Supplemental Digital Content is Available in the Text.
Key Words: HIV vertical transmission, PMTCT, HIV drug resistance
Abstract:
Antiretroviral drug resistance following pMTCT strategies remains a significant problem. With rapid advancements in next generation sequencing technologies, there is more focus on HIV drug-resistant variants of low frequency, or the so-called minority variants. In South Africa, AZT monotherapy for pMTCT, similar to World Health Organization option A, has been used since 2008. In 2010, a single dose of co-formulated TDF/FTC was included in the strategy for prevention of resistance conferred by single-dose nevirapine (sd NVP). The study was conducted in KwaZulu-Natal, South Africa, among pMTCT participants who received AZT monotherapy from 14 weeks of gestation, intrapartum AZT and sd NVP, and postpartum sd TDF/FTC. Twenty-six specimens collected at 6 weeks post-delivery were successfully sequenced using 454 ultra-deep sequencing. Non-nucleoside reverse transcriptase inhibitor (NNRTI) resistance was detected in 17 of 26 (65%) patients, 2 (7%) had Thymidine analogue mutations, and 3 (11%) had K65R. Of the 17 patients with NNRTI resistance, 11 (65%) had high-level NNRTI resistance, whereas 6 (35%) had intermediate NNRTI resistance. The levels of NNRTI resistance are much higher than would be expected, given the inclusion of antepartum AZT and postpartum TDF/FTC. This high level of NNRTI resistance could impact future NNRTI-containing treatment for a large proportion of pMTCT-exposed women. The detection of Thymidine analogue mutations highlights the need to understand the clinical impact of these on AZT-containing antiretroviral treatment in women exposed to AZT monotherapy.
INTRODUCTION
Although prevention of mother-to-child transmission (pMTCT) strategies have recently improved with the implementation of World Health Organization (WHO) options B and B plus,1 antiretroviral (ARV) drug resistance remains a significant problem in the wake of single-dose nevirapine (sd NVP)2,3 monotherapy and dual-therapy use in resource-limited settings. The current standard method for resistance testing is Sanger sequencing, or so-called population sequencing, and although widely used, it is limited since the sensitivity relies on mutations being present in 15%–20% of the HIV quasispecies.4,5 Thus, resistance conferring mutations present at low frequencies, or drug-resistant minority variants (DRMVs) will be missed by Sanger sequencing. The more sensitive technologies, commonly known as next-generation sequencing, include the Miseq and HiScan (Illumina, San Diego, CA), 454 GS-FLX and Junior (Roche Daignostics, Basel, Switzerland), Pac-Bio RS II (Pacific Biosciences, CA), and Ion-Torrent PGM (Life Technologies, Thermo Fischer Scientific, NY). Resistance testing performed using these technologies can detect DRMVs present at low frequencies.6 These DRMVs were shown to be clinically significant in studies investigating non-nucleoside reverse transcriptase inhibitor–based ART.7,8 DRMVs doubled the risk of virological failure to first-line NNRTI-containing antiretroviral treatment (ART).9
Following pMTCT exposure, DRMVs that develop may impact negatively on future ART, leading to virological failure.10 In South Africa, zidovudine (AZT) administered from the 14th week of pregnancy and intrapartum, together with sd NVP and a stat postpartum dose of co-formulated tenofovir (TDF) with emtricitabine (FTC) was used as the pMTCT strategy from 2010 to 2013.11 Using Sanger sequencing, high-level NVP resistance was detected in 34% of women in this context.12 This study therefore further aims to determine the patterns and frequency of DRMVs in this group of women, using ultra-deep sequencing (UDS).
METHODS
This study was conducted at Lwazi Clinic, Addington Hospital in Durban, South Africa. Ethical approval (BF069-09) was obtained from the University of KwaZulu-Natal Biomedical Research Ethics Committee. Ninety-seven pregnant women who did not qualify for ART as per National Guidelines,11 ie, CD4 count >350 cells per cubic millimeter were recruited for the study from August 2010 until December 2011. Data on adherence were captured at the 6-week post-delivery visit and limited to “Yes,” “No,” or “Unsure” with regard to receiving intrapartum AZT, sd NVP, and postpartum TDF/FTC. Furthermore, an EDTA whole-blood specimen for HIV-1 viral load testing was collected at recruitment and at 6 weeks post-delivery. A specimen for HIV-1 drug resistance testing was also collected at 6 weeks post-delivery.
