Abstract
MicroRNAs (miRNAs) are endogenously expressed non-coding RNAs of 20–24 nucleotides, which post-transcriptionally regulate gene expression in plants and animals. Recently it has been recognized that miRNAs comprise one of the abundant gene families in multicellular species, and their regulatory functions in various biological processes are widely spread. There has been a surge in the research activities in this field in the past few years. From the very beginning, computational methods have been utilized as indispensable tools, and many discoveries have been obtained through combination of experimental and computational approaches. In this review, both biological and computational aspects of miRNA will be discussed. A brief history of the discovery of miRNA and discussion of microarray applications in miRNA research are also included.
Key words: microRNA, interfering RNAs, biogenesis, sequence analysis, computational biology, microarray
Introduction
The emerging picture that small yet versatile RNA molecules can regulate gene expressions has been attributed as the most important advance in biology for decades (1). In this review we will focus on one kind of these small RNA molecules, the microRNA (miRNA). miRNAs are a family of small single-stranded RNAs about 19–25 nucleotides generated from endogenous hairpin-shaped transcripts with post-transcriptional activity 2., 3., 4., 5., 6., 7., 8., 9.. Depending on the degree of complementarity between miRNA and its target transcript, miRNA either leads to the degradation of the target transcript (near perfect complementarity), or the inhibition of the protein translation.
The Second RNA Revolution
Since the discovery of the DNA double-helix structure by Watson and Crick in 1953, the standard pathway of information flow in a cell from DNA to RNA to protein has been the dominant theme in molecular biology. A cell reads the information encoded in a gene’s DNA and uses DNA as a template to make a strand of RNA (a process called “transcription”). This messenger RNA, or mRNA, travels through the cell to ribosomes, sites of protein synthesis. These microscopic factories then read the mRNA to determine what amino acids to string together into a protein, a process called “translation”.
In this standard pathway of information flow, RNA was initially considered a mainly passive intermediary and has long stood in the shadow of DNA. It was thought that enzymes and other biological catalysts were exclusively proteins. Then came the first RNA revolution in 1980s as Cech discovered the enzymatic activity of RNA (10). This discovery led to the views that the origin of life on the earth might come from RNA, with DNA and protein coming later (the “RNA world” theory; ref. 11).
In the past few years the discoveries of RNA interference (RNAi) and miRNA brought up the second RNA revolution (1). RNAi is a process of silencing of gene expression by double-stranded RNA (dsRNA; ref. 12). Since its discovery it has quickly become a powerful experimental tool to decipher the function of genes. The staggering pace of discoveries on the mechanism of RNAi not only clarify the RNAi pathway itself where the mRNA of genes to be silenced is marked out by short interfering RNAs (siRNAs), but also lead to other gene silencing mechanisms, such as transcription blocking by methylation of chromatin and translation inhibition. A key finding of the biochemical mechanism of RNAi is that the exogenous dsRNAs are converted into short 21–22 nt RNA molecules (siRNA). The siRNAs then guide the cleavage and degradation of complementary mRNA targets. In experiments that were originally designed to find the distribution of siRNA generated from the input dsRNA, scientists were surprised to find endogenously encoded small RNAs, including let-7, the second member of miRNA family found in 2000. Later cloning efforts showed the widespread of miRNA in metazoans 13., 14., 15.. Now we know that the regulation of gene expressions by miRNA is a widespread natural phenomenon, with hundreds or even thousands genes in various genomes, regulating complex genetic pathways. A brief history of the discovery of miRNA is outlined in the next section.
A Brief History of miRNA Discovery
The history of the discovery of miRNA is an example of chance, serendipity, and perseverance. It is also a success story for open communication and close collaboration among scientists. The three scientists who discovered the first miRNA (lin-4) gave a moving recollection of the events and their heroic efforts that finally led them to success (16). A complementary article by the scientists who cloned the target of lin-4 (lin-14) gave an additional historical insight (17).
The first miRNA was discovered in 1993 by Lee, Feinbaum, and Ambros (18). At the beginning they were simply curious about an interesting worm mutant lin-4 discovered in Sydney Brenner’s laboratory in the mid of 1970s. No questions about non-coding RNAs or antisense regulation were in their minds at the time. The cloning of lin-4 started in the summer of 1988, and the Ambros’s group was able to pin down a 700-bp fragment. Even though they resequenced the fragment many times, there was no decent open reading frame (ORF) in the fragment. After frameshift experiments on this fragment failed to eliminate its function, they realized that lin-4 could not encode a protein, but they still did not expect it to be a transcript as short as 22 nt, so did not pay much attention to the signal at the bottom of the gels in their RNase protection experiments. It was in May of 1992 that their RNase protection experiments finally confirmed the 22-nt piece as the major gene product of lin-4.
