Abstract
A dataset of 103 SARS-CoV isolates (101 human patients and 2 palm civets) was investigated on different aspects of genome polymorphism and isolate classification. The number and the distribution of single nucleotide variations (SNVs) and insertions and deletions, with respect to a “profile”, were determined and discussed ("profile" being a sequence containing the most represented letter per position). Distribution of substitution categories per codon positions, as well as synonymous and non-synonymous substitutions in coding regions of annotated isolates, was determined, along with amino acid (a.a.) property changes. Similar analysis was performed for the spike (S) protein in all the isolates (55 of them being predicted for the first time). The ratio Ka/Ks confirmed that the S gene was subjected to the Darwinian selection during virus transmission from animals to humans. Isolates from the dataset were classified according to genome polymorphism and genotypes. Genome polymorphism yields to two groups, one with a small number of SNVs and another with a large number of SNVs, with up to four subgroups with respect to insertions and deletions. We identified three basic nine-locus genotypes: TTTT/TTCGG, CGCC/TTCAT, and TGCC/TTCGT, with four subgenotypes. Both classifications proposed are in accordance with the new insights into possible epidemiological spread, both in space and time.
Key words: SARS Coronavirus, single nucleotide polymorphism, insertions, deletions, spike protein, phylogenesis
Introduction
Severe acute respiratory syndrome (SARS), potentially fatal atypical pneumonia, first appeared in Guangdong province of China in November 2002 and soon afterward, within six months, spreaded all over the world (30 countries including China, Singapore, Vietnam, Canada, and USA), killing more than 700 people (1). In less than four weeks after the global outbreak, a novel member of Coronaviridae family, namely SARS Coronavirus (SARS-CoV), was identified in the blood of respiratory specimens and stools of SARS patients, and confirmed as the causative agent of disease according to the Koch postulates (2). Soon afterwards, first fully sequenced genomes of viral isolates were published 3., 4.. In 2005 the number of fully sequenced viral isolates exceeds one hundred (http://www.ncbi.nlm.nih.gov/entrez).
SARS-CoV probably originated due to genetic exchange (recombination) and/or mutations between viruses with different host specificities 5., 6.. Since coronaviruses are known to relatively easily jump among species, it was hypothesized that the new virus might have originated from wild animals. The analysis of SARS-CoV proteins supports and suggests possible past recombination event between mammalianlike and avian-like parent viruses (6). Common sequence variants define three distinct genotypes of the SARS-CoV: one linked with animal [palm civet (Paguma larvata)] SARS-like viruses and early human phase, the other two linked with middle and late human phases, respectively 7., 8.. SARS-CoV has a deleterious mutation of 29 nucleotides relative to the palm civet virus, indicating that if there was direct transmission, it went from civet to human, because deletions occur probably more easily than insertions (5). However, more recent reports indicate that SARS-CoV is distinct from the civet virus and it has not been answered so far whether the SARS-CoV originated from civet, or civet was infected from other species 9., 10.. The genome is relatively stable, since its mutation rate has been determined to be between 1.83×10−6 and 8.26×10−6 nucleotide substitutions per site per day (11).
The SARS-CoV genome is approximately 30 Kb positive single strand RNA that corresponds to polycistronic mRNA, consisting of 5’ and 3’ untranslated regions (UTRs), 13 to 15 open reading frames (ORFs), and about 10 intergenic regions (IGRs) 9., 12., 13.. Its genome includes genes encoding two replicate polyproteins (RNA-dependant-RNA-polymerase, i.e., pp 1a and pp 1ab), encompassing two-thirds of the genome, and a set of ORFs at 3’ end that code for four structural proteins: surface spike (S) glycoprotein (1,256 a.a.), envelope (E, 77 a.a.), matrix (M, 222 a.a.), and nucleocapsid (N, 423 a.a.) proteins. It also encodes for additional 8-9 predicted ORFs whose protein product functions are still under investigation (14; http://www.ncbi.nlm.nih.gov/entrez).
The S protein is the main surface antigen of the SARS-CoV and is involved in virus attachment on susceptible cells using mechanism similar to those of class I fusion proteins. The receptor for the SARS-CoV S protein is identified as angiotensin-converting enzyme 2 (ACE-2), which is a metallopeptidase (15). The receptor-binding domain (RBD) has been determined to lay between a.a. postions 270-625 in recent studies 16., 17., 18., 19., 20..
Several epitope sites, defined by polyclonal or monoclonal antibodies, have been identified on the S protein, depending on experimental conditions, all lying within wide or narrow regions between a.a. 12-1,192 20., 21., 22., 23., 24., 25., 26., 27., 28., 29., 30., 31.. Defining conserved immunodominant epitope regions of the S protein is of crucial importance for future anti-SARS vaccine development.
The main goal of this work was twofold: to perform mutation analysis of SARS-CoV viral genomes, with special attention to the S protein; and to group them according to different aspects of sequence similarity, eventually pointing to phylogeny and epidemiological dynamics of SARS-CoV.
Results and Discussion
Nucleotide content
Nucleotide content of SARS-CoV isolates favors T and A nucleotides. The corresponding percentages of letters in non-UTR regions of all the 96 isolates were found to be as follows: T (30.7940%), A (28.4246%), G (20.8121%), C (19.9535%), N (G, A, T, C; 0.0143%), R (Pur; 0.0005%), K (G or T; 0.0001%), M (A or C; 0.0002%), S (G or C; 0.0001%), W (A or T; 0.0002%), and Y (Pyr; 0.0004%). The overall ratio of (A,T)/(G,C) in the dataset was almost 3:2 (1.45). The ratio of Pur vs. Pyr nucleotides was almost 1 (0.97).
The distribution of nucleotides (nt) over sequences of length 250 nt is given in Figure S1 (Supporting Online Material). It exhibits three peak-regions of T nucleotide in the second quarter of the genome (ORF 1a), and rather stable behavior in the third quarter of the genome (ORF 1b), as also observed by Pyrc et al. (32) for a group of coronaviruses (HCoV-NL63, HCoV-229E, SARS-CoV, and HCoV-OC43). Deviation of percentage of nucleotides over 250-nt blocks from the corresponding percentage in the whole dataset is given in Figure S2. Except for 3’ UTR where T nucleotide is underrepresented with even about —13%, the highest excess from the average is about +10% in four peaks, which is exhibited again by T nucleotide, three of them being between positions 7,000 and 11,000 (ORF 1a), complementary with the nucleotide A represented with —10%, and the fourth one in the S protein. Otherwise the nucleotides’ offset oscillates rather regularly between —5% and +5% from the average.
Genome polymorphism
All the isolates had high degree of nucleotide identity (more than 99% pair wise). Still, they could be differentiated on the basis of their genome polymorphism, i.e., the number and sites of SNVs and insertions and deletions (INDELs). Analysis of genomic polymorphism of the isolates resulted in the following two facts (Tables 1, S1, and S2). Firstly, two isolates, HSR 1 and AS, coincided with the “profile” on all the “non-empty” positions (see Materials and Methods) up to the poly-A sequence. Secondly, three isolates had large number of undefined nucleotides (N), either as contiguous segments (Sin3408 in ORFs 8a, 8b; Sin3408L in ORF 1b), or as scattered individual nucleotides or short clusters (SinP2) (Table S2). Isolate Sin3408 was the only one that has a 34-nt longer 5’ UTR as compared with the “profile”. Thus these three isolates were not considered to be reliably compared with others.
