Abstract
To gain an enhanced understanding of the mechanism by which gibberellins (GAs) regulate the growth and development of plants, it is necessary to identify proteins regulated by GA. Proteome analysis techniques have been applied as a direct, effective, and reliable tool in differential protein expressions. In previous studies, sixteen proteins showed differences in accumulation levels as a result of treatment with GA3, uniconazole, or abscisic acid (ABA), and/or the differences between the GA-deficient semi-dwarf mutant, Tan-ginbozu, and normal cultivars. Among these proteins, aldolase increased in roots treated with GA3, was present at low levels in Tan-ginbozu roots, and decreased in roots treated with uniconazole or ABA. In a root elongation assay, the growth of aldolase-antisense transgenic rice was half of that of vector control transgenic rice. These results indicate that increases in aldolase activity stimulate the glycolytic pathway and may play an important role in the GA-induced growth of roots. In this review, we discuss the relationship among GA, aldolase, and root growth.
Key words: aldolase, gibberellin, proteome, rice
Introduction
Rice (Oryza sativa L.) is one of the most important crops in the world. It is the main staple food of more than half of the world’s population. Since rice has a genome that is significantly smaller than those of other cereals, it is an ideal model plant for genetic and molecular studies, particularly among the monocots (1). Draft sequences of rice genomes have been reported for subspecies indica (2) and japonica (3). Furthermore, the complete map-based genome sequences of chromosomes 1 (4) and 4 (5) for cultivar Nipponbare have been reported. The challenge ahead for the plant research community is to identify the functions, post-translational modifications, and the regulation of proteins encoded by the plant’s genes. Understanding the biological functions of novel genes is a more difficult proposition than merely obtaining the nucleotide or peptide sequences. This is because the existing information on amino acid sequences of known proteins in the database is derived primarily from genetic and biochemical studies, which are by nature focused and labor intensive; the use of a large number of plant species as experimental systems; and the extensive range of unique plant-produced secondary metabolites. Thus, the cumulative knowledge of functions of known and unidentified proteins does not match the wealth of nucleotide sequence information being generated through genome sequencing projects (6). The analysis of proteins using high-resolution, two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) is the most direct, first-order approach to define gene functions.
Root architecture in plants is determined by interactions between their intrinsic developmental programs and external biotic and abiotic stimuli 7., 8.. Root systems of plants growing in the field are marvelously successful at foraging for nutrients and water in a hostile, competitive environment where supplies of them are very limited, local and variable (9). The acquisition of soil resources by plant root systems is a subject of considerable interest in agriculture and ecology, as well as a complex and challenging problem in basic plant biology. Thus, root growth regulation is a critical function of terrestrial plants and is closely associated with the production, transport, and response to plant hormones. To understand the mechanism by which plant hormones regulate the root growth and development of plants, it is necessary to identify and characterize proteins regulated by plant hormones.
Gibberellins (GAs) are essential endogenous regulators of plant growth and developmental processes 10., 11.. Significant progress has been made in understanding the pathways involved in GA biosynthesis and on the mechanisms by which GA levels are regulated in plants 12., 13.. The plant response to GA applied to intact root systems appears to be concentration-dependent, and can have rather variable effects. For example, a low concentration of exogenous GA plays a role in normal elongation of roots by maintaining the extensibility of the cell wall in GA-sensitive dwarf pea (14). GA is a regulator of cell elongation in roots, and there is an apparent correlation between low levels of GA in planta and short roots (15). Since root system architecture influences water and nutrient absorption, the GA regulation of root growth is essential for plant survival. Plant cells with defects in vacuole expansion do not expand (16), and the uptake of water by expanding vacuoles and the rapid osmoregulation between cytosol and vacuole are regulated by tonoplasts (17). Vacuole expansion and cell elongation are dependent on several factors, such as rates of cell component biosynthesis, metabolite concentration, and pH gradient across the tonoplast. GA is likely to be associated with the tonoplast function.
In previous studies, proteome analysis was used to investigate the effects of GA and aldolase increases in roots treated with GA3 (18). Konishi et al. (19) have reported that increases in the expression of aldolase by exogenous GA3 were reversed in roots treated with uniconazole, an inhibitor of GA biosynthesis, and the abscisic acid (ABA) and aldolase levels were low in the roots of the GA-deficient mutant Tan-ginbozu. The GA deficiency in this mutant is due to the blocking of the metabolic steps from ent-kaurene to ent-kaurenoic acid, catalyzed by ent-kaurene oxidase (20). Since the increase of aldolase caused by GA activates the glycolytic pathway, the acceleration of root growth may be a direct result of these actions. Furthermore, antisense-transgenic rice plants were used to clarify the role of aldolase regulated by GA (21), and aldolase regulated the V-ATPase-mediated control of cell elongation, which determines root growth (22). In this review, the regulation of rice root proteins by GA as revealed by proteome analysis is discussed.
Proteome Analysis of Rice Root by 2D Gel Electrophoresis
Root protein identification using 2D electrophoresis
Proteins were extracted from roots (23) and separated by 2D-PAGE (24) using IEF and IPG tube gels (25) in the first dimension, with IEF in the low pI range (4.0–7.0) and IPG in the high pI range (6.0–10.0). After detection with CBB staining, protein spots were analyzed using Image-Master 2D Elite software. The 2D maps of the low and high pI ranges overlapped at around pI 6.0. A total of 508 proteins from rice roots were detected in the 2D-PAGE patterns. Although clear images were obtained in the range of pI 3.5–6.0 from 2D-PAGE with the IEF tube gel, including ampholytes in the first dimension, it was difficult to effectively isolate the basic proteins. Therefore, electrophoresis using IPG tube gels (pI 6.0–10.0) was carried out for improved resolution of basic proteins. About 90% of the visible proteins detected by 2D-PAGE were present in the pI 4.0–7.5 range. Proteins separated under these conditions were reproducibly observed in their relative 2D map positions. Computer analyses using Image-Master 2D Elite software revealed 508 individual rice root protein spots. After 2D-PAGE, amino acid sequences were determined by mass spectrometry and/or Edman sequencing (26).
