Abstract
The essential human enzyme O-GlcNAc transferase (OGT), known for modulating the functions of nuclear and cytoplasmic proteins through Ser/Thr glycosylation, was unexpectedly implicated in the proteolytic maturation of the cell cycle regulator host cell factor-1 (HCF-1). Here we show that HCF-1 cleavage occurs via glycosylation of a glutamate side chain followed by on-enzyme formation of an internal pyroglutamate, which undergoes spontaneous backbone hydrolysis.
Host cell factor-1 is an essential chromatin-associated protein important in cell cycle control.1,2 It contains six centrally located 26 amino acid proteolytic repeats, each consisting of a threonine-rich region and a region that undergoes cleavage (Figure 1a).3–5 Proper cell cycle regulation requires HCF-1 cleavage.6,7 The protease responsible for cleaving HCF-1 went unidentified for many years until studies implicated O-GlcNAc transferase (OGT) in the process.3,8 OGT is an essential nucleocytoplasmic glycosyltransferase best known for its role as a nutrient sensor.9–11 The discovery that OGT is involved in HCF-1 processing raised questions about how a glycosyltransferase might promote amide bond hydrolysis.
Figure 1.
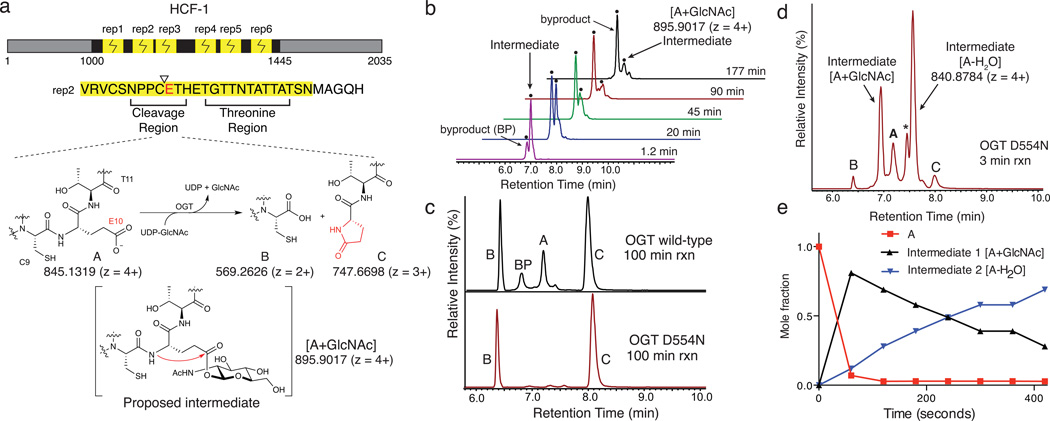
Two intermediates form during OGT-mediated cleavage of an HCF-1 repeat. (a) HCF-1 contains six similar proteolytic repeats that are cleaved between residues C9 and E10, producing a C-terminal pyroglutamate product, C, that was proposed to form via the intermediacy of a glycosyl ester.12 (b) LC traces showing EICs for m/z 895.9017 ± 5 ppm, corresponding to A+GlcNAc, from the time course shown in Supplementary Figure 1. The y-axis scaling is such that the tallest peak is 100% (axis not shown). Two peaks having the same exact mass were observed, one of which is a reaction intermediate while the other is a byproduct glycopeptide. (c) EICs for cleavage reactions of HCF-short with wild-type OGT (top) and OGTD554N (bottom) showing A, B, C and [A+GlcNAc]. (d) An LC trace showing a 3 minute time point for cleavage of HCF-short using OGTD554N. Peaks for A, B, C, the [A+GlcNAc] intermediate, and another intermediate, [A-H2O], are shown. The ‘*’ denotes a second peak having the same m/z as the [A-H2O] species. The full time course is shown in Supplementary Figure 5. (e) The temporal sequence of events was established from a time course of cleavage using a large excess of OGTD554N. Mole fractions of A, [A+GlcNAc] and [A-H2O], were plotted as a function of time. The data was truncated at 420 seconds, such that products represent a negligible mole fraction. Mole fractions were determined based on the maximal signal for each species, normalized over all species at that time point.
OGT cleavage of HCF-1 gives a product containing an N-terminal pyroglutamate (Figure 1a),12 implying that the cleavage mechanism involves an intermediate in which the glutamate side chain is activated, e.g., through formation of an ester. As UDP-GlcNAc is required for cleavage,3,12 but a non-hydrolyzable analog inhibits cleavage,12 we proposed that OGT glycosylates the glutamate side chain. A mechanism involving glutamate glycosylation would serve to link HCF-1 cleavage to nutrient status through the abundance of UDP-GlcNAc. Other enzymes that use cellular metabolites as co-substrates in amide bond cleavage reactions have been described. For example, sirtuins use NAD+ to activate the acetyl-lysine amide bond, thus linking cellular nutrition to protein deacetylation.13,14 However, we were not aware of any nucleotide-sugar glycosyltransferases that catalyzed glycosylation on glutamate, and we did not have direct evidence for this intermediate or for other intermediates that would illuminate the cleavage mechanism.
