Abstract
Gold nanostars functionalized with Gd(III) have shown significant promise as contrast agents for magnetic resonance imaging (MRI) because of their anisotropic, branched shape. However, the size and shape polydispersity of as-synthesized gold nanostars have precluded efforts to develop a rigorous relationship between the gold nanostar structure (e.g., number of branches) and relaxivity of surface-bound Gd(III). This paper describes the use of a centrifugal separation method that can produce structurally refined populations of gold nanostars and is compatible with Gd(III) functionalization. Combined transmission electron microscopy and relaxivity analyses revealed that the increased number of nanostar branches was correlated with enhanced relaxivity. By identifying the underlying relaxivity mechanisms for Gd(III)-functionalized gold nanostars, we can inform the design of high-performance MRI contrast agents.
Graphical abstract
INTRODUCTION
Control over the geometry of anisotropic gold nanoparticles (Au NPs) enables tunability over a variety of properties, such as optical response and surface characteristics.1–3 Altering the aspect ratio of Au rods or bipyramids, for example, can tune the localized surface plasmon (LSP) from the visible to near-infrared (NIR) region.4–8 Additionally, Au NP surface curvature can affect the physicochemical properties of conjugated ligands, such as the effective acid dissociation constant (pKa).9,10 As-synthesized anisotropic Au NPs are polydisperse in size and shape, however, which prevents detailed analysis of shape-dependent phenomena in ensemble measurements.11,12 Density gradient centrifugation (DGC) is an emerging postsynthetic separation method that has been used to refine structural distributions of nanomaterials.12–19 Anisotropic Au NPs that have been sorted by DGC have primarily been coated with cetyltrimethylammonium bromide (CTAB)14–16 or cetyltrimethylammonium chloride (CTAC),12 which hinder conjugation with ligands.20–22 Expanding DGC to separate anisotropic Au NPs without these strongly bound surfactants is crucial for investigating shape-dependent properties of functionalized nanoconjugates.
Gold nanostars synthesized by reducing gold salts by a biocompatible Good’s buffer (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, HEPES) are a class of anisotropic Au NPs that can be readily functionalized with thiolated biomolecules.23–26 Recently, we found that the anisotropic shape of gold nanostars had a critical role in enhancing the T1, or longitudinal, relaxivity (r1) of surface-bound MRI contrast agents. In particular, the r1 of Gd(III)-conjugated DNA (Gd(III)-DNA) bound to the surface of nanostars (DNA-Gd@stars) was 3-fold higher than that of the analogous spherical nanoconjugates (DNA-Gd@spheres).23 Since r1 is an ensemble-averaged property, however, the broad distribution of nanostar branch numbers in as-synthesized samples prevented specific identification of which structural features were primarily responsible for the observed relaxivity enhancement.
Paramagnetic Gd(III) chelates produce positive (bright) contrast in T1-weighted MRI scans by shortening the longitudinal relaxation time of nearby water molecules. The efficiency of a T1 contrast agent is quantified by r1, the longitudinal relaxation rate normalized to the concentration of the ion (e.g., Gd(III)). Typical methods to alter r1 according to Solomon–Bloemergen–Morgen (SBM) theory include changing the rotational correlation time (τr), the inner-sphere water exchange rate (τm), or the number of coordinated water molecules (q).27–31 Optimization of τm and changing q are realized by alteration of the chelate design,30,32–34 while elongation of τr is frequently achieved by coupling the Gd(III) to larger, slow-tumbling species such as a biomolecules35–41 or NPs.31,42–48 r1 contributions from second- and outer-sphere relaxivity depend on the lifetime of water molecules in these coordination spheres.30,35,49,50 Molecular confinement and the hydrophilic microenvironment have been proposed to explain second-sphere effects observed for some protein-51,52 or NP-23,45,53 bound Gd(III) contrast agents with large r1.
Although Gd(III)-conjugated NP contrast agents have shown size-dependent r1 enhancements based on τr effects,31,46,47 few studies have explicitly examined how particle shape and surface curvature affect r1. Gd(III) chelates that were selectively bound to the exterior surface of rod-shaped virus particles, for example, showed larger r1 compared to chelates that were bound to the interior surface.39 The difference was due to rigidity of the amino acids that were targeted for the site-selective attachment,39 which affects the coupling of the Gd(III) to the tumbling of the NP and therefore the effective τr.30,46 Confinement of Gd(III) chelates, gadofullerenes, and gadonanotubes within the pores of mesoporous silica particles resulted in r1 enhancements from elongated τr and reduced mobility of outer-sphere water molecules.53
Unlike T1 contrast agents, T2 contrast agents generate negative (dark) contrast in MRI scans by shorting the transverse relaxation time of nearby water molecules. Shape-dependent enhancements in T2 relaxivity (r2) of superparamagnetic NPs such as iron oxide have been observed previously.54,55 Importantly, the effects of NP shape on r2 cannot be translated to r1 because enhancements in r2 and r1 are achieved via different mechanisms. Branched octapod54 and rod-shaped55 iron oxide NPs, for example, showed increased r2 relative to spherical NPs of similar volume. In both reports, the enhancement was due to an increase in the effective radius of the superparamagnetic core, a parameter that is known to depend on morphology and directly influences r2.54,55
Previously, we reported nuclear magnetic relaxation dispersion (NMRD) analysis of as-synthesized DNA-Gd@stars.23 The exceptionally high r1 was due to a large contribution of second-sphere relaxivity related to elongated residence times of water molecules in the second coordination sphere. Although shape was the only difference between the DNA-Gd@stars and DNA-Gd@spheres, second-sphere enhancements were not observed for the spherical conjugates.23 Based on these results, we hypothesized that confinement of Gd(III)-DNA in regions of negative curvature between the nanostar branches created a hydrophilic environment that could promote second-sphere effects. Hence, we expected that that r1 would depend on the number of branches on the nanostars. Here, we explicitly examine how populations of nanostars with different branch numbers contribute to the relaxivity of surface-bound Gd(III)-DNA. The enrichment of DNA-Gd@stars as a function of branch number was achieved by performing DGC in a sucrose density gradient, where up to 2-fold differences in r1 between structurally enriched populations was found.
EXPERIMENTAL METHODS
Materials
All reagents were purchased from Sigma-Aldrich unless otherwise noted. Detailed procedures are located in the Supporting Information.