At about the same time, the Ruvkun’s group mapped the lin-14 mutation to its 3ʹ UTR (untranslated region). After they exchanged sequences of lin-4 and lin-14, Ambros and Ruvkun recognized simultaneously the antisense complementarity of lin-4 and lin-14: there are multiple sites in the 3ʹ UTR of lin-14 that are complementary to lin-4. The results of this seminal discovery were published in 1993 18., 19..
Even though the results were published in a high profile journal, due to lack of homology in other species, the finding was considered as a genetic oddity that was restricted to the species of C. elegans and “did not trigger a gold rush” 16., 17.. The situation did not change much until the second miRNA (let-7) was discovered in 2000 (20).
However, homology search using let-7 sequence against the emerging whole genome sequences of D. melanogaster and human revealed that let-7 is conserved and the sequences adjacent to the conserved sequences could also fold into stem-loop precursors as in C. elegans. In fact, it was found that let-7 is conserved in many species (21). Furthermore, the target of let-7, namely lin-41, is also conserved across species 21., 22.. At this stage the generality of miRNA is becoming clear. In 2001, three groups cloned hundreds of miRNAs from C. elegans 13., 14., 15.. Currently, there is an explosion of miRNA research activities, sometimes with new important discoveries published simultaneously by several different laboratories.
Since both lin-4 and let-7 are involved in regulating the timing of developmental transitions, these tiny RNAs are first named “small temporal RNAs” (stRNAs). Later as more miRNAs have been discovered that are involved in many other processes, the general term “microRNA” has been used to designate these small RNAs.
A registry has been set up for the assignment of miRNA gene names prior to publication (http://www.sanger.ac.uk/Software/Rfam/mirna/; ref. 23). Release 2.0 of the database contains 506 miRNA entries from six organisms. As the time of writing of this review, Release 7.0 contains 2,909 miRNA entries from dozens of species. Very recently miRNA from pig (Sus scrofa; ref. 24) has also been added to the database. A uniform system for miRNA annotation has been proposed (25).
Structure of miRNA Genes and Their Genome Locations
The genomic position of miRNA genes in human and mouse was annotated by using the latest genome sequences and transcription unit databases (26). Out of 232 known mammalian miRNAs, it was found that more than 70% are located in defined transcription units. They can be categorized into three groups 8., 26.:
1. Exonic miRNAs in non-coding transcripts. Examples are miR-21, miR-155, and the cluster miR-23a-27a-24-2.
2. Intronic miRNAs in non-coding transcripts. Example is miR-15a-16-1 cluster in the non-coding RNA gene DLEU2. Out of 232 miRNAs studied, 27 miRNAs are in this category.
3. Intronic miRNAs in protein coding transcripts. Examples are miR-26b and the cluster miR-106b-93-25. Out of 232 miRNAs studied, 90 miRNAs are in this category.
In addition, there are miRNAs that overlap with either an exon or an intron depending on the alternative splicing pattern and are labeled as “mixed” category. Furthermore, it is noticed that a small number of human miRNAs are found in the 3ʹ UTR of coding mRNAs, such as miR-198 in the 3ʹ UTR of the mRNA for human follistatin related protein (5). In mammalian, a subset of conventional miRNAs is derived from genomic repeats 27., 28..
Another feature of miRNA is that about 50% of known miRNAs are clustered and are found in close proximity to other miRNAs 13., 14., 15., 29.. These structure differences of miRNAs have profound implications for their regulations and biogenesis. For example, the clustered miRNAs are likely transcribed from a single polycistronic transcription unit, and the intronic miRNAs have different mechanisms of biogenesis (30).
Biogenesis of miRNA: Gene Transcription and Maturation
There is evidence that the transcription of miRNA genes is mediated by the RNA polymerase II (pol II). The primary transcripts of miRNAs (pri-miRNAs) contain a 5ʹ 7-methyl guanosine cap and a 3ʹ poly-A tail, modifications that are trademarks of the pol II transcription 8., 31., 32.. The association of miRNA with pol II confers the advantage that, like other genes transcribed by pol II, the miRNAs are under elaborate control of various regulation factors in different development stages and tissues. It is becoming evident that miRNA expression profiles are indeed complicated. It should bear in mind, though, that there is still possibility that a small portion of miRNAs are transcribed by pol III. And so far the DNA elements that are common to most pol II promoters such as the TATA box and the TFIIB recognition elements have not been identified for miRNAs.