In order to identify reaction intermediates, we required a cleavage-competent fragment of HCF-1 that was short enough to enable LC-MS/MS studies. A canonical 26 amino acid repeat did not undergo cleavage,15 but we found that a single repeat with a five amino acid C-terminal extension, hereafter called HCF-short, was a competent substrate. We monitored cleavage of this substrate as a function of time by high-resolution Fourier-transform mass spectrometry (FT-MS) and found that the reaction was complete within three hours (Supplementary Results, Supplementary Figures 1 and 2). The LC-MS traces revealed two low abundance peaks having a m/z of 895.9017 (4+), consistent with a glycopeptide [A+GlcNAc] (Figure 1b). One of these peaks formed earlier in the reaction and decayed over time, suggesting it was a glycopeptide intermediate; the other formed more gradually, but increased throughout the cleavage reaction, suggesting it was a glycopeptide byproduct (BP) that prevented backbone cleavage. Glycosylation on T11, the threonine adjacent to the glutamate, or on one of several threonines in the region of the peptide that binds to the tetratricopeptide (TPR) domain of OGT, could prevent cleavage by hindering glycosylation at E10. As a glycopeptide byproduct still formed when we used a HCF-short-T11V peptide, we wondered whether it would be possible to minimize glycopeptide byproduct formation using an OGT mutant impaired for Ser/Thr glycosylation, but capable of promoting cleavage. A D554H/H558D double mutant with these characteristics has been reported,15 and other studies have shown that D554 plays an important role in glycosylation.9,16,17
We tested the ability of an OGTD554N single mutant to catalyze Ser/Thr glycosylation and to effect peptide backbone cleavage using two nearly identical 151 residue polypeptides containing a single HCF-1 proteolytic repeat.12 In one polypeptide, the glutamate in the proteolytic repeat was replaced with serine. Compared to wild-type OGT, OGTD554N was dramatically impaired in its ability to glycosylate the serine-containing polypeptide; however, both enzymes cleaved the glutamate-containing polypeptide (Supplementary Figure 3). We tested the ability of OGTD554N to cleave HCF-short and found that the reaction was complete within 100 minutes, and we did not detect any glycopeptide byproduct (Figure 1c). As both the wild-type and mutant enzymes had the same cleavage requirements (Supplementary Figure 4) and gave the same products, we used OGTD554N for mechanistic studies.
We monitored OGTD554N cleavage of HCF-short over time and found that the starting material was almost entirely consumed within the first three minutes of the reaction, and two intermediates had accumulated to high levels (Figure 1d; Supplementary Figures 5–11). One of these had the same retention time and exact mass as the glycopeptide intermediate identified in the wild-type OGT cleavage reaction (Figure 1b). The other had an m/z of 840.8784 (4+), corresponding to loss of water from starting material. This species appeared as two peaks in the EIC trace (Figure 1d). We compared MS/MS fragmentation patterns for the anhydropeptide species and, except for minor differences in fragment ion intensities, they appeared identical. The fragmentation data localized the dehydration to the C9E10 region of the peptides (Supplementary Figures 12 and 13, Supplementary Tables 1–6). We detected the same anhydropeptide species in cleavage reactions with wild-type enzyme, but it accumulated to lower levels (compare Supplementary Figures 1 and 5). The LC-MS/MS data for the anhydropeptide species were consistent with formation of an internal pyroglutamate (Figure 1a), but we were not sure why there were two peaks. To rule out the possibility that one was a thiolactone resulting from cysteine participation (Supplementary Figure 14),12,18–20 we replaced Cys9 in the HCF-short peptide with Arg, an amino acid found in a proteolytic repeat from the Odobenus rosmarus divergens (walrus) HCF-1 ortholog. HCF-short-C9R was a good cleavage substrate and we again observed two peaks corresponding to an anhydropeptide intermediate in reactions with both wild-type OGT and OGTD554N (Supplementary Figures 15 and 16). We also observed two peaks for the HCF-short-T11V peptide (Supplementary Figure 15). Because two anhydropeptides are observed for three cleavage substrates having different sequences, and because the fragment ions for the two species are identical, we speculate that they are different conformational isomers that are resolvable by LC.21
To establish the temporal sequence of events, we repeated the time course using excess OGTD554N to ensure single-turnover conditions. Within 60 seconds the starting material had converted to the glycopeptide intermediate. As this species decayed, one anhydropeptide species appeared (Figure 1e; Supplementary Figures 7 and 8), and the second grew in as the reaction progressed. Both species were consumed by the end of the reaction, indicating that both are on-pathway to cleavage products. Hence, the reaction involves initial formation of a glycopeptide intermediate (Intermediate 1) followed by conversion to an internal pyroglutamate (Intermediate 2) and then backbone cleavage. We measured a rate constant of ~0.08 s−1 for the consumption of A by OGTD554N, which is approximately two orders of magnitude faster than for the wild-type enzyme (compare Supplementary Figures 2 and 8). Hence, replacement of aspartate for asparagine at position 554 dramatically accelerated formation of the glycopeptide intermediate. Overall cleavage rates for wild-type OGT and OGTD554N differed only by a factor of two to three, however, evidently because hydrolysis of the internal pyroglutamate is rate determining.