Nanostar Synthesis
Nanostars were synthesized by reduction of HAuCl4 (0.2 mM) by HEPES buffer (100 mM, pH 7.2). NP characterization was performed with UV–vis-NIR spectroscopy (Agilent Technologies), dynamic light scattering (Brookhaven Instruments), transmission electron microscopy (TEM) (JEOL), and inductively coupled plasma-mass spectroscopy (ICP-MS) (Thermo Fisher).
Gd(III)-DNA Synthesis
The Gd(III) chelate was synthesized according to previously reported protocols and contains a pendant alkyne to facilitate conjugation via click chemistry.23 This chelate had one coordinated water molecule (q = 1) and an optimized inner sphere τm (22 ns at 37 °C).23 The Gd(III)-DNA is a poly deoxythymidine (dT) oligonucleotide (24-mer) that contains a 3′ disulfide modification (for Au functionalization) and five covalently bound Gd(III) chelates per strand. Oligonucleotides were purified by reverse phase HPLC and characterized using MS-MALDI.
Conjugation of Gd(III)-DNA to Nanostars
Nanostars were functionalized with Gd(III)-DNA by deprotection of the disulfide and salt-aging. Functionalized NPs were purified by three rounds of centrifugation (10 000 rpm, 10 min) and resuspension in Milli-Q water with 0.01% TWEEN.
Stability Testing of Nanostars in Gradient Media
As-functionalized DNA-Gd@stars were prepared by functionalizing nanostars with Gd(III)-DNA and purifying by centrifugation (no exposure to gradient media). For pre-sucrose and preiodixanol functionalized conditions, these DNA-Gd@stars (8–10 nM) were then mixed into solutions of sucrose (55% w/v) or iodixanol (30% w/v) and exposed for 2 h. Then the solutions were dialyzed overnight in ultrapure deionized water to remove the excess gradient media. Particles were recovered and concentrated via centrifugation prior to analysis. For post-sucrose and post-iodixanol functionalized particles, bare nanostars (8–10 nM) were first exposed to the gradient media and dialyzed under the same conditions. Then, the sucrose-exposed and iodixanol-exposed nanostars were functionalized with Gd(III)-DNA and purified for analysis.
Density Gradient Centrifugation
Sucrose density gradients were formed using a linear density gradient maker (Hoefer, Inc.) with 9 mL starting solutions of 50% and 60% w/v (density: 1.42–1.45 g/cm3) on top of a 4 mL underlayer of 90% w/v sucrose in water. 500 µL of a concentrated solution of bare Au nanostars (8–10 nM) was loaded on top of the density gradient medium in an Ultra-Clear centrifuge tube (Beckman Coulter), then centrifuged at 4400g for 2 h at 22 °C using a Beckman SW32Ti rotor in a Beckman Optima L-80XP ultracentrifuge. The samples were fractionated at intervals of 2 mm from the meniscus (Biocomp Instruments). This centrifugation and fractionation was repeated in 26 different tubes. After each round, the corresponding fractions from the 26 sorted tubes were combined and dialyzed (20 000 MWCO, Thermo Scientific) overnight against ultrapure deionized water. The fractions from 26 tubes were combined to obtain sufficient concentrations for structural analysis and relaxivity measurements (typically 200–400 µL of 5–8 nM nanostars per fraction).
Branch Number and Volume Analysis
Structural characterization was based on analysis of TEM images of DNA-Gd@stars (Figure S1) which were taken from several areas on the grid to obtain representative populations. The branch number was manually counted from at least 10 different zoomed-out images for at least 400 particles in each fraction. Nanostar volumes were approximated from TEM images of at least 300 particles per fraction using the Analyze Particles plug-in of ImageJ. Details regarding the assumptions and calculations are in the Supporting Information.
r1 Relaxivity Measurement
The r1 was calculated by measuring the T1 relaxation times (Bruker MiniSpec60) and Gd(III) concentrations (Thermo iCap Qc ICP-MS) of 3 to 5 different dilutions of each sample at a clinically relevant temperature (37 °C) and field strength (1.41 T, 60 MHz). These data were plotted as 1/T1 versus [Gd(III)] (mM) followed by linear regression analysis. The resulting fit of the line is defined as the r1 relaxivity of the agent (Figure S6–S8). The r1 was measured twice for each fraction.
RESULTS AND DISCUSSION
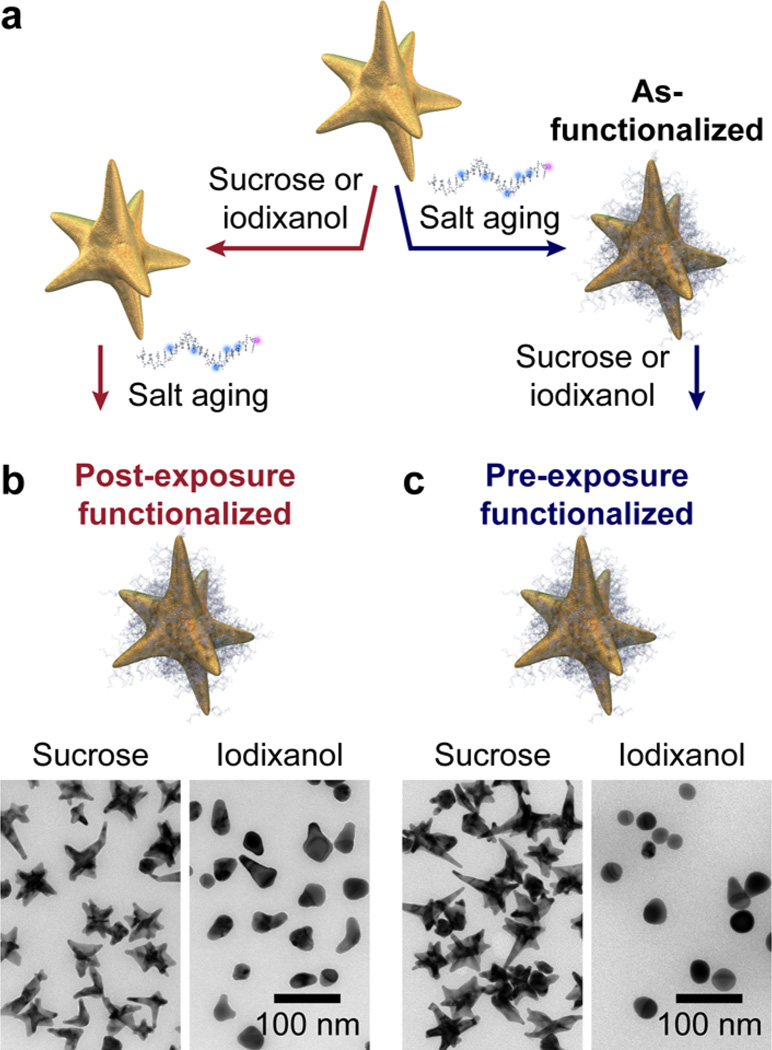
First, we explored different density gradient media (i.e., sucrose16,19,56 and iodixanol12–14,17,18) to ensure compatibility with the nanostar shape, colloidal stability, and surface functionalization with thiolated DNA (Figure 1, Figure S3–S5). These stability tests were performed without centrifugation to isolate the effects of exposure to the density-gradient media (Experimental Methods). We compared two procedures for gradient media exposure and functionalization of the nanoconjugates: (1) post-exposure functionalized nanostars were first exposed to sucrose or iodixanol and then functionalized with Gd(III)-DNA (Figure 1b); and (2) pre-exposure functionalized nanostars were first functionalized with Gd-(III)-DNA and then exposed to sucrose or iodixanol (Figure 1c). Subsequently, the gold nanostar dispersions were dialyzed to remove the gradient medium (Experimental Methods). As a control, we compared the results to as-functionalized DNA-Gd@stars that were not exposed to any gradient media. TEM (Figure 1b,c, Figure S3), optical absorbance spectroscopy (Figure S4a), and dynamic light scattering (Figure S4b) analysis revealed that sucrose had no effect on the shape or stability of the DNA-Gd@stars during either the pre- or post-sucrose functionalization methods. We also found that sucrose was compatible with nanostar surface functionalization and did not alter relaxivity of the DNA-Gd@stars (Figure S5–S6). In contrast, iodixanol induced dramatic particle reshaping, leading to reduced DNA-loading and relaxivity under all conditions tested (Figure 1b,c, Figure S3–S5).