HIV-1 Viral Load
The viral loads were performed using an automated Nuclisens EasyQ (bioMerieux) HIV-1 assay, which was later replaced by the Abbot m2000sp and Abbot m2000rt systems of extraction and real-time amplification, respectively. UDS was performed on 26 specimens which had an HIV-1 viral load of >5000 RNA copies per milliliter, with the exception of sample 3, where the viral load was 4604 RNA copies per milliliter.
Amplicon Design
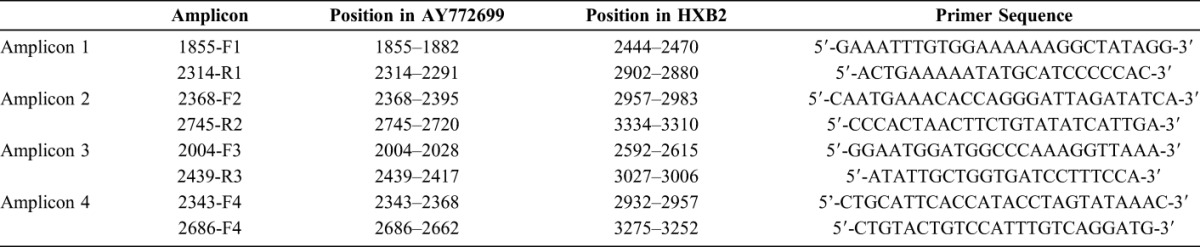
Four sets of overlapping amplicons were designed to cover the Reverse Transcriptase region of HIV-1 such that each significant codon position was interrogated by 2 separate amplicons. Primers were based on a subtype C isolate, Genbank accession no AY772699 (http://www.ncbi.nlm.nih.gov/nuccore/AY772699). Primer sequences are listed in Table 1.
TABLE 1.
Primer Sequences
RNA Preparation, Conventional Reverse Transcription Polymerase Chain Reaction, and Polymerase Chain Reaction
Two milliliter of plasma was ultracentrifuged at 14,000 rpm for 3 hours. RNA was extracted from 1 ml of plasma using the Nuclisens EasyMag HIV-1(bioMerieux, France) extraction system.
One-step reverse transcription (RT)-polymerase chain reactions (PCRs) using the SuperScript III One-Step RT-PCR System (with Platinum Taq High Fidelity; Invitrogen, Carlsbad, CA) were performed in triplicate per specimen. The triplicate RT-PCR products were then pooled and used for a second round of PCR that amplified overlapping amplicons which were also performed in triplicate per amplicon. The PCR products were pooled, purified, and sequenced.
A volume of 3.75 μl of extracted RNA was added to the RT-PCR reaction mix which had a final volume of 12.5 μl. Reagents included 2x buffer, Mg2SO4 (5 mM, final concentration of 0.6 mM), RNAse out, sterile water, SuperScript III (Invitrogen, Carlsbad, CA), and primers 1855-F1 and 2745-R2 (final concentration of 0.2mM).
A reverse transcription step at 55°C for 25 minutes was performed. Thermocycling was performed using an initial denaturation of 94°C for 2 minutes, followed by 25 cycles of 94°C for 30 seconds, 57°C for 30 seconds, 68°C for 30 seconds and a final extension step at 68°C for 1 minute. After the first round of PCR, 1 μL of the pooled PCR product was added to the second round PCR reaction mix (final volume of 50 μL) containing 10× buffer, dNTPs (200 mM), Platinum Taq High Fidelity enzyme (Invitrogen), MgS04 (50 mM, final concentration 2 mM), and DNase free water. The reaction mix was aliquoted equally into 4 separate tubes so that the relevant primers for the individual amplicons were added (F1, R1 to tube 1; F2, R2 to tube 2, etc). Conditions of cycling were the same as for the first round with omission of the RT step of 55°C for 55 minutes. After pooling, 150 μL of product was available for ultra-deep 454 sequencing. Samples were purified using the Qiagen min Elute spin columns.