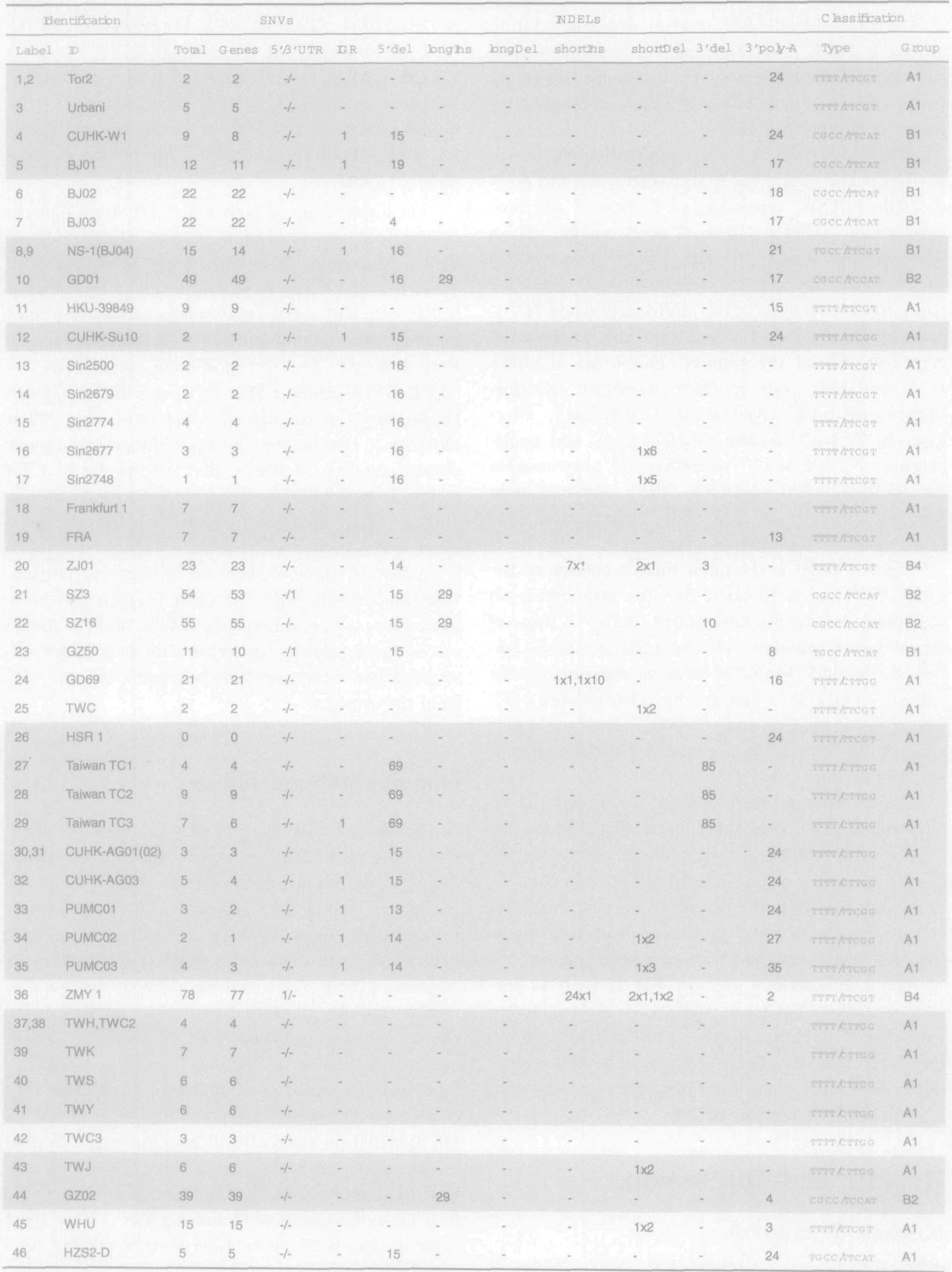
Table 1.
SARS-CoV Genome Polymorphism 20 Geno
Shaded entries correspond to annotated isolates. Identification (Label and ID) is given in accordance with the labels and identifiers from Table S1. The four SNVs columns correspond to: the total number of SNVs, the number of SNVs in genes, in 5’ and 3’ UTRs, and in IGR. The seven columns named INDELs include the number of deletions at the 5’ end (5’ del), the length of long insertions (longIns) and long deletions (longDel), the number and length of short insertions (shortIns) and short deletions (shortDel) in the form a × b where b denotes the length and a denotes the number of occurrences, the number of deletions at the 3’ end (3’ del), and the length of a poly-A sequence at the 3’ end (3’ poly-A). Classification includes two columns. The Type column corresponds to the nine-locus nucleotides that are given in the form NNNN/NNNNN and represent nucleotides at (relative to CLUSTAL X output) positions 9,420, 17,604, 222,274, 27,891 / 3,861, 9,495, 11,514, 21,773, 26,534, respectively (absolute HSR 1 positions 9,404, 17,564, 22,222, 27,827 / 3,852, 9,479, 11,493, 21,721, 26,477). The last column, Group, reflects grouping of isolates.
Nucleotide variations: single nucleotide polymorphism
There were 446 SNV sites and 1,006 SNVs in total in the dataset, with the substitution rate 1.49%, which is about three times higher (both the number of SNVs and the substitution rate) than the corresponding findings (33) for 17 isolates. An average number of SNVs per isolate was 10.48, giving an error rate of 3.6×10−4 substitutions per nucleotide copied.
There was only one site with multiple base substitutions (the original nucleotide base on that position being T): at the relative (CLUSTAL X) position 8,441 (ORF 1a), isolate ZMY 1 has the nucleotide C (absolute position 8,403), and isolates ShanghaiQXC1, ShanghaiQXC2 have the nucleotide A (absolute positions 8,312 and 7,733, respectively).
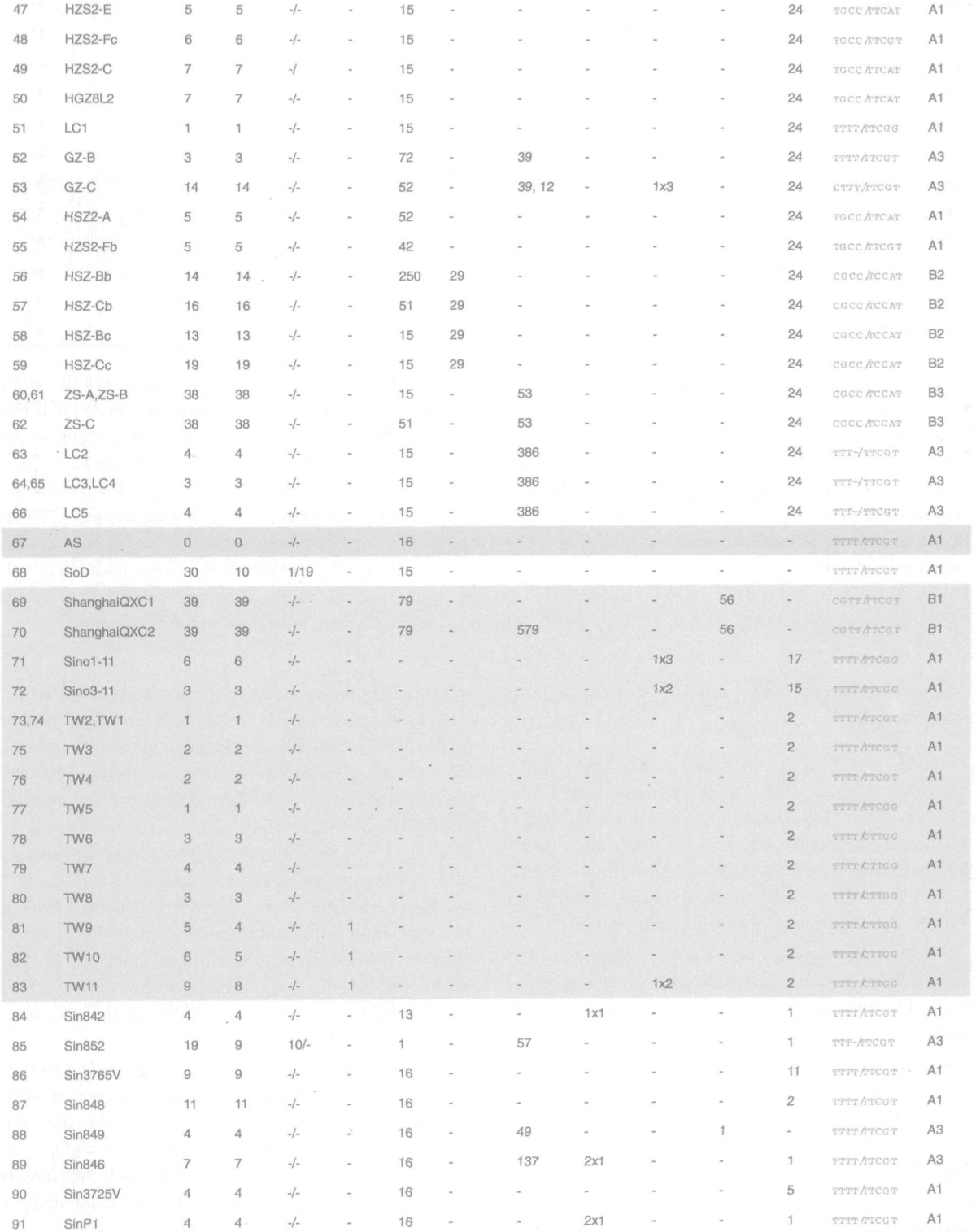
The smallest distance between the two neighboring SNV sites in the whole dataset was 1; the largest one was 23,988 (in case of TW3 and TW1), while an average distance between the neighboring SNV sites in the whole dataset was 1,987 positions (Figure S3). The distribution of isolates per SNV number (outside 5’, 3’ UTRs) showed regularity for up to 11 SNVs (almost Gaussian distribution) and irregular decrease for number of SNVs >11 (Figure S4). Thus the number of SNVs less than or equal to 11 per isolate was considered as a “small” number of SNVs, and the number of SNVs greater than 11 was considered as a “large” number of SNVs. Most SNVs are clustered within two regions in ORF la and one region at the 3’ end of the viral genome that predominantly consists of small ORFs, leaving two small regions within ORF 1a, and a region that corresponds to ORF 1b as the most conservative ones (Figure 1B).
