–710. doi: 10.1038/nature01305. [DOI] [PubMed] [Google Scholar]
- 79.Noble D. Modeling the heart—from genes to cells to the whole organ. Science. 2002;295:1678–1682. doi: 10.1126/science.1069881. [DOI] [PubMed] [Google Scholar]
- 80.Strogatz S.H. Exploring complex networks. Nature. 2001;410:268–276. doi: 10.1038/35065725. [DOI] [PubMed] [Google Scholar]
- 81.Maslov S., Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- 82.Shen-Orr S.S. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 83.Milo R. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 84.Hucka M. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 85.Hedley W.J. A short introduction to CellML. Phil. Trans. R. Soc. Lond. A. 2001;359:1073–1089. [Google Scholar]
- 86.Hucka M. The ERATO Systems Biology Workbench: enabling interaction and exchange between software tools for computational biology. Pac. Symp. Biocomput. 2002:450–461. doi: 10.1142/9789812799623_0042. [DOI] [PubMed] [Google Scholar]
- 87.Kanehisa M. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mewes H.W. MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 2002;30:31–34. doi: 10.1093/nar/30.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gough N.R. Science’s signal transduction knowledge environment: the connections maps database. Ann. N. Y. Acad. Sci. 2002;971:585–587. doi: 10.1111/j.1749-6632.2002.tb04532.x. [DOI] [PubMed] [Google Scholar]
- 90.Gallagher R., Appenzeller T. Beyond reductionism. Science. 1999;284:79. [Google Scholar]