A total of 38 N-terminal sequences of the 94 root proteins separated by 2D-PAGE were determined by Edman sequencing. A further 35 separated proteins excised from gels were analyzed by mass spectrometry. Using these approaches, at least some sequence information was determined for 73 individual proteins. The N-terminal amino acid sequences of 56 (59.6%) proteins of the total could not be determined by the Edman technique likely due to a blocking group at the N-terminus. This result is consistent with a report in which 134 rice proteins were subjected to sequencing, of which 79 (59%) were found to have blocked N-termini (27).
Functional classification of root proteins
Twenty-six percent of the identified proteins were involved in plant defense, indicating that the rice plant produces more disease-resistance and defense-related proteins than any other type. This functional category involves metallothionin, glutathione S-transferases, chitinases, NBS-LRR-type resistant proteins, antifungal protein R, thaumatin-like proteins, superoxide dismutase (Cu-Zn) [(Cu/Zn)SOD], type-1 pathogenesis-related (PR-1) protein, and SalT protein. A root-specific protein, RCc3, was expressed in the elongation and maturation zones of primary and secondary roots, as well as in root caps (28). These results may be consistent with marsh plants or helophytes, since they are genetically acclimatized to harsh soil environments and are tolerant of toxicity in the soil. In contrast, the root tip proteins of maize seedlings during hypoxic acclimation have been reported, and of 48 proteins analyzed by mass spectrometry, 46 were identified (29). Soluble metabolic enzymes were the most abundant, but no proteins involved in the defense category were identified (29). These results reflect the differences in gene expression patterns at the protein level under conditions of stress. Many root proteins were found in only one tissue in these assays, suggesting that their expressions are tissue-specific. Since these proteins are related to the primary function of the specific tissue, they can provide valuable insights into the specialized physiological function of each tissue.
About 20% of root proteins were placed in the category of “no assigned function”. The functions of these proteins will need to be identified to provide information about their roles in the physiology or anatomy of the tissue. This study presents the results of a direct differential display with 2D-PAGE for the identification of proteins that undergo alterations in concentration, and their potentially localization, under different physiological and developmental conditions in rice roots. This system readily visualizes changes in protein content, directly and rapidly extracts proteins of interest from gels, and analyzes the protein primary structure using mass spectrometry and/or Edman sequencing. Harvesting the information stored in root tissues holds great promise in helping to predict the functions of many other proteins in response to environmental challenges and for expediting the molecular cloning of genes that are critical for both understanding plant function and practical application.
Identification of Root Proteins Regulated by GA in Rice
Differential increase of four proteins in seedling roots treated with GA
Although GAs are well-characterized, essential endogenous regulators of root growth and developmental processes (14), the precise role of GA in root growth is still unclear. To assess the elongation of seminal roots, germinated seeds were grown for 72 h in soil amended with GA3. Shoots were measured to test the effects of GA3 on germinated seeds and seedlings. The result showed that shoot elongation increased with the concentration of GA3. Roots grown in 0.1 μΜ GA3 were 3.8% longer than the controls.
Root lengths regulated by GA were analyzed by comparing a normal sized cultivar, Nipponbare, with a semi-dwarf cultivar, Tan-ginbozu, and by treatment with uniconazole, an effective inhibitor of GA biosynthesis (30). Germinated seeds of Nipponbare, Ginbozu, and Tan-ginbozu with or without 0.1 μΜ GA3, 10 μΜ uniconazole, and 10 μΜ ABA were grown for 72 h in soil. Seedling root lengths were measured and compared to the control (Nipponbare). Ginbozu roots were slightly longer (0.5%) and Tan-ginbozu roots were 69.8% shorter than Nipponbare roots. Treatment with 10 μΜ uniconazole decreased root growth by 44.6%, and 10 μΜ ABA caused a 79.2% decrease in root length. Root elongation was normal in Tan-ginbozu treated with 0.1 μΜ GA3 and Nipponbare treated with 10 μΜ uniconazole or 0.1 μΜ GA3. Since root elongation was inhibited in both Tan-ginbozu and uniconazole-treated Nipponbare, and could be recovered by GA3 treatment, the proteins whose accumulation was affected by these treatments may be involved in the mechanisms for GA-induced root growth. It is well established that GA and ABA act antagonistically in many aspects of plant development (31), as illustrated by the reduction in seedling root length when seeds were germinated in 10 μΜ ABA.
To clarify the relationship between root elongation and GA, root proteins differentially accumulated in Tan-ginbozu and Nipponbare roots treated with uniconazole or ABA were investigated using a proteomics approach. Images of 2D-PAGE from 0.1 μΜ GA3-treated and control extracts were synthesized and the positions of individual proteins were evaluated automatically using Image-Master 2D Elite software. Since the same samples were subjected to IEF and IPG, additional equipment for electrophoresis followed the same procedure. Using the protein differential display analysis, it became apparent that four proteins were up-regulated due to treatment with GA3. All but one of the sequences of differentially accumulated proteins were found to be identical to proteins previously reported. The other three proteins were homologous to the beta 5 subunit of 20S proteasome, aldolase, and glyceraldehyde 3-phosphate dehydrogenase.
Aldolase is a widely distributed enzyme and a key constituent of both the glycolytic/gluconeogenic pathways and the reductive pentose phosphate cycle in plants. This enzyme plays a vital role in carbohydrate metabolism and in the production of triose phosphates in signal transduction (32). Glyceraldehyde 3-phosphate dehydrogenase is also a ubiquitous enzyme involved in glycolysis and gluconeogenesis (33). Two of the four proteins up-regulated by GA treatment are key enzymes in the glycolytic pathway, suggesting that GA signaling and root growth may be mediated by the glycolytic production of ATP.