Figure 1.
Sucrose is compatible with nanostar stability and surface functionalization. (a) Scheme of different methods of exposure to sucrose or iodixanol and surface functionalization with Gd(III)-DNA. TEM images for (b) post- and (c) pre-exposure functionalized DNA-Gd@stars.
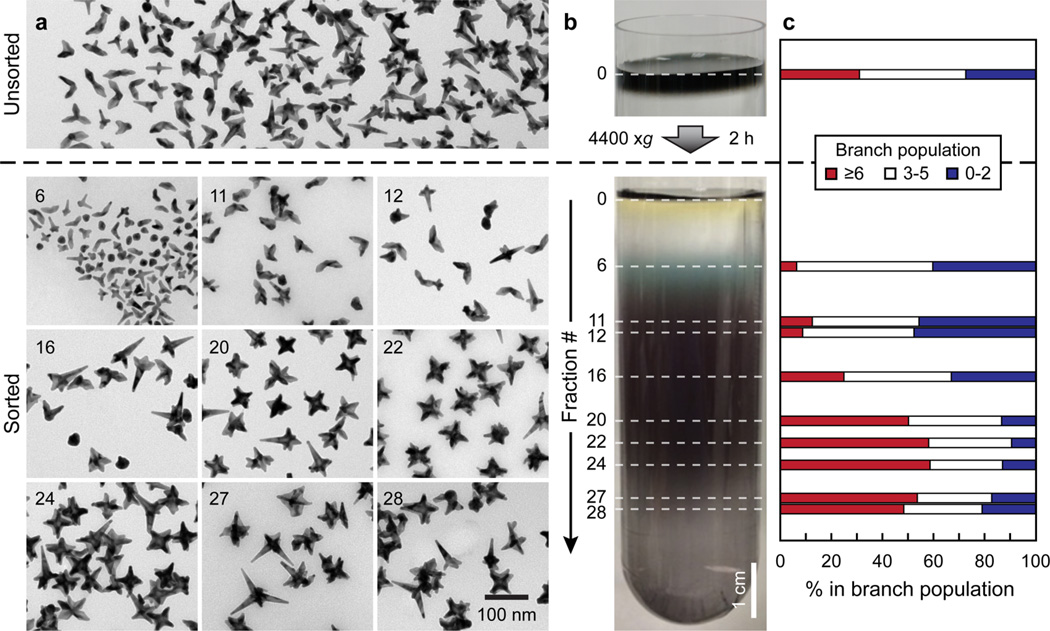
Since sucrose showed excellent compatibility with the anisotropic nanoconjugates, and the DNA loading on post-sucrose functionalized particles was higher than that of the presucrose functionalized DNA-Gd@stars (Figure S5), post-sucrose functionalization conditions were chosen for the following DGC sorting experiments. Unsorted post-sucrose functionalized DNA-Gd@stars showed a broad distribution of shapes (0–8 branches) and sizes (Figure 2a, top), thus sucrose-based DGC was used to refine the shape populations of the nanoconjugates. A linear sucrose gradient with a narrow change in density was chosen instead of a step gradient since the latter is more suitable for the separation of a few distinct populations.15,17,57 After layering the as-synthesized nanostar population on top of the gradient (Figure 2b, top) and centrifuging at 4400g for 2 h, we observed continuous spreading of nanostars throughout the centrifuge tube (Figure 2b, bottom). The formation of colored bands showed high reproducibility for 26 rounds, and the corresponding fractions from each tube were combined and dialyzed overnight in ultrapure deionized water. The separated populations were then functionalized with Gd(III)-DNA for relaxivity measurements (Supporting Information).
Figure 2.
DGC refines shape populations of DNA-Gd@stars. (a) TEM images of unsorted DNA-Gd@stars (top) and different fractions after DGC (bottom). (b) Photographs of a centrifuge tube before (top) and after (bottom) centrifugation of a concentrated nanostar solution in a sucrose linear density gradient. (c) Distribution of different DNA-Gd@star populations within each fraction based on branch counting of at least 400 particles per fraction.