Fig. 1.
Density distribution of SNVs (B), INDELs (C), mapped onto the gene map of the HSR 1 isolate, coinciding with the “profile” (A). Central region of the genome is rather conserved (lower density of SNVs is exhibited in the second third of the genome, ORF 1b), while the rest of the genome features high SNVs density. SNV peaks are present at (absolute HSR 1) positions 3,852, 9,404, 9,479, 11,493, 17,564 (ORF lab), 21,721, 22,222 (S protein), 26,477 (M protein), and 27,827 (ORF 8a).
The entropy of each genome nucleotide position was calculated, showing that the most conserved sites are the ones with the smallest entropy and that the least conserved sites are the ones with the highest entropy (34; Figure S5). The nine loci used for classification can be found among the sites with the highest entropy.
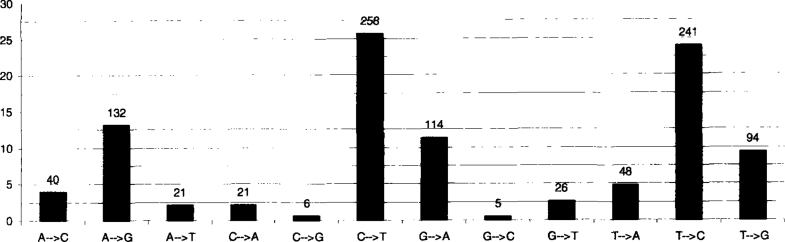
Percent of each category of substitution is given in Figure 2. There are 141 transversion sites and 306 transition sites, i.e., 31.54%:68.46%, with 261 transversions (2.72 in average per isolate) and 745 transitions (7.76 in average per isolate).
Fig. 2.
Distribution of nucleotide substitution categories. The most represented are the substitutions C↔T and the least represented are the substitutions C↔G.
Length variations, insertions and deletions
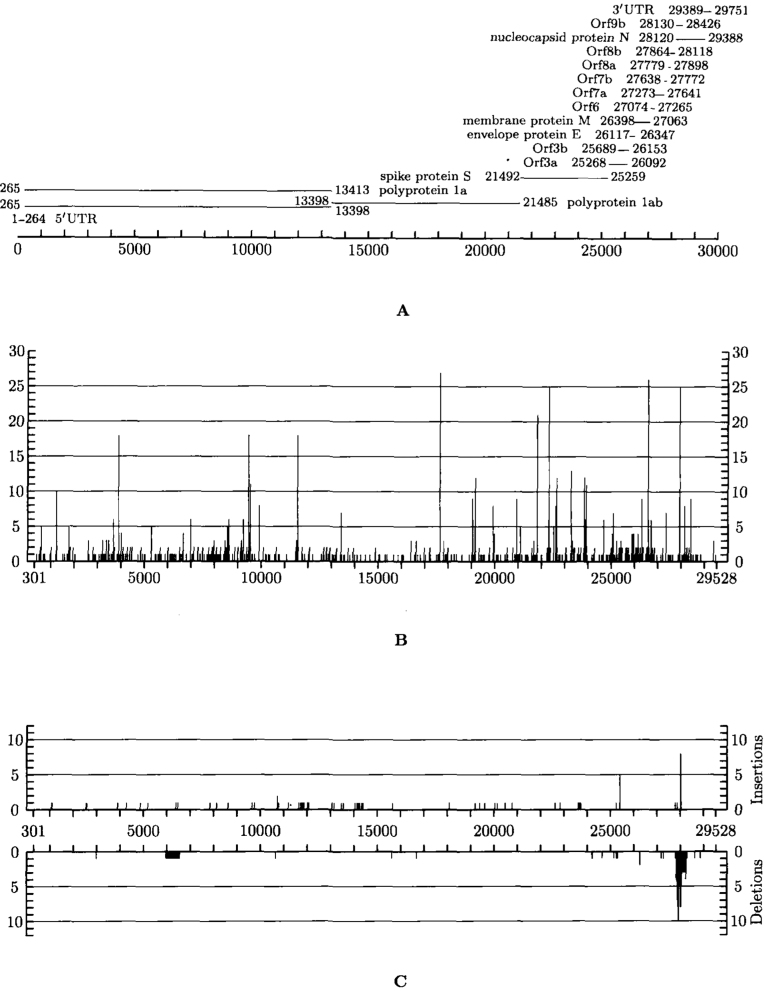
Analysis of the SARS-CoV genome showed that long INDELs were concentrated close to the 3’ end (except for the 579-nt deletion in the ShanghaiQXC2 isolate at the position 5,834, located in ORF 1a), while individual insertions were found along the whole genome, most in the second quarter, and individual deletions were quite rare. Density distribution of INDELs inside the SARS-CoV genome, and 5’ UTR, 3’ UTR length variations, are represented in Figures 1C, S6A and B, respectively. Figure S6C represents the region of the genome between positions 27,700 and 28,300 (ORFs 7b, 8a, 8b, part of N-protein, in HSR 1 annotation), which is especially abundant with INDELs. While individual INDELs are present both in longer and shorter ORFs, longer INDELs are (except for previously mentioned deletions in the ShanghaiQXC2) all located in short ORFs.