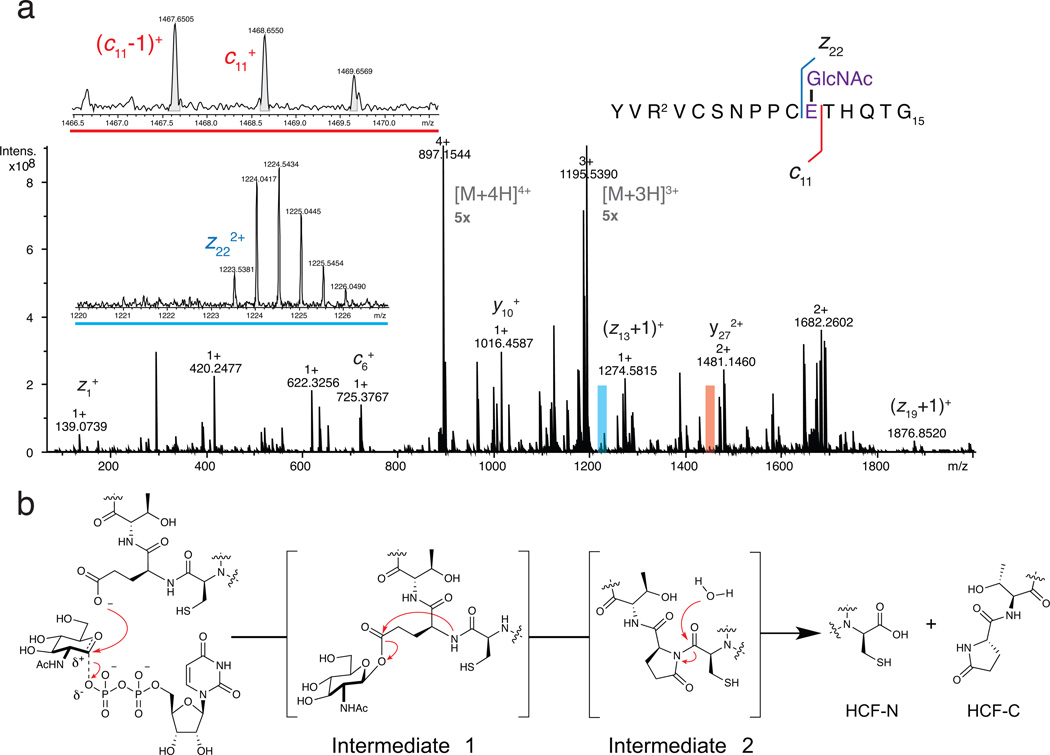
Because accelerating the first step in the reaction results in substantial accumulation of the glycopeptide intermediate, we were able to obtain sufficient material for MS/MS analysis by quenching the OGTD554N cleavage reaction at an early time point. The intermediate proved stable enough to allow for LC purification, and fragmentation by electron-capture dissociation (ECD) allowed unambiguous assignment of the GlcNAc modification to glutamate 10 (Figure 2a, Supplementary Figures 17 and 18, Supplementary Tables 7 and 8). While the stability of the glycosylated intermediate was unexpected, we note that glycosyl esters produced by protein ADP-ribosylation are also sufficiently stable to allow isolation and characterization.22,23
Figure 2.
The mechanism of HCF-1 cleavage proceeds via glycosylation on glutamate followed by decomposition to an internal pyroglutamate, which undergoes backbone hydrolysis. (a) An ECD mass spectrum identified Intermediate 1 as a glutamate-linked glycopeptide. Insets show zooms of ions c11+ and z222+ that flank the glycosylated residue. Full spectra are presented in Supplementary Figures 17 and 18. Intermediate 2 was identified as an internal pyroglutamate from MS/MS fragments (see Supplementary Figures 12 and 13). (b) Chemical mechanism of HCF-1 cleavage by OGT. Both glycosylation on glutamate and conversion to the internal pyroglutamate (Intermediate 2) occur on the enzyme (see text and Supplementary Figures 19 and 20), but hydrolysis likely occurs after dissociation.
Our studies show that OGT effects HCF-1 amide bond cleavage by the mechanism in Figure 2b. Glycosylation on glutamate is followed by conversion to an internal pyroglutamate, which is susceptible to spontaneous hydrolysis.18–20,24 Because an internal pyroglutamate can form non-enzymatically if a glutamate side chain is activated,24 we wondered whether Intermediate 1 converts to Intermediate 2 while bound to OGT. Therefore, we isolated the glycopeptide intermediate and re-subjected it to the reaction conditions, but without OGT. We observed formation of an anhydropeptide followed by hydrolytic cleavage over 90 minutes. Because the rate of conversion of glycopeptide to internal pyroglutamate was an order of magnitude slower than observed in the presence of OGT (Supplementary Figure 19), we conclude that both the first and second steps in Figure 2b occur while the peptide is bound to OGT. Hydrolysis likely occurs after dissociation from the enzyme. As only the second-formed anhydropeptide species was observed in the non-enzymatic reaction of intermediate 2, we speculate that it is a lower energy conformer that, in the case of the enzyme-catalyzed reaction, arises after dissociation (Supplementary Figure 20).