To limit random sampling error caused by the sampling of only a few viral variants in patients with low viral loads, only patient samples with viral loads >5000 copies per milliliter were used, with the exception of sample 3, where the viral load was 4604 RNA copies per milliliter. Primers were designed to target conserved regions to limit primer induced selection bias, where particular templates are amplified earlier than others, and these become overrepresented in the final amplicon pool; each sample was amplified in 3 independent PCRs, and the PCR products were pooled before sequencing to compensate for biased priming and random sampling error during the PCR; multiplex identifier adapters were added after the amplification step to avoid the selection bias induced by using fusion primers.
Ultra-Deep 454 Sequencing
UDS was performed using the Roche 454 GS-FLX at the Technology Innovation Agency, National Genomics Platform in Durban. Twenty-six samples were successfully sequenced. Amplicon lengths varied in size (Amplicon 1: 459 bases, Amplicon 2: 376 bases, Amplicon 3: 436 bases, Amplicon 4: 344 bases). Samples had to meet standard requirements for library preparation after passing quality control. Samples were tagged with multiplex identifier adapters during library preparation. After emulsion PCR, sequencing was performed fulfilling all quality criteria and using a 4-lane divider on the picotiter plate. Four standard flowgram format files were generated and used for data analysis.
Data Analysis
For statistical analysis, nonparametric methods in SPSS version 23.0 (IBM Corp, Armonk, NY), including the Mann–Whitney U test, were used.
For bioinformatics analysis, Amplicon Variant Analyzer software v2.7 (Roche Diagnostics, Basel, Switzerland) was used to analyze and obtain sequence alignments against HIV-1 subtype C reference sequence (Genbank ID: AY772699). A short sequence length filter was applied based on the amplicon design and the corresponding sequence length. Short sequences (<90% of expected sequence length) were discarded. Error-corrected consensus sequences, as obtained from Amplicon Variant Analyzer, were used for amino acid variant calling. Variants were considered valid when present in both forward and reverse directions in a balanced manner as reported elsewhere.13 To control for sample cross-contamination, phylogenetic trees were built for all amplicons and samples with evidence of interfering cross-contamination were discarded. A minimum 500×/300× depth of coverage was required to call a minor variant (≤20%) and a major variant (>20%), respectively. Depth of coverage is provided in Table S1, Supplemental Digital Content, http://links.lww.com/QAI/A854. A 1% conservative minimum threshold was defined based on internal sequencing controls and on published literature.13–16
To estimate whether sufficient viral templates were sampled, we used the formula pVL = NRNA(λ)/(VfeERNAXEcDNA) to calculate the minimal viral load required to detect minor variants at 1%, where pVL is the minimum viral load required; NRNA(λ) is the number of RNA copies that according to the Poisson distribution should be tested to detect at least 1 minor variant with a likelihood of > 99%; V, the volume of plasma (milliliter); fe, the fraction of the RNA eluent used for DNA synthesis; ERNAX, the extraction yield and EcDNA, the RT efficiency.17 Based on the following V = 1 mL, ERNAX = 0.96 and EcDNA = 0.7, using 0.5 as the fraction of the RNA eluent used for DNA synthesis, the minimum viral load required to reliably detect minor variants at 1% is 1488 copies per milliliter. Viral loads of all samples that underwent 454 sequencing were in excess of 5000 copies per milliliter, with the exception of sample 3, where the viral load was 4604 RNA copies per milliliter. Ensuring that an acceptable number of templates were sampled (Table 2).