Another identified protein, the 20S proteasome, is a proteolytic complex that is involved in removing abnormal proteins (34). The 26S proteasome is composed of a 20S core proteasome with 19S regulatory complexes on either side. GA-insensitive dwarf 2 (GID2) is a positive regulator of GA signaling, and the slender rice1 protein is degraded via the ubiquitin/26S proteasome pathway mediated by the SCFGID2 complex (35). Increases in the beta 5 subunit of the 20S proteasome by GA3 may be related to the activation of the SCFGID2-proteasome pathway.
Differential expressions of proteins from roots treated with uniconazole and ABA
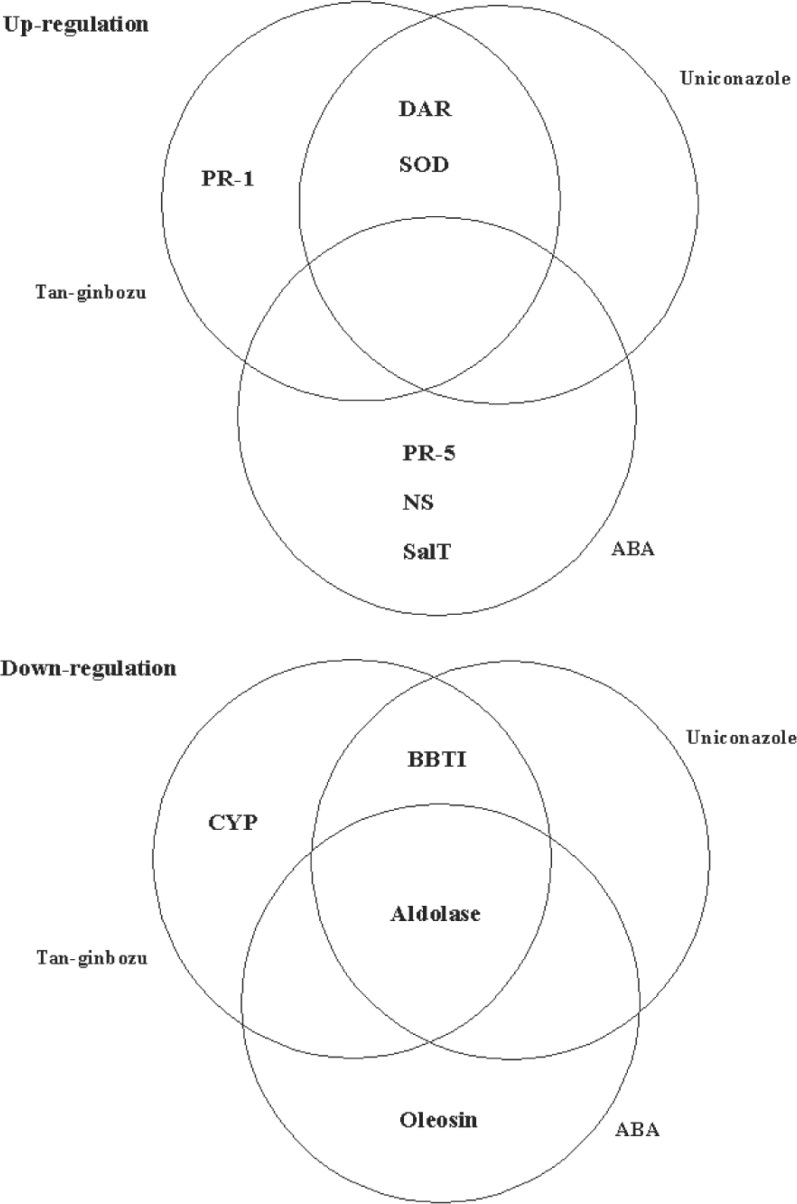
There were thirteen proteins identified that were expressed in significantly different amounts in Tan-ginbozu and Nipponbare and/or when treated with uniconazole or ABA, which had amino acid sequence similarity to other plant proteins. A scheme summarizing the regulation of the characterized proteins is shown in Figure 1. PR-1 protein accumulated in large amounts in Tan-ginbozu. GSH-dependent dehydroascorbate reductase 1 and [Mn]SOD accumulated in large amounts in Tan-ginbozu and also increased with uniconazole treatment. Antifungal protein-R, nicotianamine synthase 2, and SalT protein increased after treatment with ABA. Cyclophilin 2 accumulated in smaller amounts in Tan-ginbozu than controls. Bowman-Birk trypsin inhibitor accumulated in small amounts in Tan-ginbozu and decreased with uniconazole treatment. Oleosin decreased with ABA treatment. Aldolase accumulated to low levels in Tan-ginbozu and decreased after treatment with uniconazole or ABA.
Fig. 1.
Venn diagrams of the effects of the Tan-ginbozu semi-dwarfism, uniconazole-P, and ABA on protein accumulation. PR-1: type-1 pathogenesis-related protein; DAR: dehydroascorbate reductase; SOD: superoxide dismutase; PR-5: antifungal protein-R; NS: nicotianamine synthase; SalT: SalT protein; CYP: cyclophilin; BBTI: Bowman-Birk tripsin inhibitor.
[Modified from Konishi et al. (19)]
We propose that all of the proteins placed in the “up-regulation” category of Figure 1 are down-regulated by GA3. Plant dehydroascorbate reductase reduces oxidized ascorbate to maintain an appropriate level of ascorbate in plant cells. Dehydroascorbate reductase activity in rice seedlings is stimulated at high temperature (36). SOD catalytically scavenges superoxide radicals, providing a defense against reactive oxygen species (37). GA is likely to suppress the expression of GSH-dependent dehydroascorbate reductase and SOD. PR-1 protein, which plays a role in defense against stress, accumulated in large amounts in Tan-ginbozu. However, the correlation between PR-1 protein and root growth caused by GA is not clear. The action of ABA is unlikely to be limited strictly to antagonism of GA, and can be expected to have independent effects on protein expression. Antifungal protein R, nicotianamine synthase 2, and SalT protein increased as a result of treatment with 10 μΜ ABA in comparison with the controls, indicating that ABA induced the synthesis of these proteins. PR proteins constitute lines of defense against pathogen or herbivory, and antifungal protein R is a thaumatin-like (PR-5) protein from barley (38). Nicotianamine synthase catalyzes the trimerization of S-adenosylmethionine to form one molecule of nicotianamine, an intermediate in the biosynthetic pathway of the mugineic acid family of phytosiderophores. Iron deficiency in rice induces nicotianamine synthase gene expression in roots (39). Salt stress causes water deficit responses and a defense response reminiscent of aspects of the plant’s response to wounding and pathogen attack. Jasmonic acid, ABA, and NaCl induce accumulation of the salT transcript in roots (40).