We have shown that OGT and OGTD554N go through the same intermediates to effect amide bond cleavage, but OGTD554N catalyzes glycosylation on glutamate much faster than wild-type enzyme. It might seem curious that a mutant so impaired with respect to Ser/Thr glycosylation (Supplementary Figure 3) would be unimpaired with respect to glutamate glycosylation; however, D554 is proposed to serve as a base to shuttle the proton released from the acceptor hydroxyl out of the active site following glycosylation,9,17 and this function would not be required for glycosylation of a carboxylate. This explanation resolves a seeming paradox, but does not address how OGTD554N can catalyze glutamate glycosylation faster than the wild-type enzyme. A 2.05 Å ternary structure of OGTD554N bound to UDP and a CKII glycopeptide (see Supplementary Figure 22 and Supplementary Table 9) shows that the structure is virtually identical to that of the wild-type enzyme (0.36 Å root mean square deviation),17 implying that the difference in activity is not due to a conformational change in the protein. Instead, glutamate glycosylation may be disfavored by a repulsive interaction with the D554 side chain in wild-type OGT. In order for glutamate to attack the anomeric carbon of UDP-GlcNAc, it must approach within ~4.5 Å of the D554 carboxylate (Supplementary Figure 22d), and this unfavorable interaction is eliminated in the D554N variant.
The work reported here illuminates how the glycosyltransferase OGT catalyzes amide bond cleavage. Although the rates of cleavage of the model peptide studied here may appear slow, they are consistent with the slow rates estimated from pulse-chase data for cleavage of HCF-1 in cells.4 The relevant timescale for HCF-1 cleavage is the length of the mammalian cell cycle, and the rates of peptide cleavage measured here are fully compatible with that timescale. To the best of our knowledge, HCF-1 is the first example of a protein that undergoes a physiologically relevant backbone cleavage reaction triggered by glutamate glycosylation, but a glycosidase was recently reported to undergo in vitro cleavage and a similar mechanism was proposed.25 Hence, glutamate glycosylation may be a general way to effect amide bond hydrolysis in proteins. OGT may have other cleavage substrates, and because 1–2% of genomic space throughout all kingdoms of life is occupied by glycosyltransferases, there may exist other enzymes that catalyze glycosylation-mediated amide bond cleavage.
Online Methods
Reagents and General Methods
14C-UDP-GlcNAc was obtained from American Radiochemicals (ARC). UDP-GlcNAc was obtained from Promega as a 100 mM solution in water. THP [Tris(3-hydroxypropyl)phosphine] was obtained from Novagen as a 0.5 M solution in water. Calf alkaline phosphatase (CIP) was obtained from New England Biolabs. Peptides were obtained from Biomatik (Supplementary Table 10); their purity was evaluated by HPLC and mass spectrometry. Lyophilized peptides were dissolved in water to ~5 mM and added to reactions according to the final concentration desired. Peptide concentration for HCF-short peptides was determined by measuring A274 (λmax, tyrosine) in water, assuming an extinction coefficient of 1400 M−1 cm−1. The primary peptide used in these studies was the HCF-short, which is derived from human HCF-1 repeat 2, but contains Q rather than E at position 13. This E13Q point mutant shows no appreciable difference in rate from wild-type substrate (bearing E13), and was used so that a single system could be employed for both reaction monitoring and isotope labeling studies. Numbering starts from the second residue in the HCF-1 peptides (valine) to be consistent with numbering in the repeats.
Purification of HCF-3R constructs
HCF-3R constructs (EAA, SAA, DAA and AAA, Supplementary Table 10) were purified from E. coli BL21(DE3) as follows: LB media, supplemented with 50 µg/mL kanamycin, was warmed to 37 °C and inoculated with a 1:100 dilution of an overnight culture. Growth was monitored at OD600, and once the OD reached ~1.1 the temperature was reduced to 16 °C over 30 minutes. Overexpression was initiated by addition of IPTG to a final concentration of 0.2 mM and allowed to proceed for 16 h at this temperature. After this time, cells were harvested by pelleting at 5,000×g for 20 min (4 °C). Cell pellets were resuspended in 1×TBS (150 mM NaCl and 20 mM Tris) pH 7.4, supplemented with: 0.1 mg/mL lysozyme, 0.1 mg/mL DNAse I and 1 mM PMSF. Cells were lysed using a cell disruptor. Lysates were supplemented with imidazole to a final concentration of 40 mM and clarified by centrifugation at 15,000×g for 20 minutes (4 °C). Clarified lysates were flowed over Ni-NTA resin that had been equilibrated with 1×TBS supplemented with 40 mM imidazole and at pH 8. The flow-through was collected, and re-applied to the resin. After applying the lysate the second time, the resin was washed with 10 column volumes of 1×TBS supplemented with 50 mM imidazole, pH 8. Lastly, the protein was eluted with 4 column volumes of 1×TBS supplemented with 250 mM imidazole, pH 8. The eluate was dialyzed (twice) against 1×TBS pH 8.0 using 3.5 kDa MWCO cassettes to remove imidazole. After dialysis, samples were collected and supplemented with THP to 1 mM. Constructs were evaluated for purity by SDS-PAGE, but typically do not require additional purification. Concentrations of HCF-3R constructs were determined by DC™ (detergent compatible) protein assay (Bio-Rad). HCF-3R constructs were flash frozen and stored at −80 °C; once thawed, they were kept for up to one week at 4 °C.