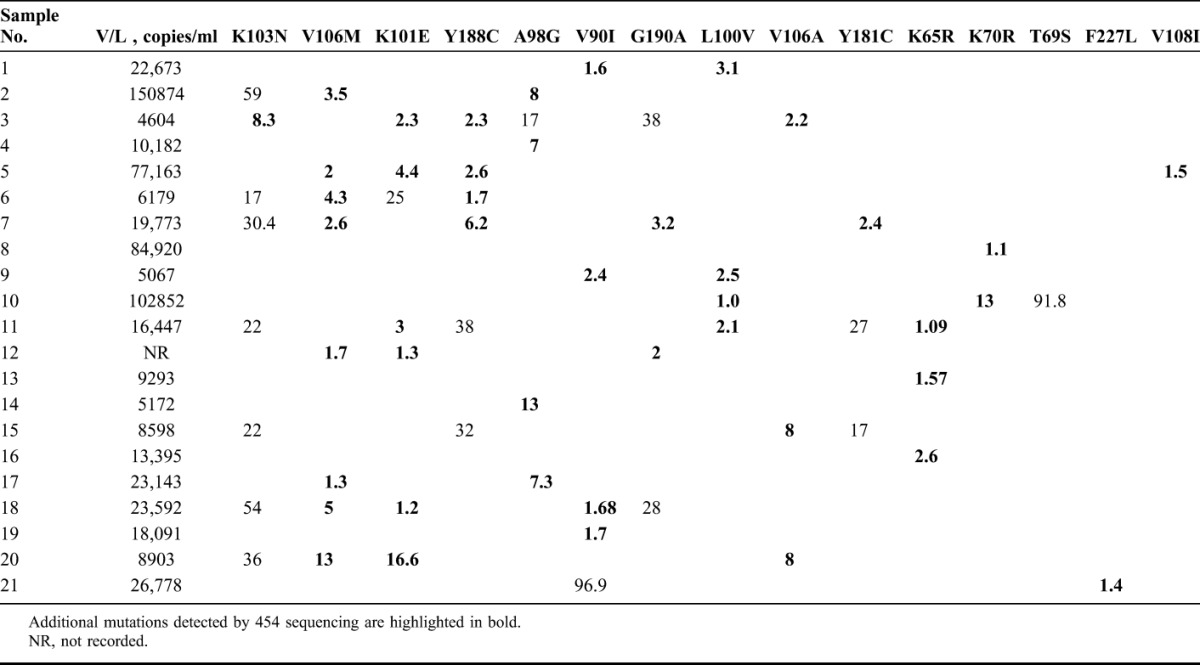
TABLE 2.
Viral Loads and Mutations Detected in Each Patient (provided as the Percentage of the Variant Within the Quasispecies)
RESULTS
There was no statistical difference in the CD4 cell count or HIV-1 viral load (at recruitment and at 6 weeks postdelivery) between those patients who developed NNRTI resistance and those who did not using the Mann–Whitney U test in SPSS version 23.0 (IBM Corp).
The median overall viral load was 17,269 copies per milliliter, with an interquartile range of 17,307 copies per milliliter (Table 2). The median viral load among patients where no Thymidine analogue mutations (TAMs) were detected was 14,921 copies per milliliter (interquartile range of 15262 copies/ml) compared with the median viral load of 93886 copies/ml in patients where TAMs were detected (P value 0.042).
The mean duration of AZT exposure overall was 16 weeks. The median duration of AZT exposure in those who developed TAMs was 20 weeks and 18 weeks (interquartile range of 8 weeks) in those who did not develop TAMs (P value 0.318).
Mutations conferring resistance to NRTIs and NNRTIs were detected at variable frequencies (Table 2). Of 26 patients, 20 patients (77%) had mutations conferring resistance. NNRTI resistance was detected in 17 of 26 (65%) patients, 2 (7%) patients had TAMs, and 3 (11%) patients had K65R. Of the 17 patients with NNRTI resistance, 11 (65%) had high-level resistance to NVP and EFV, whereas 6 (35%) had intermediate NNRTI resistance. One patient had both high-level NNRTI resistance and high-level resistance to TDF and 1 patient had both low to intermediate NNRTI resistance and K70R.
Of all mutations conferring resistance to NNRTIs, the most common were those conferring high-level NNRTI resistance such as K103N in 8 of 26 (30%), V106M in 8 of 26 (30%), Y188C in 6 of 26 (23%), G190A in 4 of 26 (15%), Y181C in 3 of 26 (11%), and V106A in 3 of 26 (11%) patients. K103N and V106M were the most common mutations detected. In patients who had K103N, it was also the predominant variant within the viral population compared to the other mutations detected as minor variants only. Mutations conferring low to intermediate NNRTI resistance included K101E in 7 of 26 (27%), A98G 5 of 26 (19%), L100V 4 of 26 (15%), V108I in 1 of 26 (3%) and F227L in 1 of 26 (3%) of patients. V90I which is associated with minimal, if any, detectable reduction in NNRTI susceptibility was found in 5 of 26 (19%) of patients.