The sedimentation rate of objects in a viscous fluid depends on both mass and shape.12,16 For example, nanorods with the same mass but different aspect ratios have been separated, but some rods and spheres with different masses may sediment at the same rate.16 Since nanostars with many branches typically have a larger mass than those with fewer branches, we expected that particles with higher branch numbers would sediment farther in the sucrose gradient. Although the distribution of both sizes and shapes leads to some competition in the sedimentation rate, representative TEM images of the sorted DNA-Gd@stars (Figure 2a bottom, Figure S1) revealed improved particle homogeneity. We note that the reproducibility and scalability of this technique is demonstrated by the uniformity of the fractions, which were combined from 26 separate rounds of DGC. The structural analysis thus incorporates the distribution of nanostars that sedimented the same distance in each sucrose gradient. To compare the shape distributions quantitatively, the branch number was manually counted on at least 400 particles per fraction on TEM images taken from several areas on the grid (Supporting Information). We defined different populations of interest based on branch number: particles with 0–2 branches are defined as “few-branched” population, and particles with ≥6 branches are defined as “many-branched” population. Nanoparticles with 3–5 branches are not discussed in detail here since these populations are mostly evenly distributed in all fractions. Few-branched particles resemble spheres (0 branches) and rods (1–2 branches), which primarily contain regions of positive and neutral surface curvature. In contrast, many-branched particles contain several regions of negative curvature between adjacent branches. Within each fraction, TEM images were analyzed to determine various structural features such as average size (Feret diameter, mass) and the percentage of many- and few-branched particles (Figure 2c, Table S1).
Unsorted DNA-Gd@stars had a relatively even distribution of branch number populations: the few-branched particles made up 27% of the batch and the many-branched particles made up 31%. In general, the average size and percentage of many-branched particles increased while the few-branched particles decreased with increasing fraction number (sedimentation distance). Fractions #22–24 showed the highest percentage of many-branched particles, which was nearly double that of unsorted DNA-Gd@stars. The maximum percentage of few-branched particles was achieved in fractions #6–12. Near the bottom of the nanostar band (fractions #27–28), the structural uniformity began to decrease (i.e., decreased ratio of many- to few-branched particles relative to fractions #22–24, Table S1). This trend can be explained by the competition between mass and shape since the average particle mass began to level off between fractions #20–28 (Table S1). This evolution of shape population was also confirmed by monitoring the ensemble optical properties of each DNA-Gd@star fraction (Figure S2).
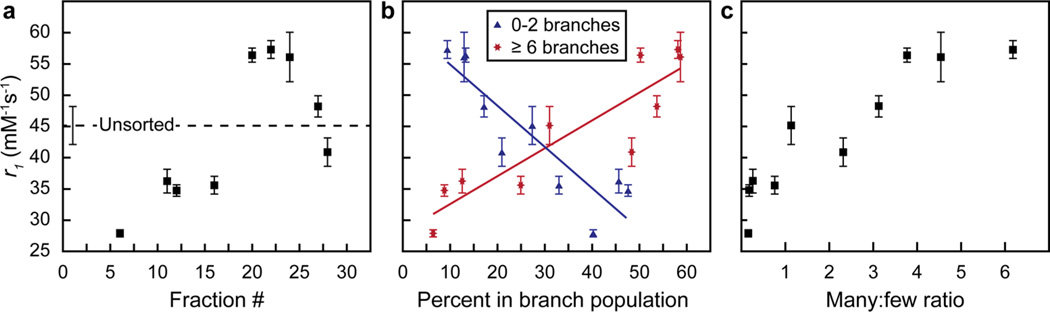
To correlate this structural analysis of the refined DNA-Gd@star populations with relaxivity, r1 was measured (37 °C, 1.41 T) on each sorted fraction and the unsorted control (Figure 3a, Figure S7). The r1 of the unsorted DNA-Gd@stars (post-sucrose functionalized) was 45.2 mM−1 s−1 (Table S1) and reflects contributions from the broad distribution of nanostar shapes. This r1 is within the standard deviation of our previously measured DNA-Gd@stars.23 After structural refinement via DGC, the relaxivity of DNA-Gd@stars increased as the percentage of many-branched particles increased, and decreased as the few-branched particles increased (Figure 3b). In particular, the fraction with the highest percentage of many-branched particles showed over a 2-fold increase in r1 (57.3 mM−1 s−1, #22) compared to the fraction with the lowest percentage (27.9 mM−1 s−1, #6) (Figure 3a). When considering only one of the branch populations, however, some deviations in this trend emerged. For example, the relaxivities of fractions #27–28 were lower than that of fraction #20 despite having a similar percentage of many-branched particles. We hypothesize that the slight increase in few-branched particles in fractions #27–28 was responsible for bringing down the ensemble r1 of these fractions. Therefore, we also analyzed the relationship between the ratio between many- to few-branched nanostars (many:few) and the relaxivity to take into account how the mixture of both populations might contribute to the ensemble r1 of each fraction (Figure 3C). A clear positive trend emerged between the many:few branch ratio and r1, which further supported our hypothesis that nanostar shape influences the relaxivity of conjugated Gd(III)-DNA.
Figure 3.
Distribution of many- and few-branched gold nanostars influences the relaxivity of Gd(III)-DNA. r1, measured at 37 °C and 1.41 T (60 MHz), (a) for each fraction, (b) as a function of the percentage of many- or few-branched DNA-Gd@stars (solid lines: linear fit), and (c) as a function of the ratio of many:few-branched particles. Error bars are the standard deviation from two r1 measurements of each sample.