Proteins in the “down-regulation” category of Figure 1 may be up-regulated by GA3. Bowman-Birk proteinase inhibitor is believed to play a role in defense: its induced gene expression follows a kinase-signaling cascade in rice (41). Cyclophilin is a specific cytosolic binding protein responsible for the accumulation of the immunosuppressant cyclosporin A by lymphoid cells, and Cyp2, a rice cyclophilin, may be preferentially translated during stress conditions (42). Bowman-Birk trypsin inhibitor and cyclophilins are likely to be related to the action triggered by GA. On the other hand, oleosin decreased in roots treated with ABA. Oil bodies are lipid storage organelles in which plants store energy as polysaccharides or lipids to support periods of active metabolism (43). The most abundant oil-body-associated proteins are oleosins, and much of the oil body surface may be covered by oleosin (44). Stored energy for active metabolism may be regulated by the balance between the action of ABA, decreasing the amount of stored energy, and GA exerting the opposite effect.
Relative aldolase mRNA and protein levels
The relationship between transcriptional regulation and translational expression is not always strictly proportional, but is a key factor in understanding the complex interactions that affect tissue growth and development. To compare the expression of aldolase protein and mRNA, root protein extracts from plants treated as above were probed with anti-aldolase antibody in an immunoblot assay. Fructose-bisphosphate aldolase accumulated in small amounts in Tan-ginbozu and levels decreased with uniconazole or ABA treatment. The enzyme accumulated at normal levels in Tan-ginbozu treated with GA3 and in Nipponbare treated simultaneously with uniconazole and GA3.
Northern blot analysis was carried out with an aldolase DNA probe. Relative amounts of mRNA were proportional to the accumulation of aldolase protein. Aldolase protein increased in GA3-treated roots, was at low levels in Tan-ginbozu roots, and levels further diminished in roots treated with uniconazole or ABA, suggesting that an adequate amount of aldolase in rice roots is required for normal root growth. The low level of aldolase protein observed in untreated Tan-ginbozu, which was likely due to the phenotypic selection for semi-dwarfism, was increased by treatment with uniconazole or ABA. Accumulation of aldolase was at “normal” levels in Tan-ginbozu treated with GA3, and in Nipponbare treated simultaneously with uniconazole and GA3. Furthermore, levels of aldolase mRNA and protein were directly proportional, indicating that aldolase may act as a mediator between GA signaling and root growth.
Characterization of Aldolase Regulated by GA in Rice Roots
Changes in aldolase mRNA and protein levels in response to GA in rice
The growth of rice seedling roots is markedly stimulated by the addition of GA3 and is correlated with a significant increase in aldolase accumulation. In rice roots, 14 proteins were up-regulated by GA3 (18). Aldolase was shown to be at a low level in Tan-ginbozu roots, and decreased in Nipponbare roots treated with uniconazole or ABA (19). In the root, GA is a regulator of cell elongation that is dependent on several factors, including the pH gradient across the tonoplast. Thus, we focused our attention on the glycolysis enzyme aldolase and its quantitative and functional regulation by GA3, because the stimulation of aldolase accumulation in GA3-treated roots is thought to modulate cell elongation and proliferation by controlling the rate of metabolism and tonoplast proton pumps.
The exogenous application of GA to rice roots was shown to affect the level of aldolase (18). To examine how GA3 changes the level of aldolase in rice roots, proteins were extracted from roots treated with GA3 at various concentrations for 0 to 72 h, and subjected to Western blot analysis with an anti-aldolase antibody. Measurable increases in aldolase accumulation were noted after 24 h exposure to GA3. By 48 h, there was a pronounced increase in aldolase protein that continued until 72 h. Aldolase levels increased in a dose-dependent manner, and treatment with 0.1 μΜ GA3 was the optimum concentration for root elongation. Changes in aldolase mRNA accumulation during treatment with GA3 were also examined for 24 h by Northern blot analysis. The transcript level of aldolase was extremely low at 0 h, and reached a maximum at 12 h. Transcript levels also increased in a dose-dependent manner and were coincident with aldolase protein levels. Root elongation of rice seedlings was accelerated by treatment with 0.1 μΜ GA3 (19). Treatment with GA3 for 48 h for aldolase protein, or 12 h for transcripts, was optimal at 0.1 μΜ. Therefore, rice plant tissues were treated with 0.1 μΜ GA3 for 48 h for protein or 12 h for mRNA in the following experiments.
To examine the organ-specific accumulation of aldolase in roots in response to GA3, proteins were extracted from organs of roots treated with 0.1 μΜ GA3 for 48 h and subjected to Western blot analysis with anti-aldolase antibody. Aldolase predominantly accumulated in roots and was detectable in leaf sheaths in the absence of GA3. Treatment with GA3 increased aldolase mRNA and protein levels in roots and leaf sheaths, but aldolase protein and transcript levels in leaf blades were very low. Furthermore, to examine the zone-specific accumulation of aldolase in roots in response to GA3, proteins extracted from the basal, medial, and apical regions of the root were subjected to Western blot analysis. Aldolase protein and transcripts accumulated prominently in the apical region, and treatment with GA3 led to an increase in aldolase protein accumulation in all three regions.