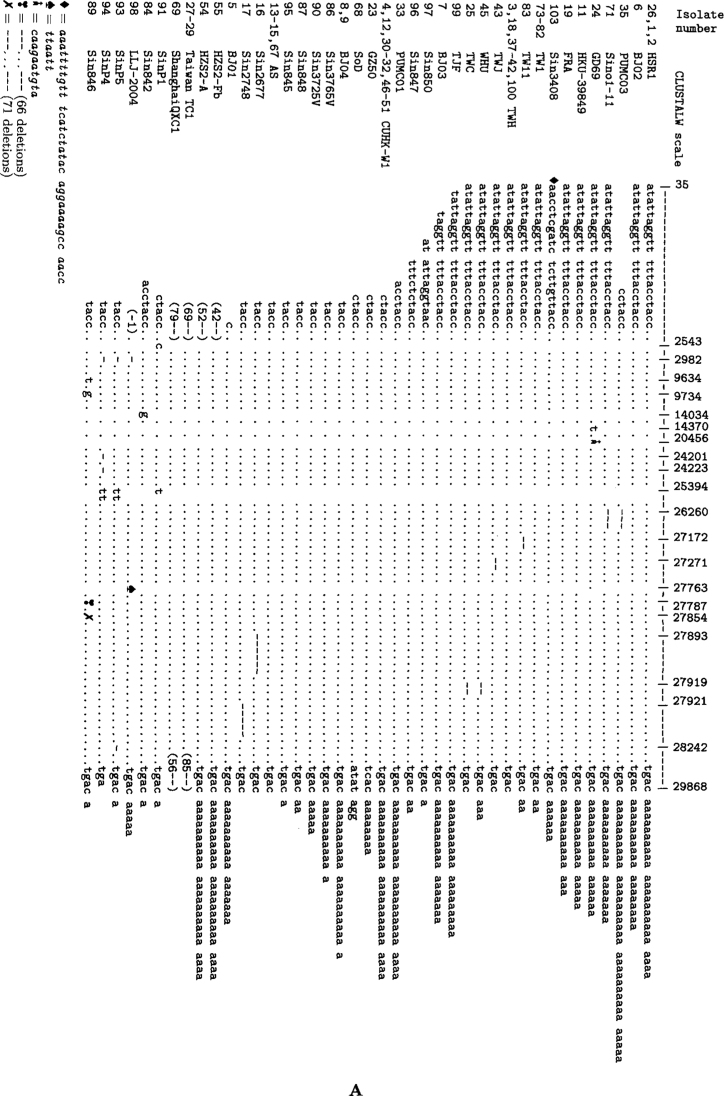
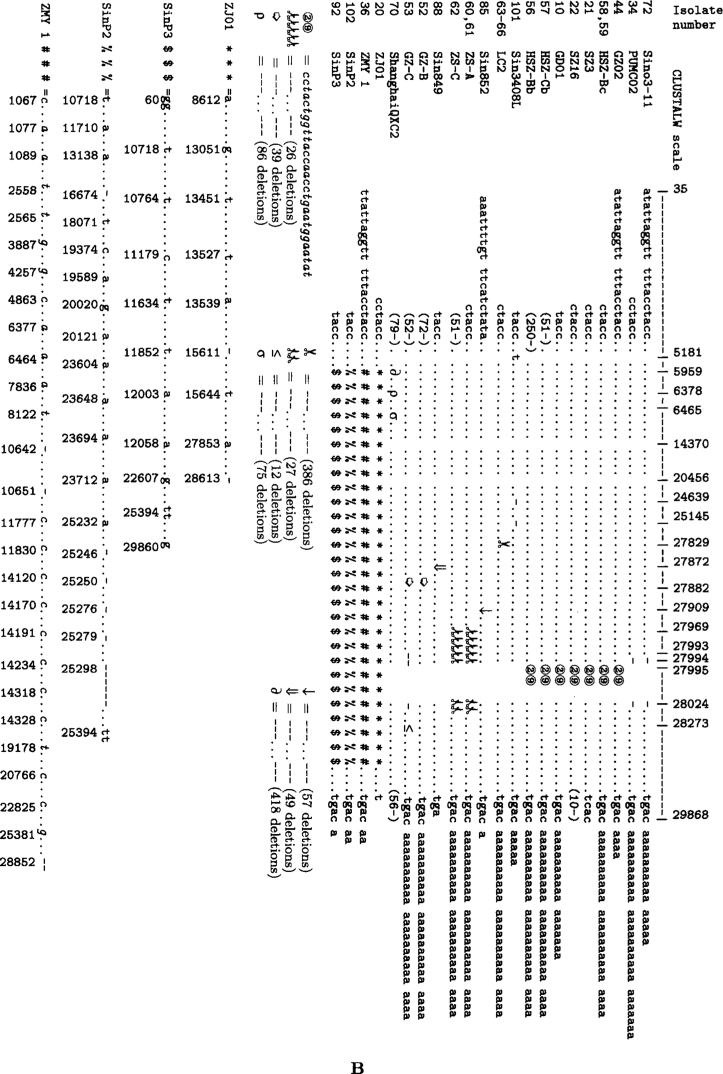
Figure 3 represents comparison results of genome primary structure of the analyzed isolates, summarizing the following facts:
Fig. 3.
Comparison of nucleotide structures of SARS-CoV complete genome isolates, represented in parts A and B of the figure according to similarity in their SNVs or INDELs positions.
Firstly, although the SARS-CoV genome has the established length of 29,727 nt (12), most isolates were shorter at the 5’ end (for the first 15 positions, majority of isolates were “empty”), and had various length “poly-A” strings at the 3’ end, or both (Table 1). Several isolates had some short deletions inside the sequence, e.g., Sin2677, Sin2748, TWC, PUMC02, PUMC03, TWJ, WHU, Sino1-11, Sino3-11, TW11, and SinP5.
Secondly, there was a group of isolates that had insertions of length 29 nucleotides (GD01, SZ3, SZ16, GZ02, HSZ-Bb, HSZ-Bc, HSZ-Cb, and HSZ-Cc) at the relative position 27,995 (absolute position 27,869 in SZ3, SZ16, HSZ-Cc, and HSZ-Bc; protein BGI-PUP GZ29-nt-Ins, ORF 8a). Two of them were isolates from palm civet (SZ3 and SZ16) and the other six were isolates from human patients. This specific insertion is also treated as a deletion in all the other isolates, evolved from this early group (10).
Thirdly, there were several groups of isolates that had long deletions: GZ-B, GZ-C (length 39 at the relative position 27,882, or absolute position 27,719 in GZ-C, ORF 7b), ZS-A, ZS-C (length 53 at the relative position 27,969, absolute 27,843 in ZS-A, ORF 8a), LC2, LC3, LC5 (length 386 at the relative position 27,829, absolute 27,704 in LC2, ORFs 7b, 8a, 8b), ShanghaiQXC2 (length 579 at the relative position 5959, absolute 5834, ORF la), Sin852, Sin849, and Sin846 (of length 57, 49, 137, respectively, at relative positions in region between 27,787 and 27,966, ORFs 7b, 8a) (Tables 1 and S2, Figure 3).
Fourthly, a large number of individual INDELs were identified in ZJ01, ZMY 1, SinP2, and SinP3 (Tables 1 and S2, Figure 3).
Mutation analysis
While the distribution of nucleotides over different distances from SNV sites did not exhibit any regularities, the distribution of different nucleotides on distance 1 left to SNV sites (−1) did exhibit significant difference from their overall percentage in the dataset. The corresponding right (+1) distance distribution of nucleotides is almost uniform (Table 2).
Table 2.
Distribution of Nucleotides on Distance 1 Left and Right to SNV Sites
| Nt | (−1)num | (−1)% | (−1)diff% | (+1)num | (+1)% | (+1)diff% |
|---|---|---|---|---|---|---|
| A | 358 | 35.59% | 7.17% | 283 | 28.13% | −0.29% |
| C | 179 | 17.79% | −2.16% | 203 | 20.18% | 0.23% |
| G | 230 | 22.86% | 2.05% | 215 | 21.37% | 0.56% |
| Τ | 238 | 23.66% | −7.13% | 302 | 30.02% | −0.77% |
The distribution of nucleotides on distance 1 left to SNV sites (−1) and right to SNV sites (+1) is presented in total number of nucleotides, percentage, and difference from their overall percentage in the dataset.
Figure S7 represents differences between the percentage of nucleotides at a given position and in the whole genome, for up to the distance 10 left and right from SNV sites. Figures S8A and B represent distribution of substitutions preceded by different nucleotide bases, and followed by different nucleotide bases, respectively. It can be seen that on the C↔T substitutions, both C→T and T→C are favored by the preceding A and the following Τ (almost 40% of all the C↔T substitutions; Figure 2), while the substitution T→C is almost prohibited by the preceding T (only 3%). Clustered substitutions of length 2 are rare (e.g., TC→AA, GA→CA).
Codon usage
Analysis of distribution of individual nucleotides over the three codon positions in annotated ORFs of all the annotated isolates showed that, except for short proteins such as E, M, and presumptive ORFs, all the codons exhibit the same tendency of T nucleotide dominating at the third codon position, and the G nucleotide dominating at the first codon position, while A and C appearing more often at the second codon position than elsewhere. Figure S9 represents distribution of nucleotides over the three codon positions in individual ORFs, and in total.
Analysis of codon usage demonstrated the same facts as the distribution of nucleotides over the three codon positions. In total, the third nucleotide favored T (40.10%) over A (24.83%), C (18.90%), and G (16.16%). It was especially true for four-codon families a.a. (Thr, Pro, Ala, Gly, and Val). The same held for four-codon subsets of six-codon families (Arg, Leu, and Ser), differring at the third codon position only. The above was true for the ORF lab, S and N proteins, but not for another two structural proteins (E and M). The codon usage for SARS-CoV genome proteins is represented by Table S3, and it is consistent with the results obtained for another human CoV genome, HCoV-NL63 (32).