Purification of ncOGT
Wild-type ncOGT used in this work corresponds to the human protein sequence spanning residues 22-1042. ncOGT D554N was prepared by Quickchange mutagenesis. hOGT4.5 (D554N), used for crystallography, was prepared as previously reported.26 For ncOGT, the construct used bears an N-terminal T7 tag followed by an octa-His and an HRV3C protease site. Sequences are provided in Supplementary Table 10. Both wild-type and D554N ncOGT were expressed and purified by the same method, described as follows: E. coli BL21(DE3) ΔArnA were grown in LB media, supplemented with 50 µg/mL kanamycin overnight. To LB pre-warmed to 37 °C, was added a 1:100 inoculum from the overnight culture. Growth was monitored at OD600, and once the OD reached ~1.1 the temperature was reduced to 16 °C over 30 minutes. Overexpression was initiated by addition of IPTG to a final concentration of 0.2 mM and allowed to proceed for 16 h at this temperature. After this time, cells were harvested by pelleting at 5,000×g for 20 min (4 °C). Cell pellets were resuspended in 1×TBS (150 mM NaCl and 20 mM Tris) pH 7.4, supplemented with: 0.1 mg/mL lysozyme, 0.1 mg/mL DNAse I and 1 mM PMSF. Cells were lysed using a cell disruptor. Lysates were supplemented with imidazole to a final concentration of 40 mM and clarified by centrifugation at 15,000×g for 20 minutes (4 °C). Clarified lysates were applied to Ni-NTA resin that had been equilibrated with 1× TBS supplemented with 40 mM imidazole and at pH 7.4. Once applied to the resin, the supernatant was incubated with end-over-end rotation for 60 minutes at 4 °C, after which the flow through was collected. The resin was washed with 10 column volumes of 1×TBS supplemented with 50 mM imidazole, pH 7.4. Lastly, the protein was eluted with 4 column volumes of 1×TBS supplemented with 250 mM imidazole, pH 7.4. The eluate was supplemented with THP to a final concentration of 1 mM. For ncOGT constructs, N-terminal tag cleavage was not required. Instead, protein was directly purified by size-exclusion chromatography (Superdex 200, GE) with an isocratic elution using 1× TBS as a mobile phase. Fractions were pooled, supplemented with THP to 1 mM and evaluated for purity by SDS-PAGE. Generally, no additional purification is required. ncOGT used in the experiments described below was quantified by nanodrop (A280) using an extinction coefficient, ε280 = 118,955 M−1 cm−1, and assumed to have a MW of 118,047 Da. OGT can be flash-frozen and stored at −80 °C. Once thawed, OGT activity does not appreciably change over a week at 4 °C. Generally, OGT from the same batch (either thawed freshly, or thawed several days prior) should always be used for experiments to be directly compared.
HCF-3R cleavage for SDS-PAGE analysis
In a 20 µL reaction volume was mixed: 1 mM UDP-GlcNAc, 1 mM THP, 100 µM HCF-3R construct, and 0.1 U CIP in a buffer of 20 mM MgCl2, 20 mM Tris and 150 mM NaCl at pH 7.4. Either ncOGT (to a final concentration of 2.5 µM) or water were added, and reactions were mixed by pipetting then incubated at 37 °C in a thermomixer to ensure uniform heating. After 6 hours, reactions were quenched by addition of an equal volume (20 µL) of 2× Laemmli loading buffer (with β-ME). Samples were boiled for 10 minutes, and spun down at 15,000×g for 10 minutes. 5 µL of sample was loaded per lane onto a 4–20% Tris-Gly gradient gel and run for 45 minutes at 200V. The gel was stained with Coomassie overnight, and destained the following day until bands were clearly visible.
HCF-3R glycosylation for SDS-PAGE (autoradiography) analysis
In a 10 µL reaction volume was mixed: 1 mM UDP-GlcNAc (100 µM hot; 900 µM cold), 1 mM THP, 55 µM HCF-3R construct, and 0.1 U CIP in a buffer of 20 mM MgCl2, 20 mM Tris and 150 mM NaCl at pH 7.4. Either ncOGT (to a final concentration of 2.5 µM) or water was added, and reactions were mixed by pipetting then incubated at 37 °C in a thermomixer to ensure uniform heating. After 5 hours, reactions were quenched by addition of an equal volume (10 µL) of 2× Laemmli loading buffer (with β-ME). Samples were boiled for 10 minutes, and spun down at 15,000×g for 10 minutes. 7 µL of sample was loaded per lane onto a 4–20% Tris-Gly gradient gel and run for 5 minutes at 90V, followed by 65 minutes at 150V. The gel was dried and placed in a cassette. A storage phosphor was placed on top and the cassette and exposed for 40 hours before the phosphor was imaged with a Typhoon imager.