These findings support the hypothesis that aldolase is implicated in the tissue growth and differentiation triggered by GA3. The accumulation of aldolase induced by GA3 was the most notable in the apical region, which includes the root meristem (45). Exogenous GA3 increased aldolase expression in all regions of the roots, although the response varied with the region. Increased levels of aldolase may stimulate the growth of all of the root cells in the roots by causing an increase in energy production. Also, metabolic activation in the roots could cause increased water uptake.
Repression of root growth in aldolase-antisense transgenic rice
Aldolase-antisense transgenic rice plants were constructed to determine the effects of aldolase function on rice root growth. An aldolase cDNA clone was introduced into rice cells in an antisense orientation under the control of the CaMV 35S promoter in a pIG121-Hm binary vector by Agrobacterium-mediated transformation. Transgenic plants selected on media containing hygromycin were transplanted to soil and grown to maturity. Following a second round of selection on hygromycin, seeds were harvested and used in the following experiments. In a root elongation assay, roots of vector-only control rice plants grew to 55 mm by day 6, reaching the bottom of the container, whereas the growth of aldolase-antisense transgenic rice lines was only about half of the wild type’s rate.
The glycolytic pathway, which catalyzes the oxidization of glucose to pyruvate, produces two ATP molecules per glucose molecule. It is essential for providing energy and a wide range of other physiological functions (46). Activation of glycolytic enzyme expression by increasing aldolase may promote root growth. Addition of glucose to cells growing on a non-fermentable carbon source results in major changes in enzymatic activities and gene expression (47). However, the induction of glycolysis occurs via a glucose-independent mechanism in potato tubers (48). The up-regulation of glycolysis may accelerate cell growth. The present study clearly demonstrated the physiological importance of aldolase in root growth using the aldolase-antisense transgenic rice plants. Aldolase activity increased with exogenous GA3 treatment in control plant roots, but the increase was marginal in aldolase-antisense transgenic plant roots.
An aldolase assay was performed to confirm whether aldolase activity in roots is affected by GA3 in aldolase-antisense transgenic plants. In roots of vector-only control plants, treatment with 0.1 μΜ GA3 caused an increase in aldolase activity of 25%. In the roots of aldolase-antisense transgenic plants, aldolase activity was 43% lower than that in vector control plants and increased very little with 0.1 μΜ GA3 treatment.
Co-immunoprecipitation of aldolase with V-ATPase
In yeast cells deficient in aldolase, the peripheral V1 domain of V-ATPase dissociates from the integral membrane V0 domain, indicating that there is a coupling of glycolysis to the proton pump (49). Since the glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase is physically associated with the aldolase-V-ATPase complex and the glycolytic enzyme complex is directly coupled to the V-ATPase proton pump (49), association of aldolase with V-ATPase may result in the location of the ATP-generating glycolytic enzyme P-glycerate kinase close to the ATP-utilizing ATPase. Aldolase deletion mutant cells display a growth phenotype similar to that observed in V-ATPase subunit deletion mutants, and the V-ATPase abnormalities shown in aldolase deletion mutant cells can be restored to normal levels by aldolase complementation (49). The binding of aldolase to V-ATPase should provide the cells with a means for localized ATP generation by glycolysis. Furthermore, cytoplasmic aldolase has been identified in the vacuolar membrane fraction isolated from rice (50), suggesting that aldolase may associate with the vacuolar membrane protein.
Immunoprecipitation analysis was performed to examine whether aldolase interacts with V-ATPase in rice roots. V-ATPase is a multi-subunit complex and two antibodies were prepared that are specific either to the 100-kDa subunit or to the subunit A of V-ATPase. The antibodies were added to the extracted proteins from roots. The immunoprecipitated proteins were detected with anti-aldolase antibody. The V-ATPase complex co-precipitated with aldolase, thus indicating that they are physically associated in rice roots. Co-immunoprecipitation was mediated by both antibodies raised against V-ATPase 100-kDa subunit and subunit A. The accumulation of osmotically active ions accompanies vacuole expansion, and tonoplast proton pumps generate the trans-tonoplast electrical and proton gradient that drives the uptake of many solutes (15). Blocking the expression of V-ATPase subunit A causes the inhibition of root elongation, confirming the importance of this proton pump in cell expansion (51). Vacuolar solute accumulators that are energized by the H+ electrochemical potential gradient also influence the expansion of root cells (52). Vacuolar expansion is important for cell elongation, and thus may also be implicated in root cell growth mediated by aldolase activity.
Effect of Ca2+ on aldolase accumulation in the roots of OsCDPK13-antisense transgenic rice
In plants, the role of cytosolic Ca2+ concentration in the coupling of stimulus and response is not completely clear. However, GA3 induces a sustained increase in cytosolic Ca2+ concentration (31), and the locus, HvCDPK1, which encodes a Ca2+-dependent kinase, regulates vacuolar function during the GA3 response in barley aleurone (53). Moreover, OsCDPK13 expression is increased in rice leaf sheath segments treated with GA3 and is highly constant in leaf sheath and root as compared with leaf blade (54). To investigate the correlation between CDPK and aldolase accumulation, OsCDPK13-antisense transgenic rice (55) was used. To assess the possible involvement of Ca2+ in aldolase accumulation, rice seedlings were treated with CaCl2 and the Ca2+ channel blocker LaCl3. In roots of vector-only control seedlings, aldolase accumulation was stimulated when 5 mM CaCl2 and 5 μΜ Ca2+ ionophore A23187 were added together to excised roots for 48 h. Under similar conditions, aldolase accumulation was dramatically inhibited by 5 mM LaCl3. In roots of OsCDPK13-antisense transgenic seedlings, the accumulation of aldolase was markedly less than in control seedlings. Aldolase levels were slightly enhanced by treatment with 5 mM CaCl2 and 5 μΜ A23187. Aldolase was undetectable in 5 mM LaCl3-treatment roots.