Changes in amino acids
Besides the number of SNVs, isolates differed in positions of SNVs, too. Table S4 represents positions where two or more SNVs occurred, for all the annotated isolates, along with nucleotides and ORFs (based on HSR 1 annotation), type of mutation (transition/transversion), a.a. position in ORF, a.a. change, a.a. property change, and nucleotide position in codon. Positions of multiple SNVs have been chosen in order to reduce the chance of erroneously determined SNV. There were 91 such SNV sites with 288 SNVs. It is interesting to notice that there were no multiple base substitutions (more than two different bases) in any of these positions. There were 227 transitions at 75 sites and 61 transversion at 16 sites, out of which 5 were in structural proteins: 2 in S, 2 in E, and 1 in M proteins. The most common mutation was C↔T mutation (45 sites or 50%), followed by A↔G (30 sites), A↔T and G↔T (7 sites each), and C↔A (2 sites). There was no mutation of the type C↔G.
There were 28 SNV sites corresponding to the first codon position (20 transitions and 8 transversions), 2 of which representing silent mutation sites (C↔T, Leu). There were 33 SNV sites corresponding to the second codon position (31 transitions and 2 transversions), all of which cause a.a. change. There were 30 SNV sites corresponding to the third codon position (25 transitions and 5 transversions), 29 of which representing silent mutation sites (the only non-silent one is G↔T, Leu ↔ Phe).
There were 31 synonymous multiple substitution sites and 60 non-synonymous ones, with substitution rate 0.31% (91/29,228) and non-synonymous substitution rate 0.21%, which is consistent with the corresponding findings for 17 SASR-CoV isolates (33). The number of multiple substitutions was for about 30% lower than the number of the overall substitutions, and so were the substitution rate and non-synonymous substitution rate.
Table S5 summarizes the above findings. It represents the number of transition and transversion sites and the number of SNVs (in the form N1/N2) per position in codon and per mutation type, as well as the percentage of SNVs, and the number of silent mutation sites and silent SNVs.
Concerning non-synonymous sites, 35 are within pp lab, 5 within ORF 3, 1 within E protein, 3 within M protein, 1 within ORF 6, 1 within ORF 8a, 1 within ORF 8b, 1 within N protein, and 11 within S protein (only for two-or-more substitution sites, and only in annotated isolates).
Mutation analysis of the S protein
The S protein is of particular interest for mutation analysis, being the key for host range determination. Multiple sequence alignment of the S protein in all the 96 SARS-CoV isolates showed that five of them, namely ZMY 1, SinP2, SinP3, SinP4, and Sin3408L, had large discrepancies with all the others due to individual insertions or deletions in them. Since such significant mismatches in the S protein sequence seemed to be the result of erroneous sequencing, we eliminated these five isolates and analyzed the S protein in the remaining 91 isolates.
There were 34 isolates without SNVs in the S protein: TW2-TW11, Sino3-ll, AS, LC1, WHU, TWC3, PUMC01-PUMC03, CUHK-AG01, CUHK-AG3, Taiwan TC1-3, TWC, Sin2748, Sin2500, Sin2677, CUHK-Su10, HKU-39849, TWH, TWJ, TWK, TWY, and HSR 1. There were 62 SNV sites with 208 SNVs in total, and no multiple mutations. Table S6 represents SNV sites and all the SNVs in the S protein of the 91 isolates, along with nucleotides, type of mutation (transition/transversion), a.a. position in the protein, a.a. change, a.a. properties change, nucleotide position in codon, and number of SNVs at each SNV site. These findings overlap with the results reported in Song et al. (Ref. 7; concerning SNVs with multiple occurrences, in 103 S protein genes, some of which being nucleotide-identical, with 80% in common with our dataset), and are consistent on the intersecting data. Table 3 summarizes the results from Table S6.
Table 3.
Mutation Analysis of the S Protein: Categories of Nucleotide Substitutions
| 1.pos | 2.pos | 3.pos | Total No. | 1.pos% | 2.pos% | 3.pos% | Total% | Silent | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transitions | A-G | A→G | 6/15 | 2/8 | 3/20 | 11/43 | 16/73 | 7.21% | 3.85% | 9.62% | 20.68% | 3/20 |
| G→A | 2/7 | 2/22 | 1/1 | 5/30 | 3.37% | 10.58% | 0.48% | 14.43% | 1/1 | |||
| C-T | C→T | 3/7 | 6/25 | 6/19 | 15/51 | 24/94 |
3.37% | 12.02% | 9.13% | 24.52% | 6/19 | |
| T→C |
2/3 |
3/29 |
4/11 |
9/43 |
1.44% |
13.94% |
5.29% |
20.67% |
4/11 |
|||
| Total | 13/32 | 13/84 | 14/51 | 40/167 | 15.38% | 40.38% | 24.52% | 80.28% | 14/51 | |||
| Transversions | A→C | A→C | 2/2 | 1/1 | 2/2 | 5/5 | 8/10 | 0.96% | 0.48% | 0.96% | 2.40% | 2/2 |
| C→A | 1/1 | 1/2 | 1/2 | 3/5 | 0.48% | 0.96% | 0.96% | 2.40% | 0 | |||
| A-T | A→T | 0 | 0 | 0 | 0 | 6/7 | 0 | 0 | 0 | 0 | 0 | |
| T→A | 2/2 | 1/1 | 3/4 | 6/7 | 0.96% | 0.48% | 1.92% | 3.37% | 1/1 | |||
| G-C | G→C | 1/1 | 0 | 0 | 1/1 | 3/5 | 0.48% | 0 | 0 | 0.48% | 0 | |
| C→G | 0 | 2/4 | 0 | 2/4 | 0 | 1.92% | 0 | 1.92% | 0 | |||
| G-T | G→T | 0 | 1/1 | 1/1 | 2/2 | 5/19 |
0 | 0.48% | 0.48% | 0.96% | 1/1 | |
| T→G |
2/14 |
0 |
1/3 |
3/17 |
6.73% |
0 |
1.44% |
8.17% |
1/3 |
|||
| Total | 8/20 | 6/9 | 8/12 | 22/41 | 9.62% | 4.33% | 5.77% | 19.72% | 5/7 | |||
| Total | 21/52 | 19/93 | 22/63 | 62/208 | 25.00% | 44.71% | 30.29% | 100% | 19/58 | |||
S proteins in 91 isolates are considered. The number of transition and transversion sites and the number of SNVs (in the form N1/N2) per position in codon and per mutation type, as well as the percentage of SNVs, and the number of silent mutation sites and silent SNVs (in the form N1/N2), are presented.
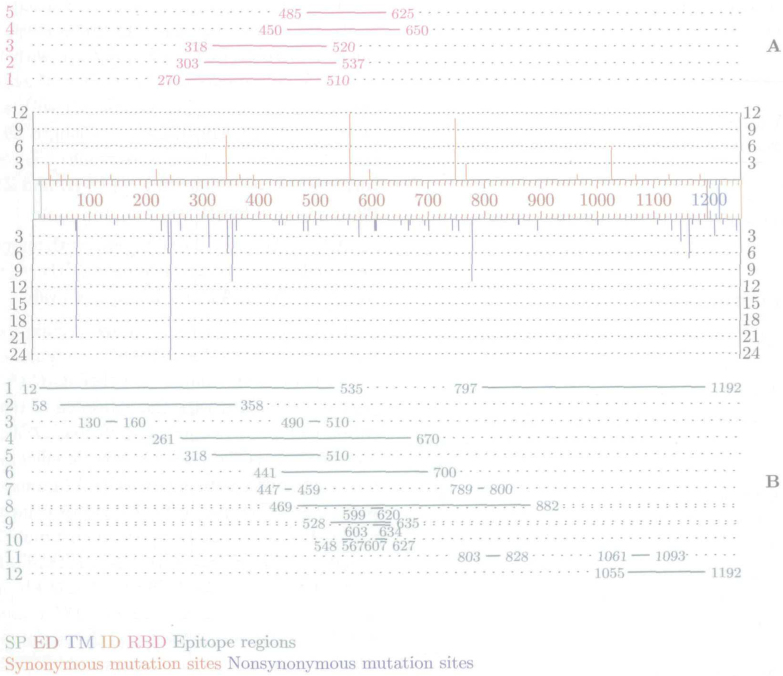
Out of 62 SNV sites, 19 were observed to be synonymous, with 58 synonymous SNVs in total, and 43 were observed to be non-synonymous substitution sites, with 150 non-synonymous SNVs in total (Table S6). Substitution rate was 1.65% (62/3,768) and non-synonymous substitution rate was 1.14% (43/3,768), which is consistent with findings for the whole genome in the enlarged dataset, and is about three times higher than the corresponding findings for 17 isolates in Bi et al. (Ref. 33; 22 substitution sites, 13 non-synonymous, substitution rates 0.58, 0.35, respectively). As represented on Figure 4, most non-synonymous a.a. substitutions are located in external domain (ED); 14 of non-synonymous substitution are in RBD, 3 of them in the most narrow intersecting range. As it concerns epitopes, 40 of non-synonymous a.a. substitutions are located in overall epitope domains determined by various researchers. Finally, one non-synonymous a.a. substitution is located in transmembrane domain (TM) and two in internal domain (ID).