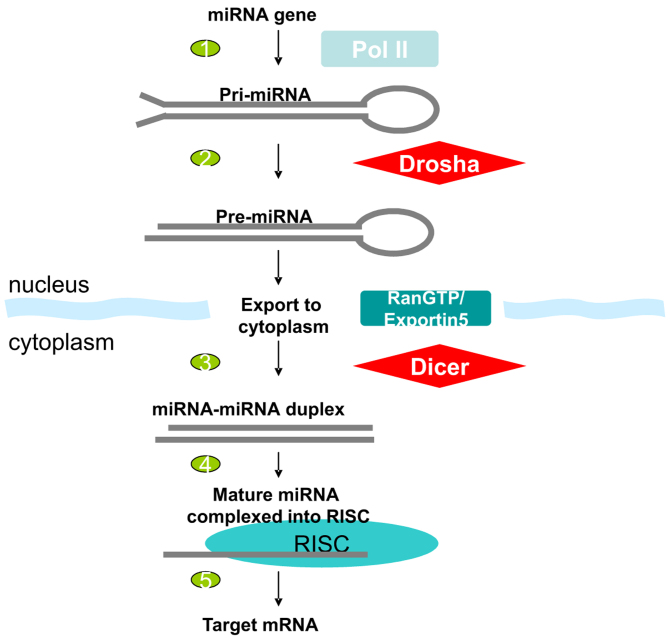
The processing and maturation of miRNAs are compartmentalized: the long primary transcripts are first processed into the hairpin intermediates (pre-miRNAs) in nucleus. The pre-miRNAs are transported into cytoplasm, where they are cleaved into mature miRNAs. In Figure 1 the predicted hairpin loops of lin-4 and let-7 are shown. Two RNase III enzymes are involved in miRNA biogenesis: Drosha and Dicer. Both enzymes contain tandem RNase III domains and a double-stranded RNA-binding domain (dsRBD) on the C-terminus. Drosha is conserved in animals, and Dicer is highly conserved in almost all eukaryotic organisms. The key steps are outlined below (Figure 2):
Fig. 1.

The predicted stem loops for lin-4 (left) and let-7 (right). The sequences of mature miRNAs are shown in red.
Fig. 2.
Model of miRNA biogenesis.
1. Pri-miRNAs (several kilobases) are processed by Drosha (Drosha-DGCR8 complex is also called the Microprocessor complex) in nucleus to yield the precursor miRNA (pre-miRNA), a stem-loop of about 60–100 nt with a 2-nt 3ʹ overhang. It is not very clear how Drosha recognizes primary RNA substrates and selects its cleavage sites, since the substrates of Drosha have no discernible sequence homology. The clue might come from the 3D structure of pre-miRNA. It was shown that human Drosha selectively cleaves RNA hairpins with a large terminal loop, greater than or equal to 10 nt. It uses the distance information to decide where to cut: from the junction of the loop and the adjacent stem, Drosha cleaves approximately two helical RNA turns into the stem to produce the pre-miRNA. In addition, the stem extension of approximately one helix turn is also essential for efficient processing (33).
2. In animals, pre-miRNAs are exported from nucleus to cytoplasm by exportin-5, with GTP-binding co-factor Ran. Exportin-5 is also responsible for tRNA exportation from nucleus to cytoplasm. Evidence comes from the cells in which exportin-5 is depleted. In such cells, pre-miRNAs and mature miRNA levels are reduced. The structure motifs for pre-miRNA export by exportin-5 is an RNA stem-loop of 16 bp or longer with a short 3ʹ overhang (34).
In contrast to mammals, it was found that miRNAs exist as single-stranded 20–21-nt molecules in the nucleus in Arabidopsis, indicating that proteins involved in miRNA biogenesis are located in the nucleus in Arabidopsis (35).
3. Pre-miRNAs are processed by Dicer in cytoplasm: Dicer was first discovered to process siRNA. Since the similarity of siRNA and miRNA, it was predicted and later proved that pre-miRNAs are also processed by Dicer. But recently it has been found that different Dicer isotypes have different functions: In D. melanogaster, Dicer-1 is required for miRNA cleavage, while Dicer-2 is required for siRNA processing (36). The same dual Dicer system was also suggested for Arabidopsis (37). Dicer interacts with other proteins, but those proteins do not seem to be required for the cleavage action.
4. Mature miRNAs are incorporated into RISC (RNA-induced silencing complex). Recent discoveries show that miRNAs and siRNAs use different complexes as their effector assemblies: miRNAs are incorporated into miRISC, and siRNAs into siRISC 38., 39..
5. miRNAs direct the RISC to their targets to downregulate gene expression, either by mRNA cleavage or translational repression: if there is sufficient complementarity between miRNA and the target mRNA, which is usually true in plants, the mRNA will be cleaved. If the complementarity is not high enough, as seen in animals, the translational of the mRNA will be inhibited, usually after the translation initiation step. There are exceptions to this rule: for example, miR-196-directed cleavage of HOXB8 was reported in mouse embryos (40). On the other hand, translational repression was reported in plants (41). Very recently, microarray experiments showed that human miRNAs can reduce the levels of many of their target transcripts, not just the amount of proteins deriving from these transcripts: when human cells are transfected by brain-specific miRNA (miR-124), the cell expression profile shifts toward that of the brain. If transfected by muscle-specific miRNA (miR-1), the profile shifts towards that of the muscle (42). It seems that the regulation mechanisms of miRNAs are more complicated than previously thought.