Accumulation of aldolase in the roots of OsCDPK13-antisense transgenic seedlings (55) was markedly decreased compared with vector-only control transformant seedlings but was increased by Ca2+. CDPK mediates aldolase expression regulated by GA3, and the cytosolic Ca2+ concentration should correlate with this signaling pathway. These results indicate that GA3 up-regulates the aldolase level through cytosolic Ca2+ concentrations and CDPK13 in rice roots. Addition of Ca2+ to CDPK13-deficient plants may activate an alternate signaling pathway to up-regulate aldolase levels.
Root growth with the addition of glucose
Aldolase is a widely distributed enzyme and a key constituent of the glycolytic pathway. To investigate whether the glucose consumption due to ATP generation by glycolysis affects root growth in rice, germinated seeds of Nipponbare with or without added glucose were grown for 72 h in soil. Root growth was influenced by concentrations of glucose ranging from 100 nM to 100 mM and was concentration-dependent. There was an approximately 9.1% increase in root elongation at 100 μΜ glucose, but root growth was inhibited at 10 and 100 mM compared with controls.
Exogenous glucose was able to inhibit root elongation at high concentrations (10 and 100 mM), and promote root elongation at 100 μΜ glucose. Growth inhibition may be due to harmful osmotic influences on root growth. Glucose is a universal nutrient preferred by most organisms, and plants use hexokinase as a glucose sensor to interrelate hormone signaling networks for controlling growth and development (56). The present study suggests that fructose-bisphosphate aldolase is involved in GA-stimulated root growth through activation of the glycolytic pathway.
Conclusion
Proteome analysis techniques have been applied as a direct, effective, and reliable tool in differential protein expressions. Sixteen proteins showed differences in accumulation levels as a result of treatment with GA3, uniconazole, or ABA, and/or the differences between Tan-ginbozu and its wild type. Among these proteins, aldolase increased in roots treated with GA3, was present at low levels in Tan-ginbozu roots, and decreased in roots treated with uniconazole or ABA. These results show that proteome analysis techniques are very useful for biological studies, which are by nature focused and require the use of a large number of plant species as experimental systems.
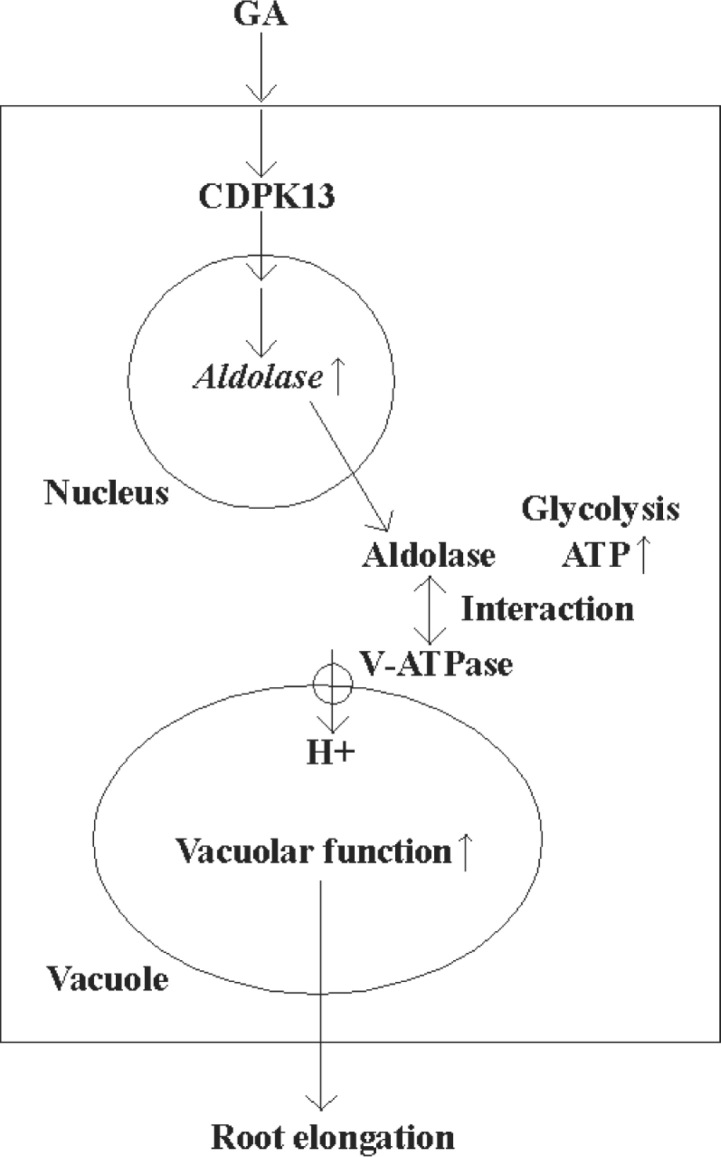
It is suggested that GA3-induced aldolase enhances the metabolic rate of glycolysis in rice roots. The present work suggests that aldolase activates V-ATPase through physical interaction. As a result, the rate of cell growth of seedling roots may be efficiently enhanced. Meanwhile, GA3 signaling for promotion of the root growth may be mediated by CDPK13 (Figure 2). A number of preliminary experiments have suggested that many modes of action may be involved in linking GA, aldolase, and root growth. More refined experiments will be needed to provide details about the exact relationships between the components of this regulation. The final result is that the growth of roots and other complex plant organs may be regulated by a sensitive matrix of interacting components, of which GA and its antagonists may play the most central role, but they act through primary metabolic intermediaries like aldolase and CDPK.
Fig. 2.
Schematic summary of the signal transduction between GA and root elongation. GA3-induced aldolase enhances the metabolic rate of glycolysis in rice roots, and aldolase activates V-ATPase through physical interactions. As a result, the rate of cell growth in seedling roots may be significantly enhanced. Also, GA3 signaling for root growth promotion may be mediated by CDPK13.