Mutations conferring NRTI resistance included K70R in 2 of 26 (7%) patients and T69S which was detected in 1 of 26 (3%) patients. Resistance to TDF (K65R) was found in 3/26 (11%) patients. No other TAMs were detected. There was a 100% correlation between the mutations detected by Sanger sequencing12 and those detected by 454 UDS in samples that underwent both methods of sequencing. In addition, 454 UDS was able to detect a significant number of mutations that were missed by Sanger sequencing as indicated in bold in Table 2.
Regarding adherence, among the patients with high-level NNRTI resistance, 1 of 11 patients said that she was unsure about receiving antepartum AZT and intrapartum TDF/FTC and 1 said that she did not receive intrapartum TDF/FTC. Among the patients where no resistance was detected, 4 of 5 patients answered “unsure” or “no” to receiving prophylactic ARVs.
DISCUSSION
Using UDS, higher rates of NNRTI resistance were detected as compared to Sanger sequencing.12 More than two-third of patients had NNRTI resistance, the majority having high-level NNRTI resistance. The most common mutations (30%) detected were K103N and V106M, which are associated with high-level NNRTI resistance. Most of the K103N mutations were detected between frequencies of 17% and 59%, making it the predominant variant in the quasispecies for those specimens (Table 2).
Resistance to sd NVP is documented to occur at an average rate of 37.5%.18 The addition of peripartum AZT19 and postpartum TDF/FTC20 was shown to reduce the rate of resistance conferred by sd NVP. An open-labeled randomized control trial in Zambia found that the addition of sd TDF/FTC reduced NNRTI resistance by half at 6 weeks post-delivery.21 However, in our study, despite the use of AZT and TDF/FTC, there was no reduction in NNRTI resistance and the rate of NNRTI resistance of 65% is significantly higher than in earlier pMTCT strategies where only sd NVP was used.18 The high rate of NVP resistance could be explained by poor adherence to the complicated overall pMTCT strategy, exposure to NVP in successive pregnancies,22 and the higher rates of transmitted NNRTI drug resistance in KZN as reported by the WHO drug resistance report of 2012.23
The clinical impact of minority NNRTI drug-resistant variants has recently become topical following advances in the next-generation sequencing technologies. Studies investigating this show that minority NNRTI-resistant variants are clinically significant and can lead to treatment failure when these patients are initiated on NNRTI-containing ARVs,7,8,13,16,24,25 Furthermore, even with 95% adherence, these variants are associated with up to 3 times the risk of virological failure.7 In addition, preexisting minority Y181C variants were associated with a risk of virological failure in patients initiated on first-line efavirenz (EFV)-containing ART24 and in EFV exposed treatment experienced patients.26 In our study, Y188C and Y181C were detected in 23% and 11%, respectively, of patients as minority variants.
The added clinical benefit of using next-generation sequencing has been demonstrated in many studies.7,26–28 Although the sensitivity is significantly better with such technologies, its inclusion for routine use faces many challenges some being the large cost factor as well as the sophisticated bioinformatics support required.
Two (7%) patients harbored the K70R mutation while no other TAMs were found. Although the rate of AZT resistance is much lower than that detected by Olson et al,29 it is possible that AZT resistant mutants may have faded by the time of sample collection in our study, ie, 6 weeks postdelivery and may also be reflective of a smaller sample size. A study in Tanzania among pMTCT recipients where a similar pMTCT strategy was used, found AZT resistance in 18% of patients by Allele-specific PCR. The higher sensitivity of Allele-specific PCR compared to deep sequencing may explain the higher rates of TAMs.30 The clinical impact of these minority AZT-resistant variants when patients initiate ART requires further investigations.
K65R was detected in 11% of patients at low frequencies (1%–2.6%). There are reports of higher levels of K65R detection in HIV-1 subtype C among patients failing first-line TDF-based ART31,32 and in ART-naive patients.33 This highlights the need to explore the impact of minority TDF drug-resistant variants in HIV-1 subtype C. The mechanism for higher levels of K65R in subtype C seems to be template specific, where a preferential pause in subtype C reverse transcription at position 65 AAG-AGG is seen.34 It is therefore important to interpret low abundance K65R mutations in subtype C with caution. In addition, PCR-induced error is an important consideration when interpreting very low-abundance variants. Varghese et al35 showed that using UDS which is PCR dependent for the sequencing of subtype C, RT KKK template may result in spurious detection of K65R.