In the two steps that involve the cleavage by RNase III enzymes, a “single processing center” model seems to apply to both Drosha and Dicer (43). In this model the single processing center is formed in the interface between two RNase III domains. More information of structural aspects of miRNA biogenesis can be found in related reviews 44., 45..
The products from Dicer are about 22-nt duplexes, which are not very stable. Usually only one strand of the duplexes are incorporated into RISC. Which strand to choose? The rules are clearer in siRNA than in miRNA. It was found that relative thermodynamic stability of the 5ʹ end of the duplex determines which strand is selected: the strand with relatively unstable 5ʹ end is selected and incorporated into RISC 46., 47.. In D. melanogaster, the protein R2D2, which has two dsRBDs, is proposed to be involved in strand selection.
Regulation of miRNA
How miRNAs are regulated is not clear. Only a few promoters of miRNAs have been identified experimentally, and very few mammalian transcription factors that regulate miRNAs have been identified. Recently it has been found that c-Myc, which encodes a transcription factor that regulates cell proliferation, activates expression of a cluster of six miRNAs on human chromosome 13 (48). It was also found that a unique set of miRNAs are expressed in human embryonic stem cells, many of which are co-regulated and clustered in the expression profile (49).
By aligning sequences upstream and downstream of orthologous nematode miRNA foldbacks, the highly conserved sequence motif CTCCGCCC upstream (about 200 bp) of almost all nematode miRNA genes was found (50). Although the biological significance of this motif in transcriptional control needs to be determined through further investigation, this finding points to the possibility that there are hidden messages in the genomes that are waiting to be discovered. So far, no such common motifs have been found in other species.
Functions of miRNA
Although lin-4 and let-7, the two founding members of the miRNA family, both are involved in the temporal regulation of development transition between larval stages [recent experiments also showed that let-7 is involved in Ras signal transduction pathway (51)], later experimental (as well as computational, see below) evidence suggests that miRNAs play important roles in many other processes. These include the determination of left/right asymmetry of neuron development by lsy-6 in C. elegans (52); the promotion of cell proliferation and suppression of apoptosis by Bantam in D. melanogaster, the first miRNA that regulates a process other than cell lineage; the promotion of haematopoietic differentiation towards the B-cell lineage by miR-181 in mouse.
In plants, experimental and computational evidence suggests that most miRNA targets are translation factors and are involved in transcriptional regulations. Examples are miR-165 and miR-166, which regulate leaf morphogenesis in Arabidopsis. A smaller number of targets that are not translation factors correspond to mRNAs encoding factors required for miRNA formation or function, such as DCL1 and AGO1. For example, DCL1 mRNA itself is an miRNA target, indicating that the miRNA apparatus in plants is regulated by a negative-feedback loop 53., 54.. Whether this kind of negative-feedback control exists in animals is not clear. It was also reported that some miRNAs might regulate other miRNAs directly (55). The roles of miRNAs in the regulation of apoptosis (programmed cell death) were reviewed by Xu et al. (56).
Computational Approaches
The small size of miRNA makes it difficult to find by cloning or other experimental efforts. This perhaps is the reason why miRNA was not found earlier, given its wide spread occurrences in different species and developmental stages. From the very beginning, computational approaches have been extensively used in the miRNA research. It was through homology searches that it was found that miRNAs are conserved in many species and the miRNA regulation of gene expression is a widespread phenomenon 13., 14., 15., 21., 22.. It was through the search for potential stem-loops around the neighboring regions of known miRNA genes that it was found that miRNA genes are usually clustered together 14., 29., 50., 57.. It was also the computational approaches that revealed the number of miRNA targets is much bigger than previous thought 58., 59., 60.. There is an increasing number of publications discussing the computational aspects of miRNAs 61., 62., 63., 64.. In the following we discuss separately the approaches used to find miRNA genes and their targets.
miRNA gene finding
Computational methods have been used to find miRNA genes in various species, including C. elegans 65., 66., 67., D. melanogaster (68), human 59., 69., 70., 71., 72., Arabidopsis and other plants 73., 74., 75., 76., and virus 77., 78.. Two of the programs developed to predict miRNA genes are MiRseeker (68) and MiRscan 50., 66., 68..
Methods to find miRNA genes computationally usually utilize the following properties of known miRNA genes:
1. The precursor of miRNAs can form stable extended stem-loop structures, with continuous helical pairing and a few internal bulges.
2. miRNAs are usually highly conserved among the genomes of related species.
3. The evolutionary divergence between orthologous miRNAs shows a characteristic pattern: the terminal loops usually have more mutations than the arms of the stem-loops, and the miRNA-coding arms are more conserved than the non-miRNA-coding ones.