[Modified from Konishi et al. (22)]
Acknowledgements
This work was supported by a grant from the Program for Promotion of Basic Research Activities for Innovative Biosciences and an MAFF Rice Genome Project grant of Japan (No. PR-1201).
References
- 1.Devos K.M., Gale M.D. Genome relationships: the grass model in current research. Plant Cell. 2000;12:637–646. doi: 10.1105/tpc.12.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu J. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 3.Goff S.A. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 4.Sasaki T. The genome sequence and structure of rice chromosome 1. Nature. 2002;420:312–316. doi: 10.1038/nature01184. [DOI] [PubMed] [Google Scholar]
- 5.Feng Q. Sequence and analysis of rice chromosome 4. Nature. 2002;420:316–320. doi: 10.1038/nature01183. [DOI] [PubMed] [Google Scholar]
- 6.Komatsu S. Rice proteomics: a step toward functional analysis of the rice genome. Mol. Cell. Proteomics. 2003;2:2–10. doi: 10.1074/mcp.r200008-mcp200. [DOI] [PubMed] [Google Scholar]
- 7.Schiefelbein J.W., Benfey P.N. The development of plant roots: new approaches to underground problems. Plant Cell. 1991;3:1147–1154. doi: 10.1105/tpc.3.11.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch J. Root architecture and plant productivity. Plant Physiol. 1995;109:7–13. doi: 10.1104/pp.109.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCully M. How do real roots work? Plant Physiol. 1995;109:1–6. doi: 10.1104/pp.109.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobs T. Why do plant cells divide? Plant Cell. 1997;9:1021–1029. doi: 10.1105/tpc.9.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kende H., Zeevaart J. The five “classical” plant hormones. Plant Cell. 1997;9:1197–1210. doi: 10.1105/tpc.9.7.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedden P., Proebsting W.M. Genetic analysis of gibberellin biosynthesis. Plant Physiol. 1999;119:365–370. doi: 10.1104/pp.119.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi S., Kamiya Y. Gibberellin biosynthesis: its regulation by endogenous and environmental signals. Plant Cell Physiol. 2000;41:251–257. doi: 10.1093/pcp/41.3.251. [DOI] [PubMed] [Google Scholar]
- 14.Tanimoto E. Interaction of gibberellin A3 and ancymidol in the growth and cell-wall extensibility of dwarf pea roots. Plant Cell Physiol. 1994;35:1019–1028. [Google Scholar]
- 15.Dolan L., Davies J. Cell expansion in roots. Curr. Opin. Plant Biol. 2004;7:33–39. doi: 10.1016/j.pbi.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Schumacher K. The Arabidopsis det3 mutant reveals a central role for the vacuolar H+-ATPase in plant growth and development. Genes. Dev. 1999;13:3259–3270. doi: 10.1101/gad.13.24.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaumont F. Characterization of a maize tonoplast aquaporin expressed in zones of cell division and elongation. Plant Physiol. 1998;117:1143–1152. doi: 10.1104/pp.117.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka N. Proteome analysis of rice tissues by two-dimensional electrophoresis: an approach to the investigation of gibberellin regulated proteins. Mol. Genet. Genomics. 2004;270:485–496. doi: 10.1007/s00438-003-0929-9. [DOI] [PubMed] [Google Scholar]
- 19.Konishi H. Identification of rice root proteins regulated by gibberellin using proteome analysis. Plant Cell Environ. 2005;28:328–339. [Google Scholar]
- 20.Sakamoto T. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004;134:1642–1653. doi: 10.1104/pp.103.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konishi H. Characterization of fructose-bisphoaphate aldolase regulated by gibberellin in roots of rice seedling. Plant Mol. Biol. 2005;56:839–848. doi: 10.1007/s11103-004-5920-2. [DOI] [PubMed] [Google Scholar]
- 22.Konishi H. Characterization of vacuolar membrane proteins changed in rice root treated with gibberellin. J. Proteome Res. 2005;4:1775–1780. doi: 10.1021/pr050079c. [DOI] [PubMed] [Google Scholar]
- 23.Zhong B. Screening of rice genes from a cDNA catalog based on the sequence data-file of proteins separated by two-dimensional electrophoresis. Breed. Sci. 1997;47:245–251. [Google Scholar]
- 24.O’Farrell P.H. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 25.Hirano H. Two-dimensional gel electrophoresis using immobilized pH gradient tube gels. Electrophoresis. 2000;21:440–445. doi: 10.1002/(SICI)1522-2683(20000101)21:2<440::AID-ELPS440>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 26.Cleveland D.W. Peptide mapping by limited proteolysis in sodium dodecyl sulphate and analysis by gel electrophoresis. J. Biol. Chem. 1977;252:1102–1106. [PubMed] [Google Scholar]
- 27.Tsugita A. Separation and characterization of rice proteins. Electrophoresis. 1994;15:708–720. doi: 10.1002/elps.1150150198. [DOI] [PubMed] [Google Scholar]
- 28.Xu Y. Characterization of a rice gene family encoding root-specific proteins. Plant Mol. Biol. 1995;27:237–248. doi: 10.1007/BF00020180. [DOI] [PubMed] [Google Scholar]
- 29.Chang W.W. Patterns of protein synthesis and tolerance of anoxia in root tips of maize seedlings acclimated to a low-oxygen environment, and identification of proteins by mass spectrometry. Plant Physiol. 2000;122:295–318. doi: 10.1104/pp.122.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Izumi K. Studies of sites of action of a new plant growth retardant (E)-1-(4-chlorophenyl)-4,4-dimethyl-2-(1,2,4-triazole-1-yl)-1-penten-3-ol (S-3307) and comparative effects of its stereoisomers in a cell-free system from Cucurbita maxima. Plant Cell Physiol. 1985;26:821–827. [Google Scholar]