The limitations of this study include the lack of baseline genotyping, limited adherence information and lack of knowledge of previous exposure to sd NVP. Owing to the increasing rates of transmitted NNRTI resistance,23 it is possible that the high levels of NNRTI resistance detected in this study is partially reflective of the transmitted NNRTI resistance. However for AZT, resistance most likely developed while on short course AZT since these patients were not exposed to ART regimens, had high HIV-1 viral loads and prolonged AZT exposure.36,37
We have demonstrated a high level of NNRTI resistance (65%), which may have serious impact on the national ART programme in South Africa. Since this regimen was part of the South African pMTCT prophylaxis from 2008 to 2013,11,38,39 approximately 1.5 million women may have been exposed to this regimen, given that about 300,000 HIV-infected women require pMTCT annually in South Africa.40 If this figure is adjusted for the average uptake of pMTCT prophylaxis in South Africa at 58.7%,41,880,000 women would have been exposed to this regimen. Therefore, more than half a million women may fail first-line NNRTI-containing ART and require a switch to a protease inhibitor–based ART regimen.
Furthermore, our extrapolation does not consider, first, the women exposed to sd NVP before 2008 who subsequently may have developed NNRTI resistance when initiated on ART. Second, the WHO reported in 2012 that transmitted NNRTI resistance is increasing in Africa. The prevalence of transmitted NNRTI resistance in KwaZulu-Natal has increased from below 5% in 2007 to 5%–15% in 2010 with the most commonly detected mutation being the K103NS.23 Third, the number of patients with NNRTI mutations among those failing NNRTI-based ART is high in rural South Africa, ie, 82% in both adults42 and children,43 with K103NS again being the most commonly detected NNRTI mutation. Finally, the underestimation of ART resistance using conventional sequencing and the rising evidence of the clinical impact of minority NNRTI mutations remains an important consideration.
These factors may consequently contribute to a higher than expected ART failure rate among patients on first-line NNRTI-containing ART. Therefore, it may be prudent to consider more rigorous monitoring for virological failure in these women to ensure good future treatment outcomes.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Columbia University-South Africa Fogarty Aids and TB Training and Research Program (AITRP); Professor Daniel Kuritzkes, Dr Jonathan Li, and Dr Athe Tsibris for their kind assistance during my AITRP traineeship at the HIV Research lab, Harvard Medical School; Technology Innovation Agency, Dawn Stephens.
Footnotes
Supported by the Medical Research Council of South Africa and the National Health Laboratory Service Research Trust (NHLSRT).
The authors have no conflicts of interest to disclose.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.jaids.com).
REFERENCES
- 1.WHO. Use of Antiretroviral Drugs for Treating Pregnancy Women and Preventing HIV Infection in Infants. 2012. Available at: http://whqlibdoc.who.int/hq/2012/WHO_HIV_2012.6_eng.pdf. Accessed July 11, 2016. [Google Scholar]
- 2.Lockman S, Shapiro RL, Smeaton LM, et al. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. N Engl J Med. 2007;356:135–147. [DOI] [PubMed] [Google Scholar]
- 3.Stringer JS, McConnell MS, Kiarie J, et al. Effectiveness of non-nucleoside reverse-transcriptase inhibitor-based antiretroviral therapy in women previously exposed to a single intrapartum dose of nevirapine: a multi-country, prospective cohort study. PLoS Med. 2010;7:e1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gianella S, Richman DD. Minority variants of drug-resistant HIV. J Infect Dis. 2010;202:657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charpentier C, Laureillard D, Piketty C, et al. High frequency of integrase Q148R minority variants in HIV-infected patients naive of integrase inhibitors. AIDS. 2010;24:867–873. [DOI] [PubMed] [Google Scholar]
- 6.Samuel R, Paredes R, Parboosing R, et al. Minority HIV-1 drug-resistant mutations and prevention of mother-to-child transmission: perspectives for resource-limited Countries. AIDS Rev. 2014;16:187–198. [PubMed] [Google Scholar]