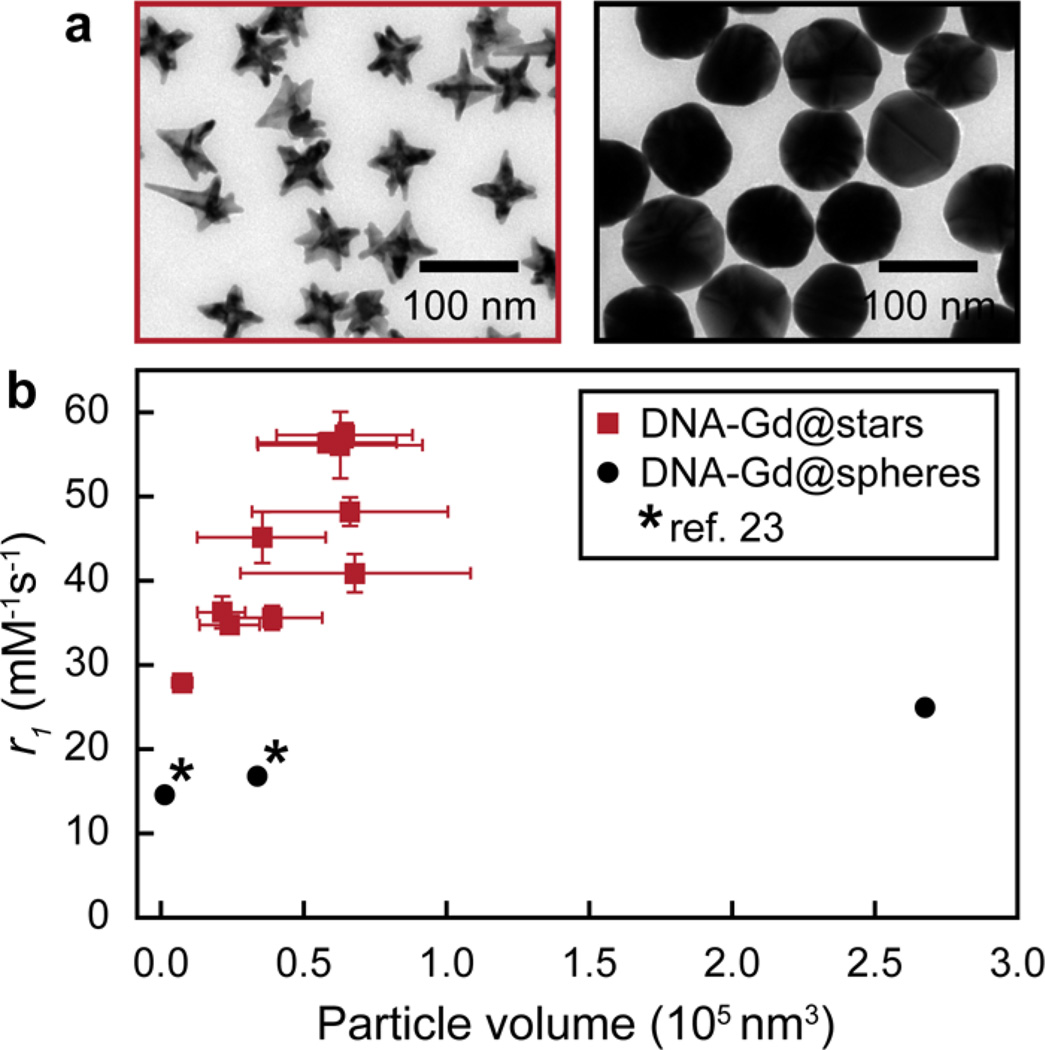
Since NP size is typically exploited to enhance r1 of conjugated Gd(III) chelates based on elongation of τr31,37,41,46 and because DGC separates particles by both shape and size,12,15 we needed to confirm that the r1 trends were not simply based on NP size. Figure 4 shows the stars fraction (red) compared to the analogous DNA-Gd@spheres of different sizes (black). We measured the r1 of large (80 nm diameter) spherical Au NPs after conjugating with Gd(III)-DNA. The r1 of 15- and 40 nm DNA-Gd@spheres was previously reported.23 The average nanostar volumes and standard deviation were determined from analysis of over 275 particles in TEM images (Supporting Information). The volumes of the spherical NPs were calculated based on the diameters reported by the manufacturers. TEM images of sorted DNA-Gd@stars (fraction #22) and 80 nm DNA-Gd@spheres are shown at the same scale in Figure 4a to visually compare the NP sizes and shapes.
Figure 4.
NP size is not the primary factor influencing r1 of DNA-Gd@star relaxivity. (a) TEM images of fraction #22 of DNA-Gd@stars (red) and 80 nm DNA-Gd@spheres (black). (b) Relationship between NP volume and r1 of DNA-Gd@stars and DNA-Gd@spheres. Error bars (x) represent the standard deviation of average NP volumes within each fraction. Error bars (y) are the standard deviation from two r1 measurements of each sample. * ref 23.
As expected, the DNA-Gd@spheres showed a modest increase in r1 (14.623 to 25.2 mM−1 s−1) with a 150-fold increase in volume (Figure 4b, Table S1). In contrast, a clear trend between size and r1 was not observed for DNA-Gd@stars. For example, the volumes of DNA-Gd@stars fractions #22–28 are similar (no statistical difference), yet the r1 ranges from 40.9 to 57.3 mM−1 s−1. Importantly, the relaxivities of all DNA-Gd@stars fractions were larger than those of DNA-Gd@spheres of any size (Figure 4b, Table S1), indicating that even few-branched nanostars could enhance the relaxivity of surface-bound Gd(III). These results support our interpretation that nanostar branch number, rather than size, is primarily responsible for influencing the r1 of DNA-Gd@stars.
Finally, for NP-based Gd(III) contrast agents, there are clear differences between the ionic (per-Gd(III)) r1 and per-particle r1. The ionic r1 is the standard way to report contrast agent performance since it reflects the average relaxation efficiency of each Gd(III) chelate and is neither a function of NP surface area nor Gd(III) loading. The per-particle r1 is often reported to encompass the high Gd(III) payload that can be achieved in or on NPs.58–60 In this manuscript, we exclusively discussed the ionic r1 to demonstrate how NP shape fundamentally influences the performance of surface-bound Gd(III) chelates. These results are significant because NP shape was not previously expected to affect the ionic r1 of T1 MRI contrast agents.
CONCLUSION
In conclusion, we have characterized the structure–property relationship between relaxivity and shape distribution of many-and few-branched DNA-Gd@stars and found that relaxivity increases with the percentage of many-branched particles. This study was enabled by a sucrose-based DGC separation protocol that maintains compatibility with subsequent biomolecule surface conjugation. The r1 of the population enriched in many-branched DNA-Gd@stars reached levels as high as 57.3 mM−1 s−1. Importantly, this relaxivity is 19-fold higher than that of clinical Gd(III) agent, ProHance61 and is among the highest reported for NP-23,45 or protein-bound35,52,62 Gd(III) chelates containing only one inner-sphere water molecule (q = 1). Furthermore, the relaxivity trends as a function of shape support the principle that negative curvature between branches facilitates a long-lived second hydration sphere surrounding the Gd(III)-DNA, leading to enhanced relaxivity. Thus, shape is a new parameter that can be tuned in the development of NP-based T1 MRI contrast agents. We anticipate that these sucrose-based DGC methods can also improve the performance of functionalized anisotropic nanoconjugates in a variety of other applications such as lowering detection limits for sensors and diagnostics or enabling new modes of self-assembly.