- 31.Gilroy S., Jones R.L. Gibberellic acid and abscisic acid coordinately regulate cytoplasmic calcium and secretory activity in barley aleurone protoplasts. Proc. Natl. Acad. Sci. USA. 1992;89:3591–3595. doi: 10.1073/pnas.89.8.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaeffer G.W. Fructose 1,6-bisphosphate aldolase activity in leaves of a rice mutant selected for enhanced lysine. Phytochemistry. 1997;46:1335–1338. doi: 10.1016/s0031-9422(97)00470-6. [DOI] [PubMed] [Google Scholar]
- 33.Duée E. Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity. J. Mol. Biol. 1996;257:814–838. doi: 10.1006/jmbi.1996.0204. [DOI] [PubMed] [Google Scholar]
- 34.Sassa H. Primary structural features of the 20S proteasome subunits of rice (Oryza sativa) Gene. 2000;250:61–66. doi: 10.1016/s0378-1119(00)00190-6. [DOI] [PubMed] [Google Scholar]
- 35.Sasaki A. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science. 2003;299:1896–1898. doi: 10.1126/science.1081077. [DOI] [PubMed] [Google Scholar]
- 36.Urano J. Molecular cloning and characterization of a rice dehydroascorbate reductase. FEBS Lett. 2000;466:107–111. doi: 10.1016/s0014-5793(99)01768-8. [DOI] [PubMed] [Google Scholar]
- 37.Fridovich I. Superoxide dismutases. Annu. Rev. Biochem. 1975;44:147–159. doi: 10.1146/annurev.bi.44.070175.001051. [DOI] [PubMed] [Google Scholar]
- 38.Hejgaard J. Two antifungal thaumatin-like proteins from barley grain. FEBS Lett. 1991;291:127–131. doi: 10.1016/0014-5793(91)81119-s. [DOI] [PubMed] [Google Scholar]
- 39.Higuchi K. Nicotianamine synthase gene expression differs in barley and rice under Fe-deficient conditions. Plant J. 2001;25:159–167. doi: 10.1046/j.1365-313x.2001.00951.x. [DOI] [PubMed] [Google Scholar]
- 40.Moons A. Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transcripts in rice roots. Plant Cell. 1997;9:2243–2259. doi: 10.1105/tpc.9.12.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rakwal R. Characterization of a rice (Oryza sativa L.) Bowman-Birk proteinase inhibitor: tightly light regulated induction in response to cut, jasmonic acid, ethylene and protein phosphatase 2A inhibitors. Gene. 2001;263:189–198. doi: 10.1016/s0378-1119(00)00573-4. [DOI] [PubMed] [Google Scholar]
- 42.Buchholz W.G. Cyclophilins are encoded by a small gene family in rice. Plant Mol. Biol. 1994;25:837–843. doi: 10.1007/BF00028878. [DOI] [PubMed] [Google Scholar]
- 43.Murphy D.J., Vance J. Mechanism of lipid-body formation. Trends Biochem. Sci. 1999;24:109–115. doi: 10.1016/s0968-0004(98)01349-8. [DOI] [PubMed] [Google Scholar]
- 44.Frandsen G.I. Oil bodies and their associated proteins, oleosin and caleosin. Physiol. Plant. 2001;112:301–307. doi: 10.1034/j.1399-3054.2001.1120301.x. [DOI] [PubMed] [Google Scholar]
- 45.Ranathunge K. Water permeability and reflection coefficient of the outer part of young rice roots are differently affected by closure of water channels (aquaporins) or blockage of apoplastic pores. J. Exp. Bot. 2004;55:433–447. doi: 10.1093/jxb/erh041. [DOI] [PubMed] [Google Scholar]
- 46.Fernie A.R. Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 2004;7:254–261. doi: 10.1016/j.pbi.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 47.Rolland F. Glucose-sensing mechanisms in eukaryotic cells. Trends Biochem. Sci. 2001;26:310–317. doi: 10.1016/s0968-0004(01)01805-9. [DOI] [PubMed] [Google Scholar]
- 48.Trethewey R.N. Expression of a bacterial sucrose phosphorylase in potato tubers results in a glucose-independent induction of glycolysis. Plant Cell Environ. 2001;24:357–365. [Google Scholar]
- 49.Lu M. The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase. J. Biol. Chem. 2004;279:8732–8739. doi: 10.1074/jbc.M303871200. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka N. Proteomics of the rice cell: systematic identification of the protein populations in subcellular compartments. Mol. Genet. Genomics. 2004;271:566–576. doi: 10.1007/s00438-004-1002-z. [DOI] [PubMed] [Google Scholar]
- 51.Gogarten J.P. The use of antisense mRNA to inhibit the tonoplast H+-ATPase in carrot. Plant Cell. 1992;4:851–864. doi: 10.1105/tpc.4.7.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng N.H. The Arabidopsis cax1 mutant exhibits impaired ion homeostasis, development, and hormonal responses and reveals interplay among vacuolar transporters. Plant Cell. 2003;15:347–364. doi: 10.1105/tpc.007385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCubbin A.G. The calcium-dependent protein kinase HvCDPK1 mediates the gibberellic acid response of the barley aleurone through regulation of vacuolar function. Plant J. 2004;39:206–218. doi: 10.1111/j.1365-313X.2004.02121.x. [DOI] [PubMed] [Google Scholar]
- 54.Yang G. OsCDPK13, a calcium-dependent protein kinase gene from rice, is induced in response to cold and gibberellin. Plant Physiol. Biochem. 2003;41:369–374. [Google Scholar]
- 55.Abbasi F. OsCDPK13, a calcium-dependent protein kinase gene from rice, is induced by cold and gibberellin in rice leaf sheath. Plant Mol. Biol. 2004;55:541–552. doi: 10.1007/s11103-004-1178-y. [DOI] [PubMed] [Google Scholar]
- 56.Moore B. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science. 2003;300:332–336. doi: 10.1126/science.1080585. [DOI] [PubMed] [Google Scholar]