- 7.Li JZ, Paredes R, Ribaudo HJ, et al. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA. 2011;305:1327–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li JZ, Paredes R, Ribaudo HJ, et al. Relationship between minority nonnucleoside reverse transcriptase inhibitor resistance mutations, adherence, and the risk of virologic failure. AIDS. 2012;26:185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cozzi-Lepri A, Noguera-Julian M, Di Giallonardo F, et al. Low-frequency drug-resistant HIV-1 and risk of virological failure to first-line NNRTI-based ART: a multicohort European case-control study using centralized ultrasensitive 454 pyrosequencing. J Antimicrob Chemother. 2015;70:930–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boltz VF, Zheng Y, Lockman S, et al. Role of low-frequency HIV-1 variants in failure of nevirapine-containing antiviral therapy in women previously exposed to single-dose nevirapine. Proc Natl Acad Sci U S A. 2011;108:9202–9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical Guidelines: PMTCT (Prevention of Mother-to-Child Tranmission). South Africa: National Department of Health; 2010. Available at: http://www.sahivsoc.org/upload/documents/NDOH_PMTCT.pdf. [Google Scholar]
- 12.Samuel R, Paredes R, Parboosing R, et al. A post-partum single-dose TDF/FTC tail does not prevent the selection of NNRTI resistance in women receiving pre-partum ZDV and intrapartum single-dose nevirapine to prevent mother-to- child HIV-1 transmission. J Med Virol. 2015;87:1662–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Todesco E, Rodriguez C, Morand-Joubert L, et al. Improved detection of resistance at failure to a teno fovir, emtricitabine and efavirenz regimen by ultradeep sequencing. J Antimicrob Chemother. 2015;70:1503–1506. [DOI] [PubMed] [Google Scholar]
- 14.Charpentier C, Lee GQ, Rodriguez C, et al. Highly frequent HIV-1 minority resistant variants at baseline of the ANRS 139 TRIO trial had a limited impact on virological response. J Antimicrob Chemother. 2015;70:2090–2096. [DOI] [PubMed] [Google Scholar]
- 15.Gianella S, Delport W, Pacold ME, et al. Detection of minority resistance during early HIV-1 infection: natural variation and spurious detection rather than transmission and evolution of multiple viral variants. J Virol. 2011;85:8359–8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pou C, Noguera-Julian M, Perez-Alvarez S, et al. Improved prediction of salvage antiretroviral therapy outcomes using ultrasensitive HIV-1 drug resistance testing. Clin Infect Dis. 2014;59:578–588. [DOI] [PubMed] [Google Scholar]
- 17.Paredes R, Marconi VC, Campbell TB, et al. Systematic evaluation of allele-specific real-time PCR for the detection of minor HIV-1 variants with pol and env resistance mutations. J Virol Methods. 2007;146:136–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arrive E, Newell ML, Ekouevi DK, et al. Prevalence of resistance to nevirapine in mothers and children after single-dose exposure to prevent vertical transmission of HIV-1: a meta-analysis. Int J Epidemiol. 2007;36:1009–1021. [DOI] [PubMed] [Google Scholar]
- 19.Farr SL, Nelson JA, Ng'ombe TJ, et al. Addition of 7 days of zidovudine plus lamivudine to peripartum single-dose nevirapine effectively reduces nevirapine resistance postpartum in HIV-infected mothers in Malawi. J Acquir Immune Defic Syndr. 2010;54:515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chi BH, Ellis GM, Chintu N, et al. Intrapartum tenofovir and emtricitabine reduces low-concentration drug resistance selected by single-dose nevirapine for perinatal HIV prevention. AIDS Res Hum Retroviruses. 2009;25:1099–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chi BH, Sinkala M, Mbewe F, et al. Single-dose tenofovir and emtricitabine for reduction of viral resistance to non-nucleoside reverse transcriptase inhibitor drugs in women given intrapartum nevirapine for perinatal HIV prevention: an open-label randomised trial. Lancet. 2007;370:1698–1705. [DOI] [PubMed] [Google Scholar]
- 22.Flys TS, Mwatha A, Guay LA, et al. Detection of K103N in Ugandan women after repeated exposure to single dose nevirapine. AIDS. 2007;21:2077–2082. [DOI] [PubMed] [Google Scholar]
- 23.WHO. HIV Drug Resistance Report. 2012. Available at: http://apps.who.int/iris/bitstream/10665/75183/1/9789241503938_eng.pdf. Accessed July 11, 2016. [Google Scholar]