Supplementary Material
Acknowledgments
This work was supported by NSF (CHE-1507790; NU MRSEC DMR-1121262), and NCI CCNE initiative at NU (U54CA151880). K.S.B.C. was supported by the DoD through the NDSEG fellowship (32 CFR 168a). TEM was performed at the NU Biological Imaging Facility. Metal analysis was performed at the NU Quantitative Bio-Element Imaging Center supported by NASA Ames Research Center (NNA06CB93G). M.W.R. thanks the Mirkin Lab for the use of lab equipment for DNA synthesis and purification.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Experimental methods, additional results and discussion, Table S1, Figures S1–S8 (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Li N, Zhao P, Astruc D. Anisotropic Gold Nanoparticles: Synthesis, Properties, Applications, and Toxicity. Angew. Chem., Int. Ed. 2014;53:1756–1789. doi: 10.1002/anie.201300441. [DOI] [PubMed] [Google Scholar]
- 2.Burda C, Chen X, Narayanan R, El-Sayed MA. Chemistry and Properties of Nanocrystals of Different Shapes. Chem. Rev. 2005;105:1025–1102. doi: 10.1021/cr030063a. [DOI] [PubMed] [Google Scholar]
- 3.Dreaden EC, Alkilany AM, Huang X, Murphy CJ, El-Sayed MA. The Golden Age: Gold Nanoparticles for Biomedicine. Chem. Soc. Rev. 2012;41:2740–2779. doi: 10.1039/c1cs15237h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgin J, Liu M, Guyot-Sionnest P. Dielectric Sensing with Deposited Gold Bipyramids. J. Phys. Chem. C. 2008;112:19279–19282. [Google Scholar]
- 5.Ye X, Gao Y, Chen J, Reifsnyder DC, Zheng C, Murray CB. Seeded Growth of Monodisperse Gold Nanorods Using Bromide-Free Surfactant Mixtures. Nano Lett. 2013;13:2163–2171. doi: 10.1021/nl400653s. [DOI] [PubMed] [Google Scholar]
- 6.Nikoobakht B, El-Sayed MA. Preparation and Growth Mechanism of Gold Nanorods (NRs) Using Seed-Mediated Growth Method. Chem. Mater. 2003;15:1957–1962. [Google Scholar]
- 7.Ye X, Jin L, Caglayan H, Chen J, Xing G, Zheng C, Doan-Nguyen V, Kang Y, Engheta N, Kagan CR, et al. Improved Size-Tunable Synthesis of Monodisperse Gold Nanorods through the Use of Aromatic Additives. ACS Nano. 2012;6:2804–2817. doi: 10.1021/nn300315j. [DOI] [PubMed] [Google Scholar]
- 8.Liu M, Guyot-Sionnest P. Mechanism of Silver(I)-Assisted Growth of Gold Nanorods and Bipyramids. J. Phys. Chem. B. 2005;109:22192–22200. doi: 10.1021/jp054808n. [DOI] [PubMed] [Google Scholar]
- 9.Walker DA, Leitsch EK, Nap RJ, Szleifer I, Grzybowski BA. Geometric Curvature Controls the Chemical Patchiness and Self-Assembly of Nanoparticles. Nat. Nanotechnol. 2013;8:676–681. doi: 10.1038/nnano.2013.158. [DOI] [PubMed] [Google Scholar]
- 10.Wang D, Nap RJ, Lagzi I, Kowalczyk B, Han S, Grzybowski BA, Szleifer I. How and Why Nanoparticle’s Curvature Regulates the Apparent pKa of the Coating Ligands. J. Am. Chem. Soc. 2011;133:2192–2197. doi: 10.1021/ja108154a. [DOI] [PubMed] [Google Scholar]
- 11.Sherry LJ, Jin R, Mirkin CA, Schatz GC, Van Duyne RP. Localized Surface Plasmon Resonance Spectroscopy of Single Silver Triangular Nanoprisms. Nano Lett. 2006;6:2060–2065. doi: 10.1021/nl061286u. [DOI] [PubMed] [Google Scholar]
- 12.Shin YJ, Ringe E, Personick ML, Cardinal MF, Mirkin CA, Marks LD, Van Duyne RP, Hersam MC. Centrifugal Shape Sorting and Optical Response of Polyhedral Gold Nanoparticles. Adv. Mater. 2013;25:4023–4027. doi: 10.1002/adma.201301278. [DOI] [PubMed] [Google Scholar]
- 13.Tyler TP, Henry A-I, Van Duyne RP, Hersam MC. Improved Monodispersity of Plasmonic Nanoantennas via Centrifugal Processing. J. Phys. Chem. Lett. 2011;2:218–222. [Google Scholar]
- 14.Tyler TP, Lin PA, Tian Y, Gao H-J, Gao XPA, Sankaran RM, Hersam MC. Centrifugal Shape Sorting of Faceted Gold Nanoparticles Using an Atomic Plane-Selective Surfactant. J. Phys. Chem. Lett. 2012;3:1484–1487. doi: 10.1021/jz3005283. [DOI] [PubMed] [Google Scholar]
- 15.Akbulut O, Mace CR, Martinez RV, Kumar AA, Nie Z, Patton MR, Whitesides GM. Separation of Nanoparticles in Aqueous Multiphase Systems through Centrifugation. Nano Lett. 2012;12:4060–4064. doi: 10.1021/nl301452x. [DOI] [PubMed] [Google Scholar]
- 16.Xiong B, Cheng J, Qiao Y, Zhou R, He Y, Yeung ES. Separation of Nanorods by Density Gradient Centrifugation. J. Chromatogr. A. 2011;1218:3823–3829. doi: 10.1016/j.chroma.2011.04.038. [DOI] [PubMed] [Google Scholar]
- 17.Sun X, Tabakman SM, Seo W-S, Zhang L, Zhang G, Sherlock S, Bai L, Dai H. Separation of Nanoparticles in a Density Gradient: FeCo@C and Gold Nanocrystals. Angew. Chem., Int. Ed. 2009;48:939–942. doi: 10.1002/anie.200805047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ko SH, Vargas-Lara F, Patrone PN, Stavis SM, Starr FW, Douglas JF, Liddle JA. High-Speed, High-Purity Separation of Gold Nanoparticle-DNA Origami Constructs Using Centrifugation. Soft Matter. 2014;10:7370–7378. doi: 10.1039/c4sm01071j. [DOI] [PubMed] [Google Scholar]
- 19.Fruhnert M, Kretschmer F, Geiss R, Perevyazko I, Cialla-May D, Steinert M, Janunts N, Sivun D, Hoeppener S, Hager MD, et al. Synthesis, Separation, and Hypermethod Characterization of Gold Nanoparticle Dimers Connected by a Rigid Rod Linker. J. Phys. Chem. C. 2015;119:17809–17817. [Google Scholar]