HCF-short cleavage for LC-MS analysis
Typical peptide cleavage reactions were conducted as follows: 20 µL reaction volume with components having the following final concentrations: 1 mM UDP-GlcNAc, 1 mM THP, 12 µM HCF-short, 0.1 U CIP and 12 µM OGT in a buffer of 20 mM MgCl2, 20 mM Tris and 150 mM NaCl at pH 7.4. ncOGT was added last and reactions were mixed by pipetting then incubated at 37 °C in a thermomixer to ensure uniform heating. After the desired reaction time, reactions were quenched by addition of four volumes (80 µL) of ice-cold methanol supplemented with 0.1% formic acid. Reactions were then centrifuged at 16,100×g for 10 minutes to remove any precipitated protein (4 °C), and transferred to fresh tubes. Samples were concentrated to dryness using a speedvac (T = 30 °C), for 30–60 minutes. Samples were re-suspended in 100 µL 0.1% formic acid, mixed by vortexing, and centrifuged for 10 minutes at 15,000×g (4 °C) to remove any insoluble material. Samples were then transferred to LC vials, and maintained at 4 °C until they were analyzed.
Excess HCF-short experiments
Were conducted as above (12 µM HCF-short), except the final concentration of ncOGT was 3 µM. This experiment is shown in Figure 1d.
Excess OGT experiments
Were conducted as above, except HCF-short was used at 3 µM, and ncOGT was used at 27.5 µM (9.1×) to ensure saturation of OGT with peptide.
General LC-MS
General measurements of peptides were performed on a q-Exactive Plus FT-mass spectrometer equipped with an Ultimate 3000 uHPLC. Reaction mixture components were separated using a 33 minute reverse phase HPLC method with a Kinetex® C18 (2.6 µm particle size, 100 Å pore size, 2.1 mm ID, 150 mm length) at a constant flow rate of 0.200 mL/minute and column temperature of 35 °C. The gradient employed is shown in Supplementary Table 11; the last half of the LC method was empirically optimized to wash the column and re-equilibrate between runs to maintain column performance. 5 µL of sample was injected, unless otherwise specified. Detection on the q-Exactive Plus mass spectrometer was performed between 300–2000 m/z, using an acquisition target of 3E6, maximum IT of 200 ms at a resolution of 70,000 for MS and 35,000 for MS/MS data. For MS/MS analysis, dynamic exclusion was turned off and a global inclusion list was used to maximize S/N for the targeted analysis. A collision energy of 30 eV and an isolation window of 6 m/z was used for higher-energy collision-induced dissociation (HCD).
For some experiments additional MS/MS information was obtained using a q-TOF (CID). These measurements were conducted on a Bruker Impact HD q-TOF mass spectrometer coupled to an Agilent 1290 uHPLC. A collision energy of 35 eV and an isolation window of 6 m/z was used for collision-induced dissociation (CID). The source and TOF parameters were optimized for the transmission of higher molecular weight ions. A counter current drying gas flow rate of 12 L/min., and a nebulizing gas pressure of 45 bar were used. Funnel 1 was set to 150 V, and funnel 2 was set to 36 V, with a hexapole RF of 300Vp-p. A calibration segment was present at the beginning and end of the run for m/z calibration with sodium formate cluster using the Bruker HPC algorithm for the curve fitting.
Glycopeptide Intermediate Purification
In order to prepare a suitable quantity of glycopeptide intermediate, we ran four parallel reactions as follows: 20 µL reaction volume with components having the following final concentrations: 1 mM UDP-GlcNAc, 1 mM THP, 12 µM HCF-short, 0.1 U CIP and 12 µM ncOGT D554N in a buffer of 20 mM MgCl2, 20 mM Tris and 150 mM NaCl at pH 7.4. OGT was added last and reactions were mixed by pipetting then incubated at 37 °C in a thermomixer to ensure uniform heating. After 40 seconds, reactions were quenched by addition of four volumes (80 µL) of ice-cold methanol supplemented with 0.1% formic acid. Reactions were then centrifuged at 16,100×g for 10 minutes to remove any precipitated protein (4 °C), and transferred to fresh tubes. Samples were concentrated to dryness using a speedvac (T = 30 °C) for about 30 minutes. Samples were combined by first re-suspending one sample in 150 µL 0.1% formic acid, vortexing, and using the solution to re-suspend the next sample, etc. The final sample was centrifuged for 10 minutes at 15,000×g (4 °C) to remove any insoluble material. The sample was then transferred to an LC vial and maintained at 4 °C. In order to simultaneously detect and collect the glycopeptide, we employed an in-line ‘T’ between the column and the mass spectrometer inlet. To compensate for this change, the flow rate was adjusted to 0.2 mL/min, and the instrument tuning was adjusted for maximum sensitivity under these conditions. For each run, 10 µL of sample was injected; each sample collected was concentrated at 30 °C by speedvac and when completely dry these were kept on ice until all had been collected. Collected fractions were combined when possible and dried by the same method until the entire initial sample had been purified. This sample was stored dry at −80 °C until analysis was performed. An aliquot of one collected fraction was re-injected (1 to 5 dilution) to evaluate purity and stability, and showed intermediate 1 to be the predominant peak in the sample, along with some starting material (which happens to elute at a close retention time) as the major contaminant. Attempts to exclude starting material from the sample resulted in a low recovery of glycopeptide intermediate, likely due to the low volumes being collected. We note that we cannot exclude the possibility that some hydrolysis of this glutamyl ester contributes to the starting material peak observed.