If used separately, none of these properties is specific enough to accurately predict miRNA genes. However, if they are used together, dependable predictions can be made. A typical algorithm (68) is first to identify conserved regions by aligning the whole genome sequences of related species. Then the annotated regions such as exons and tRNAs are eliminated. This is based on the assumption that it is not likely that miRNAs reside inside these regions, although this assumption may not be correct and has to be revised soon. In the second step, some RNA folding programs, such as Mfold or RNAfold, are used to fold and score both strands of the conserved regions from the first step. Some evaluation metrics can be developed to score the folds based on the closeness of the folds to the folding pattern of the reference miRNAs. In the next step, the divergence pattern of the high-scoring conserved stem-loops is analyzed and those stem-loops that do not show the characteristic pattern are eliminated.
Recently, by aligning sequences upstream and downstream of orthologous nematode miRNA stem-loops, it has been found that there is a highly significant sequence motif with consensus CTCCGCCC upstream of almost all independently transcribed nematode miRNA genes (50). It was reported that incorporating this feature into the MiRscan program could improve the accuracy of prediction. Such sequence motif has not been found in other species so far.
A method based on phylogenetic shadowing has been used to find many more human miRNA genes (59). By using genome sequences of phylogenetically closely related species, unambiguous sequence alignments can be obtained and more accurate conservation can be determined at single nucleotide resolution. In this study, 100 miRNA regions from 10 different primate species were aligned and compared. A characteristic profile emerges: the stem-loop regions are highly conserved and there are variations in the loop, as observed before, but there is a prominent drop of conservation immediately flanking the pre-miRNA regions. This profile was then used to identify new miRNAs in more divergent species such as human and mouse or human and rat by screening human/mouse or human/rat whole genome sequence alignments. The selected regions are then fed into RNA folding program for further filtration.
A genetic programming that automatically learns common structures of miRNAs from a set of known miRNA precursors was also developed (70). The algorithm includes three steps: First, for each miRNA precursor in the training set, the RNA Common-Structural Grammar (RCSG) of populations is optimized; second, the grammars are used to screen genomes for candidate miRNA genes; finally, the candidates are filtered with a scoring model. Some satisfactory results in term of specificity and sensitivity have been obtained.
A systematic method by comparing all miRNAs in the miRNA Registry (23) to the human and mouse genomes with additional RNA folding criteria also yields more human and mouse miRNAs (71).
The prediction of miRNA genes in plants has some specific considerations, because the lengths of miRNA precursors in plants are more variable, and the sequence similarity is lower.
miRNA target finding
In contrast to the computational miRNA gene finding, predicting miRNA targets computationally is relatively easy in plants than in animals, for the reason that in plants the complementarity is higher than that in animals [although in plants there are natural targets with up to five mismatches (79)]. Early studies used near complementarity for target predictions (80) in Arabidopsis. In the past few years progress has also been made in the prediction of targets in other species 58., 60., 73., 76., 81., 82., 83., 84., 85., 86., 87., 88..
Commonly used programs are TargetScan (81) and TargetScanS (58), miRanda 83., 84., DIANA-microT (85), RNAhybrid (86), PicTar (87), and others.
Although the programs differ in technical details, many of the programs use basically the same principles in their target finding algorithms: sequence complementarity between the miRNA and the 3ʹ UTR of the potential target gene is the major consideration, supported by evidence from free energies of RNA-RNA duplexes, and conservation of target sites in related genomes. Some studies found that the miRNA sequences tend to fold into a more stable secondary structure than randomly shuffled sequences 89., 90., 91., but the predictions based on miRNA:mRNA interactions must be viewed in the context of other potential interactions and cellular conditions, since the level of miRNA regulation also depends on the amount of mRNA and available miRNA complexes.
A recent study on the principles of miRNA-target recognition (92) classified the miRNA target sites into two broad categories: 5ʹ dominant sites and 3ʹ compensatory sites. In the first category, the target sites have high complementarity to the 5ʹ end of miRNA. This high complementarity is sufficient for the miRNA-mRNA duplex to be functional. The target sites within this category can be further grouped into two subcategories: “canonical” sites, which have good base-pairings for both 5ʹ and 3ʹ ends, and “seed” sites, which require little or no 3ʹ pairing. The “seed” sites can be still functional even if the 3ʹ pairing is below the random noise level. It was found that certain sites with as little as seven base-pairings between the 5ʹ end of miRNA and the target are sufficient to regulate target in vivo. Sites in the second category have weak 5ʹ complementarity and depend on strong base-pairing to the 3ʹ end of miRNA. It was also shown that both classes of sites are used in biologically relevant genes. Other studies of characteristics of miRNA-mRNA recognition were reviewed by Smalheiser and Torvik (93). It was found that in human, unlike in C. elegans and D. melanogaster, many miRNAs exhibit exact matches of 10 nt or more to the targets.