Fig. 4.
Positions of synonymous and non-synonymous a.a. substitutions plotted against S protein primary structure. The y-axis represents number of SNVs per positions. SP, signal peptide; ED, external domain; TM, trans-membrane domain; and ID, internal domain (http://expasy.org/). A. RBD determined by: 1. Babcock et al. (16), 2. Xiao et al. (17), 3. Wong et al. (18), 4. Zhao et al. (19), and 5. Zhou et al. (20); B. epitope regions determined by: 1. Wang et al. (31), 2. Chou et al. (29), 3. Greenough et al. (30), 4. Sui et al. (26), 5. van den Brink et al. (28), 6. Lu et al. (24), 7. Hua et al. (23), 8. Ren et al. (21), 9. He et al. (22), 10. Zhou et al. (20), 11. Zhang et al. (27), and 12. Keng et al. (25).
The coefficients Ks (number of synonymous substitutions per synonymous site) and Ka (number of non-synonymous substitutions per non-synonymous site) were calculated for all the 91 S proteins in the dataset to be Ks = 0.00135, Ka = 0.00103, and the ratio Ka/Ks had the value 0.7629 < 1, which may be interpreted as evidence for the S protein not being subjected to the Darwinian selection. These findings are consistent with the similar analysis performed for 20 SARS-CoV isolates by Hu et al. (35) giving the ratio value of 0.65. Values of the corresponding coefficients and the ratio Ka/Ks for the 89 human patient isolates only, present even stronger evidence of the S protein being subject to negative selection: Ks = 0.00121, Ka = 0.00080, Ka/Ks = 0.661. The coefficients Ka, Ks, and the Ka/Ks ratio for all the human patients’ isolates and each of the palm civet isolates as the outgroup, are represented in Table 4. These values indicated that the S gene was subjected to the Darwinian selection during virus evolution (transmission from animals to humans), which is consistent with the analysis performed by Yeh et al. (36), for 28 human isolates and the SZ3 palm civet as the outgroup, giving the corresponding ratio value of 1.657, and with the analysis performed by He et al. (8), indicating that the S gene showed the strongest positive selection pressures initially, with eventual stabilization.
Table 4.
Mutation Analysis of the S Protein: Coefficients Ka, Ks, and the Ratio Ka/Ks with An Outgroup
| Outgroup | Ka | Ks | Ka/Ks |
|---|---|---|---|
| SZ16 (AY304488) | 0.006257 | 0.004930 | 1.26935>M |
| SZ3 (AY304486) | 0.005889 | 0.003803 | 1.54856>1 |
Coefficients Ka, Ks are calculated for all the human patients’ isolates and one of the palm civet isolates as an outgroup.
Phylogenetic analysis
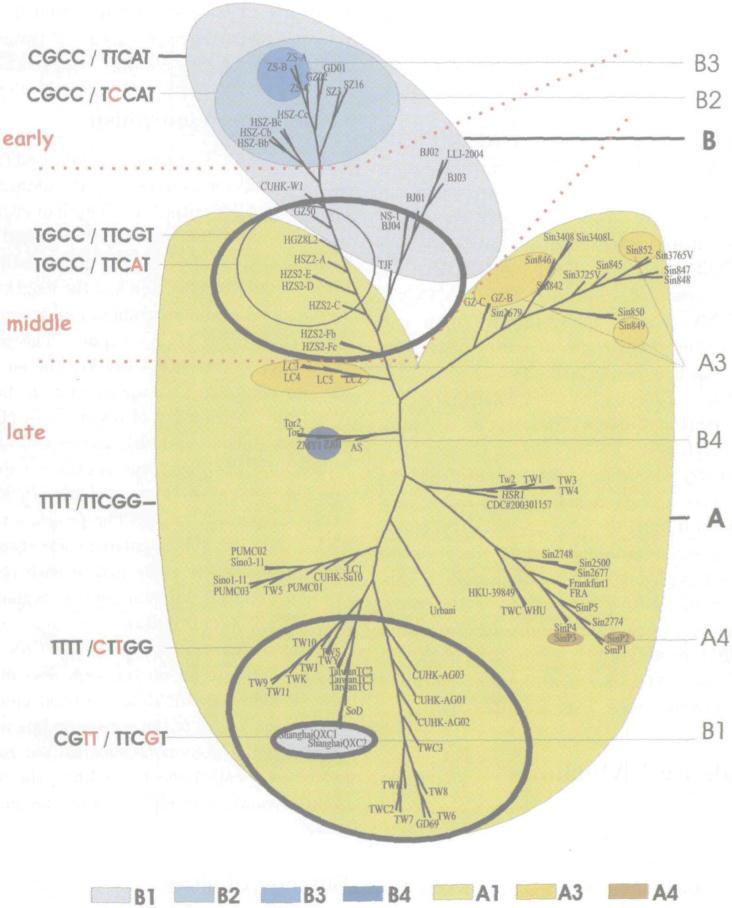
Phylogenetic tree, drawn using the PhyloDraw program for the CLUSTAL X output of aligning the 96 isolates, is represented in Figure 5. Its close relationship to the classification proposed in the paper suggests that classification of SARS-CoV isolates might be obtained by applying the computational analysis based on genome polymorphism.
Fig. 5.
Three-level classification of 103 SARS-CoV genome isolates. Grouping of isolates is based on genome polymorphism, and classification is based on nine distinguished loci, mapped onto the bootstrapped phylogenetic tree obtained using CLUSTAL X and Neighbor Joining method, and drawn using PhyloDraw programs. Bootstrapping is performed with random number generated seed 111 and number of trials in bootstrap 1000. The two basic groups, A and B, are represented in yellow and blue, respectively. Types obtained according to the nine genome loci (9,404, 17,564, 22,222, 27,827 / 3,852, 9,479, 11,493, 21,721, 26,477) are labeled along the left edge of the figure and have the form NNNN / NNNNN, where N represents any nucleotide. Different subtypes are denoted by the corresponding substituted nucleotides in red. Dotted lines distinguish between the three epidemiological phases.
All the isolates were classified according to their genome polymorphism—SNVs and INDELs, the procedure being proposed in our previous paper (57). Since SNV contents turned out to be a more distinguishable property than the presence of INDELs, as the first classification criterion we took the number and positions of SNVs. For the “profile” isolate, as the referent isolate, number of SNVs for different isolates varied from 0 (HSR 1, AS) to 78 (ZMY 1) (Table 1). All the isolates were classified into two groups based on the number of SNVs with the “profile”—those with less than or equal to 11 SNVs, and those having more than 11 SNVs. Thus, the first classification criterion resulted in two groups (Table 1):
Group A—isolates with less than or equal to 11 SNVs (Tables 1 and S2);
Group B—isolates with more than 11 SNVs relative to the “profile” isolate.
Positions of SNVs moved several isolates between the two groups (SoD from B to A, since the most of its SNVs are in 3’ UTR; CUHK-W1 from A to B, since its number of SNVs with the “profile” of the A group is larger than the one of the B group; WHU from B to A; GZ50 from A to B; GD69 from B to A; and GZ-C from B to A).
The second classification criterion was presence and position of long INDELs inside the basic A, B groups. We identified the following subgroups:
A2, B2—subgroups of the A, B groups, respectively, with long insertions. A2 remained empty, while B2 contained 8 isolates with 29-nt insertions.
A3, B3—subgroups of the A, B groups, respectively, with long deletions. A3 group consisted of 3-isolate subgroup (LC2, LC3, and LC5) with a deletion of length 386, Sin852 with a deletion of length 57, 2-isolates subgroup (GZ-B and GZ-C) with a deletion of length 39, Sin849 (deletion of length 49, embedded), and Sin846 (137, overlapping). B3 subgroup consisted of the isolates ZS-A (ZS-B) and ZS-C with the deletion of length 53.