Glycopeptide Intermediate Decomposition in the Absence of OGT
Glycopeptide intermediate was obtained as described above. The obtained dry material was dissolved in water. Equal volumes of purified Intermediate 1 was added to each of seven reaction tubes. In a 20 µL final reaction volume, glycopeptide was added to buffer containing 1 mM THP, 20 mM MgCl2, 20 mM Tris and 150 mM NaCl at pH 7.4. At specified time points, the reactions were quenched by the addition of four volumes (80 µL) of ice-cold methanol supplemented with 0.1% formic acid. Samples were treated as described above for standard LC-MS experiments, except dried down samples were reconstituted in 20 µL instead of 100 µL.
Electron-capture dissociation (ECD) MS/MS experiments
A Bruker 7.0 T Solarix Q-q-FT-ICR mass spectrometer was used for the electron capture dissociation (ECD) measurements. The dry sample of Intermediate 1 was re-suspended in 40 µL of 50% acetonitrile/water supplemented with 0.1% formic acid. The sample was introduced at a rate of 20 µL/h using a syringe pump and a Captive Spray™ nanospray source. A capillary voltage of 1000V was used to get a stable nanospray signal. The [M+4H]4+ ion of the glycosylated peptide was isolated using the quadrupole and subjected to ECD in the ICR cell using an ECD voltage of 1.0 V, and a time of 0.100 s. A two-megabyte acquisition file size was used to acquire 100 scans that were summed to obtain a single ECD mass spectrum. The data was internally calibrated using single point lock-mass correction using the predicted monoistopic m/z of 896.9017 for the precursor ion after an external multi-point linear calibration with arginine clusters. Data were manually inspected for the presence or absence of predicted ECD fragment ions within a 5 ppm mass accuracy window; RMS errors are shown in Supplementary Figure 21.
CID and HCD MS/MS experiments
Structural information about Intermediate 2 was obtained by MS/MS using both the Q-Exactive (HCD) and q-TOF (CID) instruments. In our hands these two instruments provided complementary information, with the q-TOF data sets including better data for higher molecular weight fragment ions. In order to unequivocally interpret the MS/MS data obtained for Intermediate 2 we compared it to the MS/MS spectrum of species A, obtained under identical conditions (see Supplementary Figure 11). For both peptides we observed serine/threonine dehydration in the C-terminal region as a result of CID fragmentation; only the anhydro species that forms in the reaction showed dehydration in the C9E10 region. Data were then manually inspected for predicted fragment ions within a 5 ppm mass accuracy window; RMS errors are shown in Supplementary Figure 21.
Crystallization of hOGT4.5 (D554N)
Ternary complexes were prepared by incubating hOGT4.5 D554N at 7 mg/ml with UDP-GlcNAc (1 mM) and peptide (CKII3K) (3 mM). The D554N OGT:CKII glycopeptide ternary product complex crystals were obtained by seeding the D554 hOGT4.5 protein with wild-type crystals, using the seed bead kit (Hampton) after mixing 2 µL of complex with 1 µl of reservoir consisting reservoir containing 1.6 M Li2SO4 and 0.1 M Bis Tris Propane, pH 7.0, as previously reported.17 Crystals were cryoprotected in solutions consisting of the reservoir solution plus 28% xylitol and flash frozen in liquid nitrogen.
Data Collection, Structure Determination, and Refinement
Data were collected at NSLS beamline X25 at Brookhaven National Laboratory. Data was processed using iMosflm27 and scaled with Scala or Aimless in the CCP4 crystallography suite.28 The PHENIX software package was used for molecular replacement and refinement.29 The previously determined OGT-CKII glycopeptide ternary product complex (PDB code 4GYW) was used as a search model for molecular replacement, using the program Phaser. Models were subsequently refined by rigid body refinement and multiple rounds of simulated annealing, minimization, atomic displacement parameter (ADP or B-factor) refinement and TLS refinement (determined using the TLSMD server30), with interspersed manual adjustments using Coot.31 Geometric restraints for UDP-GlcNAc were generating using PHENIX Elbow, and these restraints were used throughout refinement. Refinement statistics are presented in Supplementary Table 9. All structural figures were made with Pymol32 except for the electrostatic surface figures, which were created with CCP4mg.33 Crystallographic software was provided by SBGrid.34
Supplementary Material
Acknowledgments
JJ is a National Science and Engineering Research Council (NSERC) of Canada PGS-M and PGS-D3 fellowship recipient. Funding for this work was provided by a National Institutes of Health grant (R01 GM094263) to SW. We thank Christopher Thompson (Bruker Daltonics, Billerica, MA) for access to, and assistance with, a SolariX XR 7T q-Q-FT-ICR mass spectrometer. We thank David Vocadlo (Simon Fraser University, Vancouver, BC, Canada) for providing UDP-5SGlcNAc. X-ray diffraction data were collected at the National Synchrotron Light Source at Brookhaven National Laboratory (Beamline X25).