- 20.Wang Y, Aili D, Selegard R, Tay Y, Baltzer L, Zhang H, Liedberg B. Specific Functionalization of CTAB Stabilized Anisotropic Gold Nanoparticles with Polypeptides for Folding-Mediated Self-Assembly. J. Mater. Chem. 2012;22:20368–20373. [Google Scholar]
- 21.Khanal BP, Zubarev ER. Rings of Nanorods. Angew. Chem. Int., Ed. 2007;46:2195–2198. doi: 10.1002/anie.200604889. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi H, Niidome Y, Niidome T, Kaneko K, Kawasaki H, Yamada S. Modification of Gold Nanorods Using Phosphatidylcholine to Reduce Cytotoxicity. Langmuir. 2006;22:2–5. doi: 10.1021/la0520029. [DOI] [PubMed] [Google Scholar]
- 23.Rotz MW, Culver KSB, Parigi G, MacRenaris KW, Luchinat C, Odom TW, Meade TJ. High Relaxivity Gd(III)–DNA Gold Nanostars: Investigation of Shape Effects on Proton Relaxation. ACS Nano. 2015;9:3385–3396. doi: 10.1021/nn5070953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dam DHM, Lee H, Lee RC, Kim KH, Kelleher NL, Odom TW. Tunable Loading of Oligonucleotides with Secondary Structure on Gold Nanoparticles through a pH-Driven Method. Bioconjugate Chem. 2015;26:279–285. doi: 10.1021/bc500562s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dam DHM, Lee RC, Odom TW. Improved in Vitro Efficacy of Gold Nanoconstructs by Increased Loading of G-Quadruplex Aptamer. Nano Lett. 2014;14:2843–2848. doi: 10.1021/nl500844m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Puig H, Tam JO, Yen C-W, Gehrke L, Hamad-Schifferli K. Extinction Coefficient of Gold Nanostars. J. Phys. Chem. C. 2015;119:17408–17415. doi: 10.1021/acs.jpcc.5b03624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solomon I. Relaxation Processes in a System of Two Spins. Phys. Rev. 1955;99:559–565. [Google Scholar]
- 28.Bloembergen N, Morgan LO. Proton Relaxation Times in Paramagnetic Solutions. Effects of Electron Spin Relaxation. J. Chem. Phys. 1961;34:842–850. [Google Scholar]
- 29.Caravan P, Ellison JJ, McMurry TJ, Lauffer RB. Gadolinium(III) Chelates as MRI Contrast Agents: Structure, Dynamics, and Applications. Chem. Rev. 1999;99:2293–2352. doi: 10.1021/cr980440x. [DOI] [PubMed] [Google Scholar]
- 30.Caravan P, Farrar CT, Frullano L, Uppal R. Influence of Molecular Parameters and Increasing Magnetic Field Strength on Relaxivity of Gadolinium- and Manganese-Based T1 Contrast Agents. Contrast Media Mol. Imaging. 2009;4:89–100. doi: 10.1002/cmmi.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruckman MA, Yu X, Steinmetz NF. Engineering Gd-Loaded Nanoparticles to Enhance MRI Sensitivity via T1 Shortening. Nanotechnology. 2013;24:462001. doi: 10.1088/0957-4484/24/46/462001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manus LM, Strauch RC, Hung AH, Eckermann AL, Meade TJ. Analytical Methods for Characterizing Magnetic Resonance Probes. Anal. Chem. 2012;84:6278–6287. doi: 10.1021/ac300527z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hermann P, Kotek J, Kubíček V, Lukeš I. Gadolinium (III) Complexes as MRI Contrast Agents: Ligand Design and Properties of the Complexes. Dalton Trans. 2008:3027–3047. doi: 10.1039/b719704g. [DOI] [PubMed] [Google Scholar]
- 34.Xu J, Franklin SJ, Whisenhunt DW, Raymond KN. Gadolinium Complex of Tris[(3-Hydroxy-1-Methyl- 2-Oxo-1,2-Didehydropyridine-4-Carboxamido)Ethyl]-Amine: A New Class of Gadolinium Magnetic Resonance Relaxation Agents. J. Am. Chem. Soc. 1995;117:7245–7246. [Google Scholar]
- 35.Caravan P, Parigi G, Chasse JM, Cloutier NJ, Ellison JJ, Lauffer RB, Luchinat C, McDermid SA, Spiller M, McMurry TJ. Albumin Binding, Relaxivity, and Water Exchange Kinetics of the Diastereoisomers of MS-325, a Gadolinium(III)-Based Magnetic Resonance Angiography Contrast Agent. Inorg. Chem. 2007;46:6632–6639. doi: 10.1021/ic700686k. [DOI] [PubMed] [Google Scholar]
- 36.Prasuhn DE, Yeh RM, Obenaus A, Manchester M, Finn MG. Viral MRI Contrast Agents: Coordination of Gd by Native Virions and Attachment of Gd Complexes by Azide-Alkyne Cycloaddition. Chem. Commun. 2007:1269–1271. doi: 10.1039/b615084e. [DOI] [PubMed] [Google Scholar]
- 37.Song Y, Xu X, MacRenaris KW, Zhang X-Q, Mirkin CA, Meade TJ. Multimodal Gadolinium-Enriched DNA–Gold Nanoparticle Conjugates for Cellular Imaging. Angew. Chem., Int. Ed. 2009;48:9143–9147. doi: 10.1002/anie.200904666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caravan P. Protein-Targeted Gadolinium-Based Magnetic Resonance Imaging (MRI) Contrast Agents: Design and Mechanism of Action. Acc. Chem. Res. 2009;42:851–862. doi: 10.1021/ar800220p. [DOI] [PubMed] [Google Scholar]
- 39.Bruckman MA, Hern S, Jiang K, Flask CA, Yu X, Steinmetz NF. Tobacco Mosaic Virus Rods and Spheres as Supramolecular High-Relaxivity MRI Contrast Agents. J. Mater. Chem. B. 2013;1:1482–1490. doi: 10.1039/C3TB00461A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liepold L, Anderson S, Willits D, Oltrogge L, Frank JA, Douglas T, Young M. Viral Capsids as MRI Contrast Agents. Magn. Reson. Med. 2007;58:871–879. doi: 10.1002/mrm.21307. [DOI] [PubMed] [Google Scholar]
- 41.Villaraza AJL, Bumb A, Brechbiel MW. Macromolecules, Dendrimers, and Nanomaterials in Magnetic Resonance Imaging: The Interplay between Size, Function, and Pharmacokinetics. Chem. Rev. 2010;110:2921–2959. doi: 10.1021/cr900232t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kielar F, Tei L, Terreno E, Botta M. Large Relaxivity Enhancement of Paramagnetic Lipid Nanoparticles by Restricting the Local Motions of the GdIII Chelates. J. Am. Chem. Soc. 2010;132:7836–7837. doi: 10.1021/ja101518v. [DOI] [PubMed] [Google Scholar]