A4 and B4 were the subgroups with many individual INDELs (Table 1). The rests of the A, B groups were denoted as A1 and B1, respectively.
It can be noted that proposed grouping of 96 isolates, based on SNV and INDEL contents, conserved the earlier classification T-T-T-T/C-G-C-C (38), and partially coincided with the extension of this classification 39., 40.. The four loci (9,404, 17,564, 22,222, and 27,827), as the basis for this classification, fitted nicely into our grouping (basically A1 group coincided with T-T-T-T type, while B1 group coincided with C-G-C-C type), expressing two inter-types: T-G-C-C [isolates GZ50, HZS2-D, HZS2-E, HZS2-C, HGZ8L2, HSZ2-A, NS-1(BJ04), HZS2-Fc, HZS2-Fb, and TJF] and C-G-T-T (isolates ShanghaiQXC1 and ShanghaiQXC2) (Figure 5). We found that another five loci, which are among the most represented SNVs’ loci (positions 3,852, 9,479, 11,493, 21,721, and 26,477; Figure 1), further refined our classification providing for sub-classification of the basic types.
There were two basic nine-locus types: TTTT/TTCGG and CGCC/TTCAT, mostly coinciding with the A1, B1 groups, and the two inter-groups: an inter-(A-B)-group had the inter-type TGCC/TTCGT, and a subgroup of the group B1 (two Shanghai isolates) represented another inter-type CGTT/TTCGT (Figure 5). The proposed extension to the two main sequence variants (TTTT, CGCC) for an enlarged set of isolates, is in accordance with the new insights into possible epidemiological spread, both in space and time 36., 38., 41.. Namely, positions 3,852 and 11,493 differentiated between the two subgroups of the group A1 (all of the TTTT type): Taiwan epidemic (nucleotides C, T) from the other late epidemic isolates (nucleotides T, C) (41), i.e., isolates closer to a Hong Kong virus unrelated to Hotel M (nucleotides C, T: isolates TW6-TW10, Taiwan TC1-TC3), and the others from the Hotel M lineage [nucleotides T, C: isolates from Canada (Tor2), Singapore (all Sin’s), Frankfurt (FRA Fr 1), Taiwan (TW1-TW5), Hong Kong (HKU 39849), Italy (HSR 1), China (ZJ01), etc.] (36). Position 9,479 decomposed the B group [differentiated subgroup B1 (T) from the subgroups B2, B3 (C)], position 21,721 distinguished the group A from the group B. Precise characterization based on the nine loci, for all the isolates, is given in Table 1 and Figure 5.
As compared to genotype clustering of SARS-CoV covering the epidemics from 2002 to 2004 7., 8., it can be noticed that the grouping we proposed was at most in accordance with it. Namely, the following correspondence between the two grouping schemes may be established:
Firstly, genotype class CGCC/TCCAT (covering B2 and B3 subgroups), corresponded to human patients’ isolates from the early phase 2002-2003 (ZS, HSZ, GD01, GZ02—Guangzhou, China), and palm civet isolates (SZ3, SZ16—Hong Kong).
Secondly, genotype class TGCC/TTCGT, TGCC/TTCAT (small part of A1 group), as well as CGCC/TTCAT (B1 group), corresponded to human patients middle phase 2002-2003 (positions 3,852, 9,479, 11,493, 26,477 determined this subclass); Beijing (BJ01-BJ04), and Hong Kong (CUHK W1).
Thirdly, genotype classes TTTT/NNNNN, CGTT/NNNNN (almost the entire A group and Shanghai part of the B1 group) corresponded to human patients late phase 2002-2003 (Figure 5)—Singapore (Sin s), Taiwan (TW1-11), Shanghai (QX1, QX2), Italy (HSR 1), Canada (Tor2), Hanoi (Urbani), Hong Kong (HKU39849, CUHK-AG0x), China (ZJ01, WHU, PUMC0x), Frankfurt (FRA, Frankfurt 1), etc.
The two basic groups, A and B, were rather contiguously mapped onto the phylogenetic tree, showing a high degree of accordance among the proposed grouping and the phylogenetic relationships. Exceptions represented the two isolates of the B4 group, with large number of SNVs and individual insertions (ZMY 1, ZJ01), as well as the two isolates of the Bl group (Shanghai QXC1 and QXC2), all of which being at large root-distances (Figure S10).
Materials and Methods
Dataset
Nucleotide sequences of 103 SARS-CoV complete genomes were taken from the PubMed NCBI Entrez database (http://www.ncbi.nlm.nih.gov/entrez) in GenBank and FASTA formats. Since there were 7 pairs of nucleotide-identical isolates, we considered the dataset to consist of 96 isolates (Tables 1 and S1). All the sequences are between 29,013 (ShanghaiQXC2) and 29,767 (Sin3408) in length. Out of all, 19 sequences have ambiguous nucleotide codes, i.e., N, M, R, Y, W, K, S (Table Si). Out of 96 isolates, 42 are fully or partially annotated. All the annotated isolates have the S protein annotated of length. 3,768 and the N protein of length 1,269 nucleotides, 4 isolates do not have the E protein (CUHK-Su10, PUMC01, PUMC02, PUMC03) of length 231 (except for Sino1-11 of length 228 nucleotides), 1 isolate does not have the M protein (CUHK-Su10) of length 666 nucleotides. Out of 42 annotated isolates, 13 have 5’ UTR and 12 have 3’ UTR determined. All the isolates are human sourced except for two isolated from palm civet (Paguma larvata), SZ3 and SZ16-
Genome polymorphism
The CLUSTAL X program, version 1.83 (42) has been applied to all the isolates from the dataset. The overall CLUSTAL X output had length of 29,903 nt. Then 5’ UTR and 3’ UTR were identified based on positions in annotated isolates. Coding region encompassed the interval (301, 29,528), and had the length of 29,228 nt.
We developed a program in Perl language for analysis of a CLUSTAL X output. The program first calculated an “average” isolate, the so called “profile”, by counting, for each position in the CLUSTAL X output, the number of occurrences of each different letter (including dash), and by choosing the most represented one; positions containing dashes in the “profile” are called “empty positions”, all the others being “non-empty” ones. The program then counted SNVs, INDELs, and calculated their absolute and relative positions, for every isolate with respect to the “profile”, and for different genome regions (ORFs, 5’ UTR, 3’ UTR, and IGRs).
Substitution rate for the SARS-CoV genome and for the S protein for all the sequences in the dataset was calculated by dividing the total number of SNV sites by the length of the corresponding nucleotide sequence; non-synonymous substitution rate for the S protein was calculated by dividing the total number of non-synonymous SNV sites by the length of the S protein.
Entropy of sites
The entropy of each site has been calculated based on number of SNVs at that site, in order to estimate the sites’ conserveness. If we denote by p(b)—probability of occurrence of the nucleotide b (b being A, C, G, or T), and under assumption of sites being independent, we calculated the entropy of positions by the following formula (43): E = - sum p(6)* log[p(6)] (sum over b). In this definition, p(b)* log[p(6)] is taken to be zero if p(6) = 0.
Phylogenetic investigations
The first type of classification was performed the same way as in Pavlovic-Lazetic et al. (37). It is based on genome polymorphism (SNVs and INDELs). The distribution of isolates per SNV numbers (outside 5’, 3’ UTRs) was analyzed and the isolates were primarily classified into two groups—isolates with “small” number of SNVs and isolates with “large” number of SNVs. The isolates “close to border” were further tested (on the number of SNVs) against the profile isolates of each of the two groups, resulting in some isolates changing the group. A sub-classification was then performed on the presence of long or short INDELs inside each of the two groups.
The second type of classification was performed based on contents of the most represented SNV sites. Except for earlier identified positions (9,404, 17,564, 22,222, 27,827) classifying isolates into TTTT/CGCC genotypes 38., 39., some other positions (genotypes) were identified as potential bases for sub-classification.
In order to compare the two classification schemes developed, with the existing programming systems for phylogenetic analysis, phylogenetic bootstrapped tree was produced using CLUSTAL X program and the Neighbor Joining (NJ) method. The NJ method, as well as parsimony and the probabilistic models, produces unrooted trees. In order to produce the consensus tree, bootstrapping is performed with random number generated seed 111 and number of trials in bootstrap 1000. The tree is drawn using the PhyloDraw program (44) and the proposed classification schemes were mapped onto it.