Footnotes
Accession codes
Protein Data Bank: Coordinates and structure factors have been deposited under accession code 5HGV.
Author Contributions
J.J. and S.W. designed research and analyzed data; J.J. performed LC/MS(/MS) experiments with assistance from S.A.T.; M.B.L. performed X-ray crystallography experiments and solved the structure of hOGT4.5 (D554N); J.J. performed all other experiments reported in the paper; J.J. and S.W. wrote the manuscript with input from all authors.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Parker JB, Yin H, Vinckevicius A, Chakravarti D. Cell Rep. 2014;9:967–982. doi: 10.1016/j.celrep.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tyagi S, Chabes AL, Wysocka J, Herr W. Mol. Cell. 2007;27:107–119. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 3.Capotosti F, et al. Cell. 2011;144:376–388. doi: 10.1016/j.cell.2010.12.030. [DOI] [PubMed] [Google Scholar]
- 4.Wilson AC, Peterson MG, Herr W. Genes Dev. 1995;9:2445–2458. doi: 10.1101/gad.9.20.2445. [DOI] [PubMed] [Google Scholar]
- 5.Wilson AC, LaMarco K, Peterson MG, Herr W. Cell. 1993;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- 6.Julien E, Herr W. EMBO J. 2003;22:2360–2369. doi: 10.1093/emboj/cdg242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangone M, Myers MP, Herr W. PLoS One. 2010;5:e9020. doi: 10.1371/journal.pone.0009020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daou S, et al. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2747–2752. doi: 10.1073/pnas.1013822108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janetzko J, Walker S. J. Biol. Chem. 2014;289:34424–34432. doi: 10.1074/jbc.R114.604405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bond MR, Hanover JA. J. Cell. Biol. 2015;208:869–880. doi: 10.1083/jcb.201501101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardiville S, Hart GW. Cell Metab. 2014;20:208–213. doi: 10.1016/j.cmet.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lazarus MB, et al. Science. 2013;342:1235–1239. doi: 10.1126/science.1243990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verdin E. Science. 2015;350:1208–1213. doi: 10.1126/science.aac4854. [DOI] [PubMed] [Google Scholar]
- 14.Feldman JL, Dittenhafer-Reed KE, Denu JM. J. Biol. Chem. 2012;287:42419–42427. doi: 10.1074/jbc.R112.378877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhuiyan T, Waridel P, Kapuria V, Zoete V, Herr W. PLoS One. 2015;10:e0136636. doi: 10.1371/journal.pone.0136636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schimpl M, et al. Nat. Chem. Biol. 2012;8:969–974. doi: 10.1038/nchembio.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazarus MB, et al. Nat. Chem. Biol. 2012;8:966–968. doi: 10.1038/nchembio.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erickson BW, Khan SA. Ann. N. Y. Acad. Sci. 1983;421:167–177. doi: 10.1111/j.1749-6632.1983.tb18107.x. [DOI] [PubMed] [Google Scholar]
- 19.Khan SA, Erickson BW. J. Biol. Chem. 1982;257:11864–11867. [PubMed] [Google Scholar]
- 20.Khan SA, Sekulski JM, Erickson BW. Biochemistry. 1986;25:5165–5171. doi: 10.1021/bi00366a027. [DOI] [PubMed] [Google Scholar]
- 21.Meyer V. Pitfalls and Errors of HPLC in Pictures. Weinheim: John Wiley & Sons; 2013. Double Peaks from Stable Conformers. [Google Scholar]
- 22.Hassa PO, Haenni SS, Elser M, Hottiger MO. Microbiol. Mol. Biol. Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao Z, Gao P, Liu HW. J. Am. Chem. Soc. 2009;131:14258–14260. doi: 10.1021/ja906135d. [DOI] [PubMed] [Google Scholar]
- 24.Nalbone JM, Lahankar N, Buissereth L, Raj M. Org. Lett. 2016;18:1186–1189. doi: 10.1021/acs.orglett.6b00317. [DOI] [PubMed] [Google Scholar]
- 25.Kotzler MP, Withers SG. J. Biol. Chem. 2016;291:429–434. doi: 10.1074/jbc.C115.698696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 26.Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. Nature. 2011;469:564–567. doi: 10.1038/nature09638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. Acta Crystallogr. D. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collaborative Computational Project, N. Acta Crystallogr. D. 1994;50:760–763. [Google Scholar]
- 29.Adams PD, et al. Acta Crystallogr. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Painter J, Merritt EA. J. App. Cryst. 2006;39:109–111. [Google Scholar]
- 31.Emsley P, Lohkamp B, Scott WG, Cowtan K. Acta Crystallogr. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.8. 2015. [Google Scholar]
- 33.McNicholas S, Potterton E, Wilson KS, Noble MEM. Acta Crystallogr. D. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morin A, et al. Elife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.