- 43.Lee S-M, Song Y, Hong BJ, MacRenaris KW, Mastarone DJ, O’Halloran TV, Meade TJ, Nguyen ST. Modular Polymer-Caged Nanobins as a Theranostic Platform with Enhanced Magnetic Resonance Relaxivity and pH-Responsive Drug Release. Angew. Chem. 2010;122:10156–10160. doi: 10.1002/anie.201004867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Endres PJ, Paunesku T, Vogt S, Meade TJ, Woloschak GE. DNA–Tio2 Nanoconjugates Labeled with Magnetic Resonance Contrast Agents. J. Am. Chem. Soc. 2007;129:15760–15761. doi: 10.1021/ja0772389. [DOI] [PubMed] [Google Scholar]
- 45.Courant T, Roullin VG, Cadiou C, Callewaert M, Andry MC, Portefaix C, Hoeffel C, de Goltstein MC, Port M, Laurent S, et al. Hydrogels Incorporating GdDOTA: Towards Highly Efficient Dual T1/T2MRI Contrast Agents. Angew. Chem., Int. Ed. 2012;51:9119–9122. doi: 10.1002/anie.201203190. [DOI] [PubMed] [Google Scholar]
- 46.Botta M, Tei L. Relaxivity Enhancement in Macromolecular and Nanosized Gd(III)-Based MRI Contrast Agents. Eur. J. Inorg. Chem. 2012;2012:1945–1960. [Google Scholar]
- 47.Na HB, Hyeon T. Nanostructured T1MRI Contrast Agents. J. Mater. Chem. 2009;19:6267–6273. [Google Scholar]
- 48.Yang C-T, Padmanabhan P, Gulyás BZ. Gadolinium(III) Based Nanoparticles for T1-Weighted Magnetic Resonance Imaging Probes. RSC Adv. 2016;6:60945–60966. [Google Scholar]
- 49.Aime S, Botta M, Fasano M, Terreno E. Prototropic and Water-Exchange Processes in Aqueous Solutions of Gd(III) Chelates. Acc. Chem. Res. 1999;32:941–949. [Google Scholar]
- 50.Jacques V, Dumas S, Sun W-C, Troughton JS, Greenfield MT, Caravan P. High Relaxivity MRI Contrast Agents Part 2: Optimization of Inner- and Second-Sphere Relaxivity. Invest. Radiol. 2010;45:613–624. doi: 10.1097/RLI.0b013e3181ee6a49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aime S, Botta M, Fasano M, Crich SG, Terreno E. Gd(III) Complexes as Contrast Agents for Magnetic Resonance Imaging: A Proton Relaxation Enhancement Study of the Interaction with Human Serum Albumin. JBIC, J. Biol. Inorg. Chem. 1996;1:312–319. [Google Scholar]
- 52.Aime S, Frullano L, Geninatti Crich S. Compartmentalization of a Gadolinium Complex in the Apoferritin Cavity: A Route to Obtain High Relaxivity Contrast Agents for Magnetic Resonance Imaging. Angew. Chem., Int. Ed. 2002;41:1017–1019. doi: 10.1002/1521-3773(20020315)41:6<1017::aid-anie1017>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 53.Ananta JS, Godin B, Sethi R, Moriggi L, Liu X, Serda RE, Krishnamurthy R, Muthupillai R, Bolskar RD, Helm L, et al. Geometrical Confinement of Gadolinium-Based Contrast Agents in Nanoporous Particles Enhances T1 Contrast. Nat. Nanotechnol. 2010;5:815–821. doi: 10.1038/nnano.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao Z, Zhou Z, Bao J, Wang Z, Hu J, Chi X, Ni K, Wang R, Chen X, Chen Z, et al. Octapod Iron Oxide Nanoparticles as High-Performance T2 Contrast Agents for Magnetic Resonance Imaging. Nat. Commun. 2013;4:2266. doi: 10.1038/ncomms3266. [DOI] [PubMed] [Google Scholar]
- 55.Mohapatra J, Mitra A, Tyagi H, Bahadur D, Aslam M. Iron Oxide Nanorods as High-Performance Magnetic Resonance Imaging Contrast Agents. Nanoscale. 2015;7:9174–9184. doi: 10.1039/c5nr00055f. [DOI] [PubMed] [Google Scholar]
- 56.Lee S, Salunke B, Kim B. Sucrose Density Gradient Centrifugation Separation of Gold and Silver Nanoparticles Synthesized Using Magnolia Kobus Plant Leaf Extracts. Biotechnol. Bioprocess Eng. 2014;19:169–174. [Google Scholar]
- 57.Chen G, Wang Y, Tan LH, Yang M, Tan LS, Chen Y, Chen H. High-Purity Separation of Gold Nanoparticle Dimers and Trimers. J. Am. Chem. Soc. 2009;131:4218–4219. doi: 10.1021/ja900809z. [DOI] [PubMed] [Google Scholar]
- 58.Turner JL, Pan D, Plummer R, Chen Z, Whittaker AK, Wooley KL. Synthesis of Gadolinium-Labeled Shell-Crosslinked Nanoparticles for Magnetic Resonance Imaging Applications. Adv. Funct. Mater. 2005;15:1248–1254. [Google Scholar]
- 59.Huang C-H, Tsourkas A. Gd-Based Macromolecules and Nanoparticles as Magnetic Resonance Contrast Agents for Molecular Imaging. Curr. Top. Med. Chem. 2013;13:411–421. doi: 10.2174/1568026611313040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun H, Yuan Q, Zhang B, Ai K, Zhang P, Lu L. GdIII Functionalized Gold Nanorods for Multimodal Imaging Applications. Nanoscale. 2011;3:1990–1996. doi: 10.1039/c0nr00929f. [DOI] [PubMed] [Google Scholar]
- 61.Mastarone DJ, Harrison VSR, Eckermann AL, Parigi G, Luchinat C, Meade TJ. A Modular System for the Synthesis of Multiplexed Magnetic Resonance Probes. J. Am. Chem. Soc. 2011;133:5329–5337. doi: 10.1021/ja1099616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Min J, Jung H, Shin H-H, Cho G, Cho H, Kang S. Implementation of P22 Viral Capsids as Intravascular Magnetic Resonance T1 Contrast Conjugates via Site-Selective Attachment of Gd(III)-Chelating Agents. Biomacromolecules. 2013;14:2332–2339. doi: 10.1021/bm400461j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.