With the improved annotation and whole-genome alignment of the human, mouse, and rat genomes, as well as the recent availability of the chicken and dog genomes, it was found that some criteria in miRNA target finding algorithms can be relaxed or omitted without sacrifice of specificity. In the latest version of TargetScan (TargetScanS; ref. 58), the miRNA target finding algorithm looks for target sequence conservation in all of the five genomes (human, mouse, rat, chicken, and dog). The criteria were simplified to use 6-nt match (nt positions 2–7 of the miRNA) instead of 7-nt (nt positions 2–8) match, and the original requirements of TargetScan of multiple sites in each target and the filtration using thermodynamic stability were omitted. It was also found that the immediately downstream position of the seed match is highly conserved and is often an adenosine (A). In most cases this position base-pairs with the U of the first nucleotide of the miRNA, but it is conserved in the cases where the first nucleotide of the miRNA is not a U. Incorporating this “A anchor” into the target finding algorithm increases the sensitivity. By using these simplified criteria (6-nt base-pairing followed by the “A anchor”) the authors found many new miRNA targets. In human, over one third genes appear to be regulated by miRNAs.
The field of miRNA target prediction is a fast developing field and the accuracy of prediction will increase over time. One important question is whether we shall look at places other than 3ʹ UTR for targets. Ambros suggested that 5ʹ UTR should also be checked (3).
High-throughput miRNA Profiling
Although Northern blot analysis and cloning techniques are still the method of choice for confirmation of miRNAs (8), these procedures are laborious and have limited sensitivity, making it difficult to analyze miRNA expression pattern in a high-throughput fashion to study hundreds of miRNAs in a single experiment. Microarrays have been becoming a powerful and popular method to study gene expression profiles, and they have also emerged as a tool of choice for high-throughput miRNA profiling. In these experiments, a large number of oligonucleotides encoding the antisense sequences for the miRNAs are attached to glass slides and these slides are then hybridized to RNA samples, which contain fluorescently tagged miRNAs isolated from tissues of interest. In 2003, 44 oligonucleotides of 54–72 nt antisense were spotted to miRNAs nylon membrane to form an oligonucleotide DNA array (94). The array was used for the simultaneous analysis of miRNA expression profiles from the brain. The early report of using microarray to investigate genome-wide miRNA profiling in human and mouse tissues was published in 2004 (95). Although the results are confirmed by assessment of expression by Northern blots, real-time RT-PCR, and literature search, there are concerns with the microarray method because it measures the miRNA precursor (a 70-nt hairpin) instead of the mature miRNA (22 nt).
Genome-wide miRNA expression profiling has since been improved and applied to human B cell chronic lymphocytic leukemia (CLL; ref. 96), land plants (97), and various other species and tissues 98., 99., 100., 101., 102., 103., 104., 105., 106., 107., 108..
New methods designed specifically for using microarray for miRNA profiling have been developed. In one method, called RAKE (the RNA-primed, array-based Klenow enzyme assay; ref. 99), DNA oligonucleotide probes comprised of a universal spacer sequence on the 5ʹ end and the antisense miRNA on the 3ʹ end are attached to a glass slide. There are three thymidine nucleotides between the spacer and the antisense miRNA. When the RNA sample is hybridized to the microarray, the miRNAs base-pair with the corresponding antisense oligos. For those oligos that do not have corresponding miRNAs, they will be degraded in the following exonuclease I enzyme treatment. Subsequent addition of the Klenow fragment of DNA polymerase I generates a double-stranded fragment with tagged nucleotides that can be detected. For this method, miRNAs can be isolated and profiled from formalin-fixed paraffin-embedded tissue, which opens up new opportunities for the analysis of small RNAs from archival human tissues.
miRNA-responsive “sensor” transgenes (109) is a technique for visualizing detailed miRNA expression patterns. The sequence complementary to the miRNA under study is inserted in the 3ʹ UTR of a lacZ reporter. In cells lacking the complementary miRNA, β-gal is constitutively expressed, while in cells expressing the miRNA, the lacZ is targeted for degradation by the RNAi pathway. The technique was used in mouse embryos to reveal correlation between miRNA and mRNA expressions in various vertebrate tissues.
An oligonucleotide microarray for miRNA expression analysis was developed based on labeling RNA with quantum dot and nanogold probe, in which miRNAs were directly labeled at the 3ʹ terminus with biotin, and the hybridization to complementary oligo DNA probes was detected by measuring fluorescence of quantum dots bound to miRNAs through streptavidin-biotin interactions. The device was used to profile 11 miRNAs from the leaf and root of rice (Oryza sativa L. ssp. indica) (101). To avoid using high-cost detection equipment, colorimetric detection, a method based on nanogold probe coupled with silver enhancement, was also successfully introduced into miRNA profiling microarray detection. Other methods, such as LNA (locked nucleic acid)-modified oligonucleotides, a new method for the highly efficient detection of miRNAs by Northern blot analysis (110), have also been developed.