Annotation and analysis of the S protein
All the S protein sequences (those extracted from annotated isolates and the others we annotated using the publicly available program from PubMed tools for data mining; http://www.ncbi.nlm.nih.gov/gorf/gorf.html) have been aligned using CLUSTAL X program. Then the S protein was analyzed using the same methods as for the complete isolates.
Mutation analysis of the S protein
Non-synonymous nucleotide substitution per non-synonymous site (Ka) and synonymous nucleotide substitution per synonymous site (Ks) were calculated using the DnaSP 4.0 program (45). It is based on a method defined by Nei and Gojovori (46) that estimates the numbers of synonymous and non-synonymous nucleotide substitutions between two DNA sequences by counting the number of such substitutions in the corresponding pairs of codons. It also takes into account different evolutionary pathways between pairs of codons. The DnaSP program may run with or without an outgroup. The ratio Ka/Ks is considered as a selection parameter (Ka/Ks > 1 is usually interpreted as an indicator of positive selection). The coefficients Ka, Ks, as well as the ratio Ka/Ks were calculated first for the S protein in all the isolates in the dataset, without an outgroup. Since among the 91 isolates there were 89 human patients’ isolates and 2 palm civet isolates (SZ3, SZ16), we then calculated the Ka and Ks coefficients and the ratio Ka/Ks for the 89 human patients’ isolates only, without an outgroup, too. Eventually, we ran the program for all the human patients’ isolates and each of the palm civet isolates as the outgroup, in order to test the hypothesis that the S gene was subjected to positive selection during virus transmission from animals to humans.
Acknowledgements
This work was supported by the Ministry of Science and Technology, Republic of Serbia, Project No. 1858.
Supporting Online Material
http://www.gpbjournal.org/journal/pdf/GPB3(2)-05.pdf
Figures S1-S10, Tables S1-S6
References
- 1.Peiris J.S. Severe acute respiratory syndrome. Nat. Med. 2004;10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fouchier R.A. Aetiology: Koch’s postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rota P.A. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.Marra M.A. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 5.Guan Y. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 6.Stavrinides J., Guttman D.S. Mosaic evolution of the severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:76–82. doi: 10.1128/JVI.78.1.76-82.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song H.D. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He J.F. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. (Chinese SARS Molecular Epidemiology Consortium) [DOI] [PubMed] [Google Scholar]
- 9.Stadler K. SARS—beginning to understand a new virus. Nat. Rev. Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu R.W. Tracing SARS-coronavirus variant with large genomic deletion. Emerg. Infect. Dis. 2005;11:168–170. doi: 10.3201/eid1101.040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vega V.B. Mutational dynamics of the SARS coronavirus in cell culture and human populations isolated in 2003. BMC Infect. Dis. 2004;4:32. doi: 10.1186/1471-2334-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziebuhr J. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 2004;7:412–419. doi: 10.1016/j.mib.2004.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groneberg D.A. Molecular mechanisms of severe acute respiratory syndrome (SARS) Respir. Res. 2005;6:8. doi: 10.1186/1465-9921-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan Y.J. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antiviral Res. 2005;65:69–78. doi: 10.1016/j.antiviral.2004.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babcock G.J. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J. Virol. 2004;78:4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao X. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong S.K. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao J.C. Prokaryotic expression, refolding, and purification of fragment 450-650 of the spike protein of SARS-coronavirus. Protein Expr. Purif. 2005;39:169–174. doi: 10.1016/j.pep.2004.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou T. An exposed domain in the severe acute respiratory syndrome coronavirus spike protein induces neutralizing antibodies. J. Virol. 2004;78:7217–7226. doi: 10.1128/JVI.78.13.7217-7226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren Y. A strategy for searching antigenic regions in the SARS-CoV spike protein. Geno. Prot. Bioinfo. 2003;1:207–215. doi: 10.1016/S1672-0229(03)01026-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He Y. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J. Immunol. 2004;173:4050–4057. doi: 10.4049/jimmunol.173.6.4050. [DOI] [PubMed] [Google Scholar]
- 23.Hua R. Identification of two antigenic epitopes on SARS-CoV spike protein. Biochem. Biophys. Res. Commun. 2004;319:929–935. doi: 10.1016/j.bbrc.2004.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu L. Immunological characterization of the spike protein of the severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;42:1570–1576. doi: 10.1128/JCM.42.4.1570-1576.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keng C.T. Amino acids 1055 to 1192 in the S2 region of severe acute respiratory syndrome coronavirus S Protein induce neutralizing antibodies: implications for the development of vaccines and antiviral agents. J. Virol. 2005;79:3289–3296. doi: 10.1128/JVI.79.6.3289-3296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sui J. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. USA. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H. Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J. Virol. 2004;78:6938–6945. doi: 10.1128/JVI.78.13.6938-6945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van den Brink E.N. Molecular and biological characterization of human monoclonal antibodies binding to the spike and nucleocapsid proteins of severe acute respiratory syndrome coronavirus. J. Virol. 2005;79:1635–1644. doi: 10.1128/JVI.79.3.1635-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chou C.F. A novel cell-based binding assay system reconstituting interaction between SARS-CoV S protein and its cellular receptor. J. Virol. Methods. 2005;123:41–48. doi: 10.1016/j.jviromet.2004.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenough T.C. Development and characterization of a severe acute respiratory syndrome-associated coronavirus-neutralizing human monoclonal antibody that provides effective immunoprophylaxis in mice. J. Infect. Dis. 2005;191:507–514. doi: 10.1086/427242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S. Identification of two neutralizing regions on the severe acute respiratory syndrome coronavirus spike glycoprotein produced from the mammalian expression system. J. Virol. 2005;79:1906–1910. doi: 10.1128/JVI.79.3.1906-1910.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pyrc K. Genome structure and transcriptional regulation of human coronavirus NL63. Virol. J. 2004;1:7. doi: 10.1186/1743-422X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bi S. Complete genome sequences of the SARS-CoV: the BJ group (Isolates BJ01-BJ04) Geno. Prot. Bioinfo. 2003;1:180–192. doi: 10.1016/S1672-0229(03)01023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mooney S.D., Klein T.E. The functional importance of disease-associated mutation. BMC Bioinformatics. 2002;3:24. doi: 10.1186/1471-2105-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu L.D. Mutation analysis of 20 SARS virus genome sequences: evidence for negative selection in replicase ORF1b and spike gene. Acta Pharmacol. Sin. 2003;24:741–745. [PubMed] [Google Scholar]
- 36.Yeh S.H. Characterization of severe respiratory syndrome coronavirus genomes in Taiwan: molecular epidemiology and genome evolution. Proc. Natl. Acad. Sci. USA. 2004;101:2542–2547. doi: 10.1073/pnas.0307904100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pavlovic-Lazetic G.M. Bioinformatics analysis of SARS coronavirus genome polymorphism. BMC Bioinformatics. 2004;5:65. doi: 10.1186/1471-2105-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruan Y.J. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chim S.S. Genomic sequencing of a SARS coronavirus isolate that predated the Metropole Hotel case cluster in Hong Kong. Clin. Chem. 2004;50:231–233. doi: 10.1373/clinchem.2003.025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z.G. Molecular biological analysis of genotyping and phylogeny of severe acute respiratory syndrome associated coronavirus. Chin. Med. J (Engl.) 2004;117:42–48. [PubMed] [Google Scholar]
- 41.Lan Y.C. Phylogenetic analysis and sequence comparison of structural and non-structural SARS coronavirus proteins in Taiwan. Infect. Genet. Evol. 2005;5:261–269. doi: 10.1016/j.meegid.2004.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson J.D. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cover T.M., Thomas J.A. John Wiley & Sons, Inc.; New York, USA: 1991. Elements of Information Theory. [Google Scholar]
- 44.Choi J.H. PhyloDraw: a phylogenetic tree drawing system. Bioinformatics. 2000;16:1056–1058. doi: 10.1093/bioinformatics/16.11.1056. [DOI] [PubMed] [Google Scholar]
- 45.Rozas J. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 46.Nei M., Gojovori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotde substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1-S10, Tables S1-S6