- 24.Paredes R, Lalama CM, Ribaudo HJ, et al. Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J Infect Dis. 2010;201:662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casadella M, Manzardo C, Noguera-Julian M, et al. Clinical value of ultradeep HIV-1 genotyping and tropism testing in late presenters with advanced disease. AIDS. 2015;29:1493–1504. [DOI] [PubMed] [Google Scholar]
- 26.Halvas EK, Wiegand A, Boltz VF, et al. Low frequency nonnucleoside reverse-transcriptase inhibitor-resistant variants contribute to failure of efavirenz-containing regimens in treatment- experienced patients. J Infect Dis. 2010;201:672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson JA, Li JF, Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 2008;5:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simen BB, Simons JF, Hullsiek KH, et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis. 2009;199:693–701. [DOI] [PubMed] [Google Scholar]
- 29.Olson SC, Ngo-Giang-Huong N, Beck I, et al. Resistance detected by pyrosequencing following zidovudine monotherapy for prevention of HIV-1 mother-to-child-transmission. AIDS. 2015;29:1467–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hauser A, Sewangi J, Mbezi P, et al. Emergence of minor drug-resistant HIV-1 variants after Triple antiretroviral prophylaxis for prevention of vertical HIV-1 transmission. PLoS One. 2012;7:e32055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sunpath H, Wu B, Gordon M, et al. High rate of K65R for antiretroviral therapy-naive patients with subtype C HIV infection failing a tenofovir-containing first-line regimen. AIDS. 2012;26:1679–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Zyl GU, Liu TF, Claassen M, et al. Trends in genotypic HIV-1 antiretroviral resistance between 2006 and 2012 in South African patients receiving first- and second-line antiretroviral treatment regimens. PLoS One. 2013;8:e67188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li JF, Lipscomb JT, Wei X, et al. Detection of low-level K65R variants in nucleoside reverse transcriptase inhibitor-naive chronic and acute HIV-1 subtype C infections. J Infect Dis. 2011;203:798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coutsinos D, Invernizzi CF, Xu H, et al. Template usage is responsible for the preferential acquisition of the K65R reverse transcriptase mutation in subtype C variants of human immunodeficiency virus type 1. J Virol. 2009;83:2029–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varghese V, Wang E, Babrzadeh F, et al. Nucleic acid template and the risk of a PCR-Induced HIV-1 drug resistance mutation. PLoS One. 2010;5:e10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen C, Gotzsche PC, Nielsen CM, et al. Development of resistance to zidovudine in HIV strains isolated from CD4+ lymphocytes and plasma during therapy. Antivir Res. 1992;18:303–316. [DOI] [PubMed] [Google Scholar]
- 37.Welles SL, Pitt J, Colgrove R, et al. HIV-1 genotypic zidovudine drug resistance and the risk of maternal–infant transmission in the women and infants transmission study. The Women and Infants Transmission Study Group. AIDS. 2000;14:263–271. [DOI] [PubMed] [Google Scholar]
- 38.Policy and Guidelines for the Implementation of the PMTCT Programme. Pretoria, South Africa: National Department of Health; 2008. [Google Scholar]
- 39.Updates on Revised Antiretroviral Treatment Guidelines 2013. South Africa: National Department of Health; 2013. Available at: http://www.sahivsoc.org/upload/documents/FDC%20Training%20Manual%2014%20March%202013(1).pdf. [Google Scholar]
- 40.Unicef. Available at: http://www.unicef.org/southafrica/survival_devlop_343.html. Accessed September 7, 2015.
- 41.MRC. Evaluation of the Effectiveness of the National Prevention of Mother-to-Child Transmission (PMTCT) Programme on Infant HIV Measured at Six Weeks Postpartum in South Africa. 2010. Available at: http://www.mrc.ac.za/healthsystems/SAPMTCTE2010.pdf. Accessed July 11, 2016. [Google Scholar]
- 42.Manasa J, Lessells RJ, Skingsley A, et al. High-Levels of Acquired drug Resistance in adult patients failing first-line antiretroviral Therapy in a rural HIV treatment Programme in KwaZulu-Natal, South Africa. PLoS One. 2013;8:e72152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pillay S, Bland RM, Lessells RJ, et al. Drug resistance in children at virological failure in a rural KwaZulu-Natal, South Africa, cohort. AIDS Res Ther. 2014;11:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