Microarray profiling of miRNAs reveals their important and interesting expression patterns. For example, frequent coexpression with neighboring miRNAs and host genes of 175 human miRNAs across 24 different human organs has been reported (98), implying that miRNAs separated by <50 Kb typically derive from a common transcript. Intronic miRNAs are usually coordinately expressed with their host gene mRNAs, implying that they also generally derive from a common transcript.
As mentioned before, microarray analysis shows that some miRNAs downregulate large numbers of target mRNAs (42). This indicates that in animals, miRNA functions not only on translation levels, but also on transcript levels.
Acknowledgements
This work was supported by grants R-146-050-071-133, R-146-050-071-101, R-154-050-248-133, and R-154-050-248-101 from National University of Singapore.
References
- 1.Novina C.D., Sharp P.A. The RNAi revolution. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Cullen B.R. Transcription and processing of human microRNA precursors. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Lai E.C. MicroRNAs: runts of the genome assert themselves. Curr. Biol. 2003;13:R925–R936. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 7.Carrington J.C., Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 8.Kim V.N. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 9.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 10.Zaug A., Cech T. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986;231:470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]
- 11.Gesteland R.F., Atkins J.F., editors. The RNA world. Cold Spring Harbor Laboratory Press; New York, USA: 1993. [Google Scholar]
- 12.Fire A. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 13.Lagos-Quintana M. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 14.Lau N.C. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 15.Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 16.Lee R. A short history of a short RNA. Cell. 2004;116:S89–S92. doi: 10.1016/s0092-8674(04)00035-2. [DOI] [PubMed] [Google Scholar]
- 17.Ruvkun G. The 20 years it took to recognize the importance of tiny RNAs. Cell. 2004;116:S93–S96. doi: 10.1016/s0092-8674(04)00034-0. [DOI] [PubMed] [Google Scholar]
- 18.Lee R.C. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 19.Wightman B. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern-formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 20.Reinhart B.J. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 21.Pasquinelli A.E. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 22.Slack F.J. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 23.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wernersson R. Pigs in sequence space: a 0.66X coverage pig genome survey based on shotgun sequencing. BMC Genomics. 2005;6:70. doi: 10.1186/1471-2164-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambros V. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smalheiser N.R. EST analyses predict the existence of a population of chimeric microRNA precursor-mRNA transcripts expressed in normal human and mouse tissues. Genome Biol. 2003;4:403. doi: 10.1186/gb-2003-4-7-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smalheiser N.R., Torvik V.I. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Altuvia Y. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ying S.Y., Lin S.L. Intronic microRNAs. Biochem. Biophys. Res. Commun. 2005;326:515–520. doi: 10.1016/j.bbrc.2004.10.215. [DOI] [PubMed] [Google Scholar]
- 31.Lee Y. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai X. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng Y. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. Embo J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng Y., Cullen B.R. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park M.Y. Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2005;102:3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y.S. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 37.Finnegan E.J. Posttranscriptional gene silencing is not compromised in the Arabidopsis CARPEL FACTORY (DICER-LIKE1) mutant, a homolog of Dicer-1 from Drosophila. Curr. Biol. 2003;13:236–240. doi: 10.1016/s0960-9822(03)00010-1. [DOI] [PubMed] [Google Scholar]
- 38.Okamura K. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem. Sci. 2005;30:106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 40.Yekta S. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 41.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim L.P. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 43.Han J. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krol J., Krzyzosiak W.J. Structural aspects of microRNA biogenesis. IUBMB Life. 2004;56:95–100. doi: 10.1080/15216540410001670142. [DOI] [PubMed] [Google Scholar]
- 45.Lingel A., Sattler M. Novel modes of protein-RNA recognition in the RNAi pathway. Curr. Opin. Struct. Biol. 2005;15:107–115. doi: 10.1016/j.sbi.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 46.Schwarz D.S. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 47.Khvorova A. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 48.O’Donnell K.A. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 49.Suh M.R. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 50.Ohler U. Patterns of flanking sequence conservation and a characteristic upstream motif for microRNA gene identification. RNA. 2004;10:1309–1322. doi: 10.1261/rna.5206304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson S.M. RAS is regulated by the let-7 MicroRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 52.Johnston R.J., Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 53.Xie Z. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 54.Vaucheret H. The action of ARG-ONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lai E.C. Complementary miRNA pairs suggest a regulatory role for miRNA: miRNA duplexes. RNA. 2004;10:171–175. doi: 10.1261/rna.5191904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu P. MicroRNAs and the regulation of cell death. Trends Genet. 2004;20:617–624. doi: 10.1016/j.tig.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 57.Seitz H. A large imprinted microRNA gene cluster at the mouse Dlkl-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lewis B.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 59.Berezikov E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 60.Xie X. Systematic discovery of regulatory motifs in human promoters and 3ʹ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sachidanandam R. RNAi as a bioinformatics consumer. Brief. Bioinform. 2005;6:146–162. doi: 10.1093/bib/6.2.146. [DOI] [PubMed] [Google Scholar]
- 62.Brown J.R., Sanseau P. A computational view of microRNAs and their targets. Drug Discov. Today. 2005;10:595–601. doi: 10.1016/S1359-6446(05)03399-4. [DOI] [PubMed] [Google Scholar]
- 63.Bengert P., Dandekar T. Current efforts in the analysis of RNAi and RNAi target genes. Brief. Bioinform. 2005;6:72–85. doi: 10.1093/bib/6.1.72. [DOI] [PubMed] [Google Scholar]
- 64.Lai E.C. Predicting and validating microRNA targets. Genome Biol. 2004;5:115. doi: 10.1186/gb-2004-5-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ambros V. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- 66.Lim L.P. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grad Y. Computational and experimental identification of C. elegans microRNAs. Mol. Cell. 2003;11:1253–1263. doi: 10.1016/s1097-2765(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 68.Lai E.C. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lim L.P. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 70.Nam J.W. Computational methods for identification of human microRNA precursors. Lect. Note Artif. Intell. 2004;3157:732–741. [Google Scholar]
- 71.Weber M.J. New human and mouse microRNA genes found by homology search. FEBS J. 2005;272:59–73. doi: 10.1111/j.1432-1033.2004.04389.x. [DOI] [PubMed] [Google Scholar]
- 72.Legendre M. Profile-based detection of microRNA precursors in animal genomes. Bioinformatics. 2005;21:841–845. doi: 10.1093/bioinformatics/bti073. [DOI] [PubMed] [Google Scholar]
- 73.Jones-Rhoades M.W., Bartel D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 74.Bonnet E. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. USA. 2004;101:11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adai A. Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res. 2005;15:78–91. doi: 10.1101/gr.2908205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang X.J. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pfeffer S. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 78.Couturier J.P., Root-Bernstein R.S. HIV may produce inhibitory microRNAs (miRNAs) that block production of CD28, CD4 and some interleukins. J. Theor. Biol. 2005;235:169–184. doi: 10.1016/j.jtbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 79.Schwab R. Specific effects of microRNAs on the plant transcriptome. Dev. Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 80.Rhoades M.W. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 81.Lewis B.P. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 82.Stark A. Identification of Drosophila microRNA targets. PLoS Biol. 2003;1:397–409. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Enright A.J. MicroRNA targets in Drosophila. Genome Biol. 2004;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.John B. Human microRNA targets. PLoS Biol. 2004;2:1862–1879. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kiriakidou M. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rehmsmeier M. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krek A. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 88.Robins H. Incorporating structure to predict microRNA targets. Proc. Natl. Acad. Sci. USA. 2005;102:4006–4009. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hofacker I.L. Prediction of locally stable RNA secondary structures for genome-wide surveys. Bioinformatics. 2004;20:186–190. doi: 10.1093/bioinformatics/btg388. [DOI] [PubMed] [Google Scholar]
- 90.Bonnet E. Evidence that microRNA precursors, unlike other noncoding RNAs, have lower folding free energies than random sequences. Bioinformatics. 2004;20:2911–2917. doi: 10.1093/bioinformatics/bth374. [DOI] [PubMed] [Google Scholar]
- 91.Doench J.G., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brennecke J. Principles of microRNA-target recognition. PLoS Biol. 2005;3:404–418. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Smalheiser N.R., Torvik V.I. A population-based statistical approach identifies parameters characteristic of human microRNA-mRNA interactions. BMC Bioinformatics. 2004;5:139. doi: 10.1186/1471-2105-5-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Krichevsky A.M. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu C.G. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Calin G.A. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Axtell M.J., Bartel D.P. Antiquity of microRNAs and their targets in land plants. Plant Cell. 2005;17:1658–1673. doi: 10.1105/tpc.105.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baskerville S., Bartel D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nelson P.T. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods. 2004;1:155–161. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- 100.Barad O. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liang R.Q. An oligonucleotide microarray for microRNA expression analysis based on labeling RNA with quantum dot and nanogold probe. Nucleic Acids Res. 2005;33:e17. doi: 10.1093/nar/gni019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thomson J.M. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 103.Sioud M., Rosok O. Profiling microRNA expression using sensitive cDNA probes and filter arrays. Biotechniques. 2004;37:574–576. doi: 10.2144/04374ST01. 578-580. [DOI] [PubMed] [Google Scholar]
- 104.Stolc V. Identification of transcribed sequences in Arabidopsis thaliana by using high-resolution genome tiling arrays. Proc. Natl. Acad. Sci. USA. 2005;102:4453–4458. doi: 10.1073/pnas.0408203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sun Y.Q. Development of a microarray to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res. 2004;32:e188. doi: 10.1093/nar/gnh186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Esau C. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 107.Babak T. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miska E.A. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mansfield J.H. MicroRNA-responsive “sensor” transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat. Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- 110.Valoczi A. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]